Translate this page into:
Transcriptional regulatory signatures of systemic diseases in periodontitis with dyslipidemia
⁎Corresponding author at: College of Dental medicine, Roseman University OF Health Sciences, South Jordan UTAH- 84095, USA. spatil@roseman.edu (Shankargouda Patil)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University. Production and hosting by Elsevier.
Abstract
Background
Periodontitis is a chronic bacterial infection of tooth that increases the risk of systemic diseases like diabetes, cancer, and cardiovascular diseases. Although it has been have found altered lipids in periodontitis patients, its gene regulation is largely unknown. This study aimed to examine the lipid meditated regulatory network in periodontitis that may helpful in early detection of periodontal mediated systemic diseases.
Methods
We employed a high-throughput gene expression data of 1) patients with periodontitis (n = 6); 2) the periodontitis patients with dyslipidemia (n = 6); and 3) healthy control group (n = 6). The over represented (DEGs) genes in SET-A (control vs. periodontitis) and SET-B (control vs. periodontitis with dyslipidemia) was identified. The protein interaction network was generated for the over represented genes in both the conditions. The constructed network was dissected into multiple regulatory clusters, containing over expressed transcription factors with its interacting proteins. Further the behavior of the clusters was determined through gene ontology and molecular pathways.
Results
On expression analysis, 751 in SET-A and 561 in SET-B were over expressed compared to healthy control. Using over expressed genes, protein interaction networks were constructed for SET-A and SET-B, respectively. Topological analysis revel the difference in the complexity of both the network. Four regulatory clusters (ESR1, FOS, RUNX2, and SP1) from SET-A and six (ESR1, ESR2, FOS, JDP2, PBX1, and TAL1) from the SET-B network was extracted. Each cluster displayed a variety of molecular mechanism associated with immune system, cell cycle, and signal processing. Clusters from SET-B showed diverse regulatory pattern in associated with cancer, neurological, psychiatric and metabolic diseases.
Conclusion
Our findings demonstrate difference in regulatory patterns between periodontitis and periodontitis with dyslipidemia. These finding may provide evidence for dyslipidemia mediated periodontitis contribute to progressive systemic diseases. Further experiments are required to validate these regulators as biomarkers and drug targets.
Keywords
Dyslipidemia
Periodontitis
Systemic diseases
1 Introduction
Chronic infection of bacterial pathogens in teeth is considered the initial step of periodontal disease (PD). Most oral pathogens are gram-negative bacteria that form biofilms at the gum line of teeth. Among various gram-negative bacteria, Porphyromonasgingivalis and Aggregatibacteractinomycetemcomitans have been connected to the aetiology of periodontal disease (How et al., 2016). Gingivitis occurs at the early stages of infection, characterised by swelling, redness, and bleeding of the gums. Persistent infection triggers an inflammatory response that leads to periodontal ligament degradation, alveolar bone deterioration/resorption, and tooth loss. Some of the most common clinical features of periodontal disease are clinical attachment loss (CAL), gingival bleeding on probing (BOP), alveolar bone loss, and pocket depth (Mdala et al., 2014). Although all destructive forms of periodontal disease share clinical signs, relative contributions of genetic variables play a vital role in the aetiology of periodontitis(Kinane et al., 2017). Furthermore, evidence is mounting that suggests a link between periodontitis and systemic diseases such as cancer, cardiovascular disease, and type II diabetes (T2DM)(Priyamvara et al., 2020)(Kavarthapu and Gurumoorthy, 2021)(Wu et al., 2020). However, the association between these diseases and periodontitis is largely unclear. We believe that there exists a common pathological event that ties up these diseases with periodontitis.
Despite the fact that chronic inflammation is known to play a role in pathogenesis, dyslipidemia is still a major contributor to PD and other systemic diseases (Gomes-Filho et al., 2022). Majority of research has found increased lipid molecules in patients with periodontitis (Nepomuceno et al., 2017). According to data from the Third National Health and Nutrition Examination Survey (NHANES), there is a link between periodontal disease and serum cholesterol (Griffiths and Barbour, 2010). In T2DM, the percentage of sites with BOP has been linked to higher total cholesterol, LDL-cholesterol, and triglycerides(Dhir et al., n.d.). A Japanese studyshowed a link between PD and increased plasma TG (>149 mg/dl) was found to have an odds ratio of 2.26(Griffiths and Barbour, 2010). Also, Suvanet al. found a similar link in chronic periodontitis patients with a body mass index (Suvan et al., 2014). Notably, total cholesterol, LDL-cholesterol, and triglycerides were increased with periodontitis (Nepomuceno et al., 2017). Song et al. evaluated the association of serum lipids with dental health (Song et al., 2020). Significant association was noticed between high-density lipoprotein cholesterol and triglyceride in periodontitis patients (Song et al., 2020). Similar relationships were observed with the characteristics of plaque index, pocket depth, bleeding on probing, with lipid status in periodontitis (Awartani and Atassi, 2010)(Anirudhan et al., 2021). Fentoğlu Ö et al. found interconnectivity between lipoprotein mediators and periodontitis (Fentoğlu et al., 2020). Interestingly, studies have found lower levels of antiatherogenic HDL in periodontitis patients (Ljunggren et al., 2019). However, the underlying putative effect of the lipid mechanism in PD and its associated gene regulation leading to systemic diseases is largely unknown.
In this study, we implement a computational method that enables the integration of data sources into a protein interaction network in order to identify essential network features in A) periodontitis and B) periodontitis with dyslipidemia. The evaluation of both networks permits the identification of critical regulatory clusters encompassing transcription factors (TF), co-regulators of transcription factors (coTFs) and genes that are shared and distinct between these conditions. Specifically, the unique regulatory clusters from patients with dyslipidemia demonstrated several important dyslipidemia associated regulatory mechanisms in periodontitis. Our identified regulatory molecules may enable in developing prognostic markers and new therapeutic targets for periodontitis mediated systemic diseases.
2 Materials and Methods
2.1 Data resources and analysis
The NCBI GEO database (https://www.ncbi.nlm.nih.gov.geodata) was used to collect gene expression datasets with the accession number GSE156993. The GSE156993 dataset includes variety of experimental gene expression profiles performed with the Affymetrix Human Genome U133 Plus 2.0 Array platform. We collected only the gene expression data executed in peripheral blood mononuclear cells (PBMCs) samples that describes, periodontitis induvidual (n = 6), periodontitis with dyslipidemia (n = 6) and healthy control (n = 6). All samples included in our study were reported with their gender, age, total cholesterol, and triglyceride levels (Table.1). The differentially expressed genes (DEGs) in periodontitis (SET-A) and periodontitis with dyslipidemia (SET-B) were determined using the R-Package-limma tool by comparing with the experimental control (Anirudhan et al., 2021). The change in gene expression along with the statistical significance (p-value < 0.05) were computed to determine the DEGs.
Parameter
Healthy Control (Mean ± SD)
Periodontitis
(Mean ± SD)
Periodontitis with dyslipidemia
(Mean ± SD)
Age (Years)
41 ± 3
45 ± 7
47 ± 8
Gender
Male:2; Female: 4
Male:2; Female: 4
Male:4; Female: 2
Total Cholesterol (mg/dl)
162 ± 13.1
151 ± 17.4
262.5 ± 28.7
Triglycerides
81.5 ± 40.2
73.3 ± 25.1
221.6 ± 58.2
2.2 Protein–protein interaction networks
Next, a protein interaction (PPI) network was constructed from the significant up-regulated genes of SET-A and SET-B analyses, respectively. We use our indigenous databases containing the large set of interaction data was collected from the major resources (Nakajima et al., 2021) for network construction. The up-regulated genes encoding proteins were inputted to construct the protein network that aids in the understanding of the functional proteome of SET-A and SET-B. Cytoscape software (https://www.cytoscape.org) version 3.8.2 was used to construct the PPI network. Further, the network properties such as path length, centralization, clustering coefficient, network density, average number of neighbours, topological coefficient, and average node distribution were assessed for each network using the network topology module in Cytoscape 3.8.2.
2.3 Construction of regulatory hubs
The network (https://www.networkanalyst.ca/) analyst database containing transcription factors was used to annotate each network. The over-represented transcription factors (TFs) in the microarray analysis were identified and extracted along with their interacting proteins from the network. These extracted transcription factors with their connected proteins were termed “clusters”. Each cluster was retained, containing at least two connected proteins with TF for functional enrichment analysis.
2.4 Functional enrichment
The molecular behaviour of a selected cluster was determined by functional enrichment analysis using the Enricher tool ( https://maayanlab.cloud/Enrichr/). Enrichr provides gene ontology (OG) information such as biological processes (BP) and molecular functions (MF) for the inputted proteins. In particular, based on the OG terminology by the Gene Ontology Consortium (https://geneontology.org), the cluster proteins related to co-regulators of transcription factors (coTFs) such as transcription coactivator binding (GO:0001223), transcription coregulator binding (GO:0001221), transcription corepressor binding (GO:0001222), and nuclear receptor coactivator activity (GO:0030374) were identified. Simultaneously, the reactome pathway (https://reactome.org/) database was used to investigate the molecular pathways associated with the clusters. The significance of pathway-related clusters was assessed based on the threshold P-value < 0.05. Last, DAVID (https://david.abcc.ncifcrf.gov/: Database for Annotation, Visualization, and Integrated Discovery) was used to find out how the coTFsthecomponnet of the regulatory clusters were linked to diseases.
3 Results
3.1 Differential genes in periodontitis with and without dyslipidemia complication
Using the limma R-program, we identified 1331 DEGs in the SET-A. Of which, 751 were up-regulated and 580 were down-regulated, with a p-value ≤ 0.05. Simultaneously, the analysis of SET-B showed 561 genes were up-regulated and 915 were down-regulated from the overall 1476 DEGs with p ≤ 0.05. Among the DEGs, only the up-regulated genes from SET-A and B were used to construct protein–protein interaction networks.
3.2 Periodontitis interactome with and without dyslipidemia
The PPI networks for the up-regulated genes from SET-A and B were independently generated using the in-house interactome database. The SET-A network presents the complex interaction with 1471 extended proteins that form 13,568 interconnected edges (Fig. 1). Likewise, the SET-B forms a network with 10,875 interactions that came from 1458 proteins (Fig. 2). The complexity of each network was assessed using a network analyzer, which showed differences in network properties including path length, centralization, clustering coefficient, network density, average number of neighbours, node distribution, and topological coefficient between SET-A and SET-B (Table 2).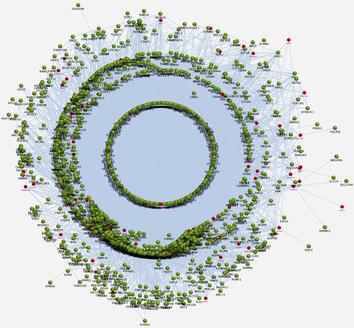
Protein-protein interaction network of periodontitis present the complex interaction with 1471 extended proteins that attribute to form 13,568 interconnected edges. Red color node represent seed DEGs encoding proteins and green color nodes represent the extended proteins of DEGs. The connectivity between the represent (gray line) the protein interaction..
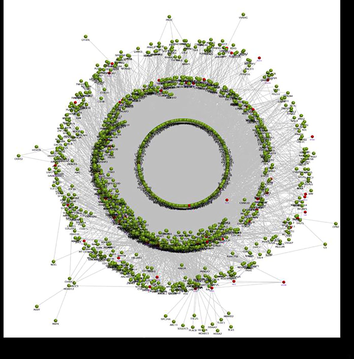
The SET-B network of periodontitis with dyslipidemia forms a network with 10,875 interaction edges that arrived from 1458 proteins Fig. 3 Molecular dynamic simulation of RdRp - lupeol complex presenting RMSD trajectory till 200 ns. (green: cα backbone of Protein and pink: lupeol).
Topological Parameter
Periodontitis (SET-A)
Periodontitis with Dyslipidemia (SET-B)
Number of nodes
1471
1458
Shortest path length
3.46
3.085
Network centralization
0.4
0.146
Clustering coefficient
0.032
0.185
Network density
0.023
0.010
Average number of neighbours
16.837
14.926
Topological coefficient
0.312
0.204
Average node distribution
8.342
6.394
3.3 Crucial cluster with transcription factors
Each constructed network was dissected into multiple clusters based on the over-expressed transcription factors as mentioned in the methodology section. In SET-A, four clusters were extracted containing TF such as ESR1, FOS, RUNX2, and SP1 (Supplementary Table 1). All four TFs showed up-regulation during SET-A gene expression analysis. Likewise, from the SET-B, six clusters were extracted based on the over-expressed ESR1, ESR2, FOS, JDP2, PBX1, and TAL1 transcription factors (Supplementary Table 2). Each cluster was represented by a transcription factor with its immediate interacting proteins. For instance, the SET-A cluster contains Runt-related transcription factor 2 (RUNX2) showed interaction with KAT6B, SMURF1, HDAC6, YAP1, MAP3K4, CDK1, SMAD2, SMAD3, EP300, RB1, PRKCD, ETS1, XRCC6, HDAC5, CEBPB, AXIN1, DVL2, HDAC3, SMAD1, TLE1, LEF1, HDAC4, KAT2B, STUB1, SMAD5,SMAD6 and XRCC5. Likewise, the JDP2 transcription factor of the SET-B cluster showed interaction with ATF7, IRF2BP1, UBC, MAPK8, JUN, CREBBP, MAPK14, ATF2, EP300, KAT2B, JUND, JUNB, SNW1, PGR, and MAPK9.
3.4 Clusters with co-regulators of transcription factors (coTFs)
Each cluster was enriched using the Enrichr ontology tool. Most clusters from the SET-A network showed involvement in regulating cell proliferation, apoptotic processes, interferon production, and cellular response to oxidative stress (Supplementary File 1). Whereas the SET-B clusters showed an almost similar mechanism, having clusters with different transcription factors except for FOS and ESR1 that were common between SET-A and B (Supplementary File 1). However, the biological processes of all the clusters from both the networks influence transcription regulation (Fig. 3). Additionally, clusters of SET-A (Fig. 4) and SET-B (Fig. 5) were pathway enriched with the reactome database, showing involvement in a variety of pathways associated with the immune system, signal transduction, gene expression, cell cycle, cell death, and cellular response to stimuli (Supplementary File 1). Although these mechanisms truly reflect the pathogenesis of periodontitis, our prime interest is to look for the coTFs within the cluster based on the GO annotation. It is noteworthy that within the cluster, a few proteins that interact with the transcription factors are recognised as coTFsthat assist TF in the transcription process. In the SET-A clusters, AHR, AR, CTNNB1, ELK1, ETS1, HDAC3, HDAC6, HMGA1, LMO2, MED1, MED12, MED13, MED17, NCOA1, NCOA2, NCOA3, NCOA6, PGR, PPARGC1A, RELA, RNF4, SMAD3, SMAD4, THRB, UBE2I, and ZBTB16 were noticed as coTFs. Similarly, in the SET-B clusters, AR, SMAD3, THRB, PGR, AHR, RELA, UBE2I, ZBTB16, CTNNB1, MED12, NCOA1, MED1,NCOA2, MED13, NCOA6, NCOA3, RNF4, PPARGC1A, MED17, SMAD4, HDAC3, HDAC6, CDK9, and LMO2 were recognised as coTFs. Further, analysis using the DAVID tools showed most coTFs from SET-B clusters were associated with cancer, renal and neurological diseases (Table 3).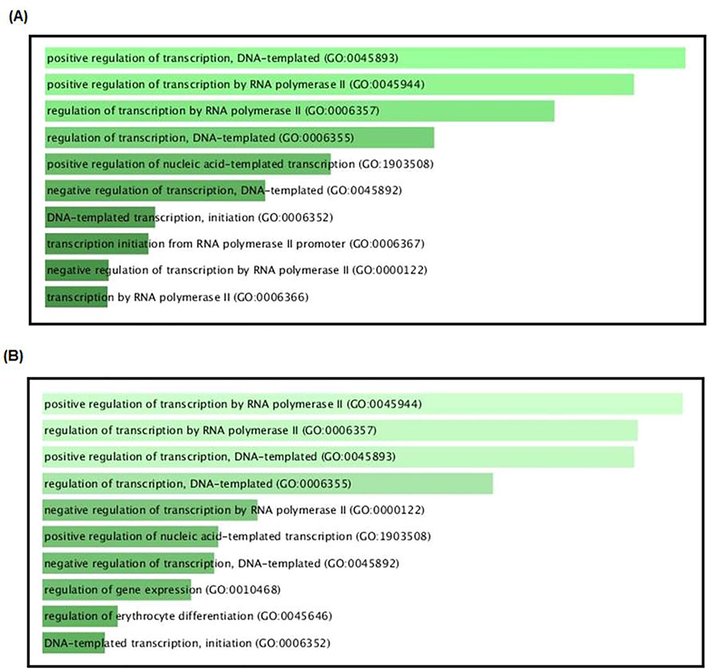
Most contributed biological process by the clusters from both the networks (A) SET-A and (B) SET-B.
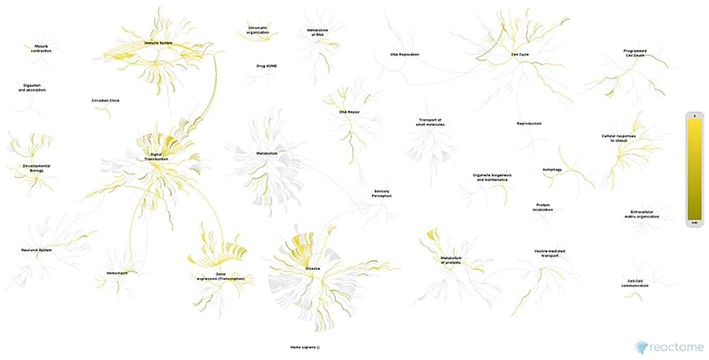
Pathway enrichment of clusters from SET-A network (periodontitis) using reactome database.
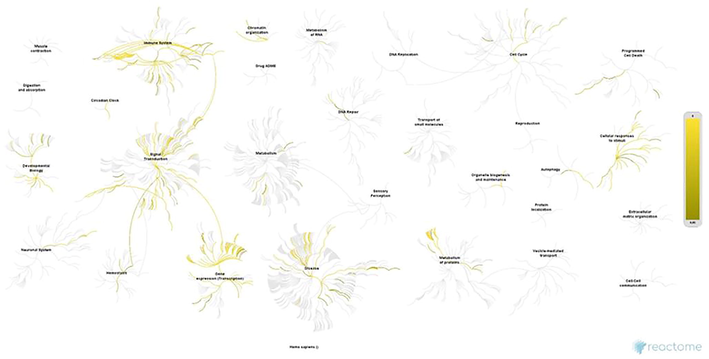
Pathway enrichment of clusters SET-B network (periodontitis with dyslipidemia) using reactome database.
Disease Term
Count
P-Value
SET-A clusters
PHARMACOGENOMIC
17
1.76E-06
CANCER
18
2.46E-06
METABOLIC
19
6.68E-04
OTHER
10
0.00146
NORMALVARIATION
6
0.002219
DEVELOPMENTAL
9
0.004205
UNKNOWN
9
0.006659
CHEMDEPENDENCY
13
0.018744
REPRODUCTION
6
0.028805
IMMUNE
11
0.032688
SET-B clusters
CANCER
22
9.74E-09
PHARMACOGENOMIC
20
4.76E-08
NORMALVARIATION
8
6.73E-05
OTHER
12
1.87E-04
METABOLIC
21
4.37E-04
DEVELOPMENTAL
11
5.48E-04
UNKNOWN
11
9.94E-04
CHEMDEPENDENCY
16
0.002445
REPRODUCTION
8
0.00286
IMMUNE
14
0.003677
PSYCH
11
0.004619
NEUROLOGICAL
12
0.024173
4 Discussion
Periodontitis is a chronic inflammatory condition that leads to the elevation of pro-inflammatory mediators in both local and systemic environments. Periodontitis affects adults and has a significant negative impact on quality of life(Ferreira et al., 2017). Systemic diseases that affect the role of neutrophils, monocytes, macrophages, and lymphocytes result in changes in the development or activity of inflammatory mediators produced by the host (Choubaya et al., 2021). Thus, there is a nexus between periodontal destruction and systemic diseases. Recent studies demonstrate the influence of dyslipidemia, which contributes to systemic diseases including cardiovascular disease, autoimmune diseases, and cancers. Furthermore, evidence of an increase in plasma cholesterol, triglycerides, LDL-cholesterol, and low levels of HDL-cholesterol is mounting to support the notion that dyslipidemia is one of several pathogenesis associated with periodontitis and its systemic diseases(Dhir et al., n.d.)(Suvan et al., 2014)(Song et al., 2020)(Awartani and Atassi, 2010)(Fentoğlu et al., 2020)(Ljunggren et al., 2019). However, the mechanism associated with dyslipidemia in periodontitis that leading to systemic diseases is still under investigation. Understanding the control mechanism for dyslipidemia in periodontitis will open up new ways to make biomarkers for the future of systemic diseases caused by periodontitis.
Researchers have made significant progress in determining the levels of lipids and lipid-related molecules in the pathogenesis of periodontitis. The periodontal modifications are most likely the result of a number of biological properties shared by a variety of lipid fractions. It is common knowledge that a state of hyperlipidemia can lead to phenotypic alterations in the microcirculation. These changes are characterised by an increase in oxidative stress, dysfunction in endothelial cells, a decrease in the bioavailability of vascular nitric oxide, and the formation of a highly proinflammatory state (Stapleton et al., 2010). Latter it is characterised by an excessive amount of neutrophil priming as well as an elevation of proinflammatory cytokines like TNF-a and IL-1b from monocytes and polymorphonuclear leukocytes that cause tissue damage (Ramadan et al., 2020)(Cekici et al., 2014). Such damage is caused by the extracellular release of reactive oxygen species, tissue-destructive enzymes such as elastase and myeloperoxidase, and the inhibition of protective factors such as a1-antitrypsin (Montecucco et al., 2009). It is anticipated that there will be a dramatic acceleration in the loss of periodontal tissue as a direct consequence. In a recent study, Scardina et al.(Scardina et al., 2011)used videocapillaroscopy to explore the morphologic and parametric aspects of the periodontal microcirculation in patients who suffered from hyperlipidemia. According to the Scardina et al., findings, patients with high levels of serum cholesterol had significant morphologic alterations in the microcirculation of the periodontium(Scardina et al., 2011).
Considering the enormous molecular link between the lipid and periodontitis, there is a lack of understanding of the transcriptional regulatory mechanism linked to the lipid and periodontitis. In this study, we employed high-throughput microarray PBMC gene expression data to detect DEGs between the analysed groups. We generated a interaction network from the up-regulated genes of both conditions. The constructed network was divided into clusters, containing TFs with their interacting proteins. Next, the behaviour of the clusters was functionally annotated for the presence of coTFs. Further coTFs were used to determine their association with the systemic diseases. As described, the microarray gene expression analysis of SET-A revealed 751 highly expressed genes, which were used in the construction of the protein network consisting of 13,568 nodes. In a similar manner, the complex network was produced from the 561 genes that were up-regulated in SET-B. The developed networks present a crucial behaviour of the molecules under periodontitis and periodontitis patient with dyslipidemia. Additionally, the analysis of network features based on the topology suggest that periodontitis network is highly complex than the network from periodontitis patient with dyslipidemia. Although SET-B network is less complex, the versatility of genes within a network play a significant role in pathogenesis.
The protein interaction network was dissected into multiple clusters, each containing a transcription factor as a regulatory component. The periodontitis (SET-A) four-cluster contains transcription factors such as ESR1, FOS, RUNX2, and SP1 that regulate multiple genes. Likewise, six clusters were selected with transcription factors such as ESR1, ESR2, FOS, JDP2, PBX1, and TAL1 were assessed from the SET-B network. Although a similar approach was adopted in both the SETs, the extracted clusters with TF and coTFs were noticed to be different between both conditions, except for the cluster with ESR1 and FOS transcription factor. This results demonstrate that the clusters with ESR1 and FOS may be the fundamental for the pathogenesis in periodontitis with or without dyslipidemia. For instance, the ESR1 transcription factor is an oestrogen receptor 1, highly expressed in periodontitis gingiva, associated with various oral conditions (Shang et al., 2016)(Arid et al., 2019)(Dalledone et al., 2019). Genetic polymorphisms ESR1 have been linked to chronic periodontitis (Musacchio et al., 2022). Herein, ESR1 was found to be overexpressed in both the conditions that interact with 163 proteins involved in signal transduction, immune, and gene regulation processes. Particularly, MyD88-independent TLR4 cascade, PI5P, PP2A and IER3 Regulate PI3K/AKT Signaling. PTEN Regulation, Regulation of beta-cell development, TGF-beta receptor signalling activates SMADs, Toll Like Receptor 9 (TLR9) Cascade, TP53 Regulates Transcription of DNA Repair Genes were associated with periodontitis. Likewise, FOS plays an essential function in regulating TGF- signalling (Ghafouri-Fard et al., 2022). In addition, FOS functions in the regulation of the formation of cells destined to form and maintain the skeletal and immune systems(Ghafouri-Fard et al., 2022). Herein, FOS was found interact with 90 proteins involved in cellular response to stimuli, signal transduction, inflammation, cell cycle and protein metabolism based on the reactome database. Notably these critical mechanisms are well associated with periodontitis progression. The immune system plays a important role in activating an innate response against microbial infection. The activation of innate immunity and the emergence of acquired immunity specific to an antigen were caused by the stimulation of various Toll-like receptors, which led to specific patterns of gene expression. (Anirudhan et al., 2020). Moreover, In order to initiat cellular processes like proliferation, differentiation, and development, a number of regulatory molecules in the cytoplasm and the nucleus were eventually activated through the participation of MAPK signal molecules in the amplification and specificity of the transmitted signals. (Li et al., 2012; Plotnikov et al., 2011).Although the clusters with ESR1 and FOS shows the fundamental process of periodontitis, our result also suggests that diverse regulatory pattern coTFsin patients with dyslipidemia may leading to systemic diseases. In particular, most coTFs from SET-B clusters (ESR1, FOS, JDP2, PBX1, TAL1 andESR2)showed an association with cancer, metabolic, psychiatric and neurological diseases, which were the notable systemic diseases associated with periodontitis(Nazir, n.d.)(Makkar et al., 2018). For instance, NgoziNwizu et al., 2020 examined the potential association between periodontitis with cancer types (Nwizu et al., 2020). Similarly, SadayukiHashioka et al., 2019, establish the possible causal relationship between periodontitis and neurological and psychiatric diseases(Hashioka et al., 2019). Likewise, the Meta-Analysis of Gobin et al. (Gobin et al., 2020) declares the association between periodontitis and the risk of metabolic diseases. Overall, our study shows the potential role of transcription factors and their molecules that regulate lipid-mediated systemic disease in periodontitis. These identified regulating molecules can pave a way for novel therapeutic strategies.
5 Conclusion
Our findings suggest regulatory targets for future research with major coTFs that could be potential indicators for lipid-mediated systemic illnesses in periodontitis. Our method utilizes a variety of data resources into a protein interaction network in order to identify the network's important features. We discovered multiple molecular regulators that were shared and unique between A) periodontitis and B) periodontitis with dyslipidemia. Few clusters with TF, coTFs and genes, were shown to be common among the conditions, contributing to a key process in periodontitis. Alternatively, periodontitis patients with dyslipidemia were shown to have distinct clusters that led to critical pathways and were connected with a variety of systemic disorders. Among these clusters, few of the coTFs may enable in developing prognostic markers and new therapeutic targets for periodontitis mediated systemic diseases.
Declarations
Funding:There is no external source of funding.
Ethical approval: Not applicable.
Acknowledgment: None.
Informed consent: Not applicable.
CRediT authorship contribution statement
Padalugu Devi Navya: Conceptualization, Data curation, Writing – original draft, Funding acquisition. Gurumoorthy Kaarthikeyan: Conceptualization, Writing – original draft. Ahmed Alamoudi: Validation, Investigation. Maha A Bahammam: Formal analysis. Samar Saeed Khan: Validation. Khalid J. Alzahrani: Methodology, Writing – original draft. Ibrahim F. Halawani: Software, Resources, Writing – review & editing. Fuad M. Alzahrani: Software, Data curation, Funding acquisition. Khalaf F Alsharif: Methodology, Validation, Writing – original draft. A.Thirumal Raj: Resources. Hosam Ali Baeshen: Formal analysis, Resources, Writing – review & editing. Shankargouda Patil: Investigation, Data curation, Writing – original draft, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References:
- Temporal changes of NF-κB signaling pathway genes in bacterial stimulated whole blood- a host mechanism associated with sepsis. Microb. Pathog.. 2020;147:104415
- [CrossRef] [Google Scholar]
- RPL6: A Key Molecule Regulating Zinc- and Magnesium-Bound Metalloproteins of Parkinson’s Disease. Front. Neurosci.. 2021;15
- [Google Scholar]
- Oestrogen receptor alpha, growth hormone receptor, and developmental defect of enamel. Int. J. Paediatr. Dent.. 2019;29:29-35.
- [CrossRef] [Google Scholar]
- Evaluation of periodontal status in subjects with hyperlipidemia. J. Contemp. Dent. Pract.. 2010;11:033-040.
- [Google Scholar]
- Inflammatory and immune pathways in the pathogenesis of periodontal disease. Periodontol.. 2014;2000(64):57-80.
- [CrossRef] [Google Scholar]
- Expression of Inflammatory Mediators in Periodontitis Over Established Diabetes: an Experimental Study in Rats. Med. Arch. (Sarajevo, Bosnia Herzegovina). 2021;75:436-443.
- [CrossRef] [Google Scholar]
- Estrogen receptor gene is associated with dental fluorosis in Brazilian children. Clin. Oral Investig.. 2019;23:3565-3570.
- [CrossRef] [Google Scholar]
- Dhir, S., Wangnoo, S., Kumar, V., n.d. Impact of Glycemic Levels in Type 2 Diabetes on Periodontitis. Indian J. Endocrinol. Metab. 22, 672–677. https://doi.org/10.4103/ijem.IJEM_566_17.
- Is the relationship between periodontitis and hyperlipidemia mediated by lipoprotein-associated inflammatory mediators? J. Periodontal Implant Sci.. 2020;50:135-145.
- [CrossRef] [Google Scholar]
- Impact of periodontal disease on quality of life: a systematic review. J. Periodontal Res.. 2017;52:651-665.
- [CrossRef] [Google Scholar]
- Downregulation of oxytocin-related genes in periodontitis. Front. Mol. Neurosci.. 2022;15:950919
- [CrossRef] [Google Scholar]
- Periodontal Diseases and the Risk of Metabolic Syndrome: An Updated Systematic Review and Meta-Analysis. Front. Endocrinol. (Lausanne).. 2020;11:336.
- [CrossRef] [Google Scholar]
- Gomes-Filho, I.S., Oliveira, M.T., Cruz, S.S. da, Cerqueira, E. de M.M., Trindade, S.C., Vieira, G.O., Couto Souza, P.H., Adan, L.F.F., Hintz, A.M., Passos-Soares, J. de S., Scannapieco, F.A., Loomer, P.M., Seymour, G.J., Figueiredo, A.C.M.G., 2022. Periodontitis is a factor associated with dyslipidemia. Oral Dis. 28, 813–823. https://doi.org/10.1111/odi.13779
- Lipoproteins and lipoprotein metabolism in periodontal disease. Clin. Lipidol.. 2010;5:397-411.
- [CrossRef] [Google Scholar]
- The Possible Causal Link of Periodontitis to Neuropsychiatric Disorders: More Than Psychosocial Mechanisms. Int. J. Mol. Sci.. 2019;20
- [CrossRef] [Google Scholar]
- Porphyromonas gingivalis: An Overview of Periodontopathic Pathogen below the Gum Line. Front. Microbiol.. 2016;7
- [CrossRef] [Google Scholar]
- Linking chronic periodontitis and oral cancer: A review. Oral Oncol.. 2021;121:105375
- [CrossRef] [Google Scholar]
- Periodontal diseases. Nat. Rev. Dis. Prim.. 2017;3:17038.
- [CrossRef]
- MAPK usage in periodontal disease progression. J. Signal Transduct.. 2012;2012:308943
- [CrossRef] [Google Scholar]
- Modified lipoproteins in periodontitis: a link to cardiovascular disease? Biosci. Rep.. 2019;39
- [CrossRef] [Google Scholar]
- Periodontal, metabolic, and cardiovascular disease: Exploring the role of inflammation and mental health. Pteridines. 2018;29:124-163.
- [CrossRef] [Google Scholar]
- Comparing clinical attachment level and pocket depth for predicting periodontal disease progression in healthy sites of patients with chronic periodontitis using multi-state <scp>M</scp> arkov models. J. Clin. Periodontol.. 2014;41:837-845.
- [CrossRef] [Google Scholar]
- Chlorhexidine prevents hypochlorous acid-induced inactivation of alpha1-antitrypsin. Clin. Exp. Pharmacol. Physiol.. 2009;36:e72-e77.
- [CrossRef] [Google Scholar]
- Bone-related polymorphisms and dental status in older men and women. Results of the longitudinal Pro.V.A. study. J. Dent. Sci.. 2022;17:528-534.
- [CrossRef] [Google Scholar]
- Databases for Protein-Protein Interactions. Methods Mol. Biol.. 2021;2361:229-248.
- [CrossRef] [Google Scholar]
- Nazir, M.A., n.d. Prevalence of periodontal disease, its association with systemic diseases and prevention. Int. J. Health Sci. (Qassim). 11, 72–80.
- Serum lipid levels in patients with periodontal disease: A meta-analysis and meta-regression. J. Clin. Periodontol.. 2017;44:1192-1207.
- [CrossRef] [Google Scholar]
- Periodontal disease and cancer: Epidemiologic studies and possible mechanisms. Periodontol.. 2020;2000(83):213-233.
- [CrossRef] [Google Scholar]
- The MAPK cascades: signaling components, nuclear roles and mechanisms of nuclear translocation. Biochim. Biophys. Acta. 2011;1813:1619-1633.
- [CrossRef] [Google Scholar]
- Periodontal Inflammation and the Risk of Cardiovascular Disease. Curr. Atheroscler. Rep.. 2020;22:28.
- [CrossRef] [Google Scholar]
- Cytokines and Chemokines in Periodontitis. Eur. J. Dent.. 2020;14:483-495.
- [CrossRef] [Google Scholar]
- Periodontal alteration of the microcirculation and hypercholesterolemia: a possible correlation? South. Med. J.. 2011;104:116-120.
- [CrossRef] [Google Scholar]
- Relationship between estrogen receptor 1 gene polymorphisms and postmenopausal osteoporosis of the spine in Chinese women. Genet. Mol. Res.. 2016;15
- [CrossRef] [Google Scholar]
- Oral health and changes in lipid profile: A nationwide cohort study. J. Clin. Periodontol.. 2020;47:1437-1445.
- [CrossRef] [Google Scholar]
- Hypercholesterolemia and microvascular dysfunction: interventional strategies. J. Inflamm. (Lond). 2010;7:54.
- [CrossRef] [Google Scholar]
- Body mass index as a predictive factor of periodontal therapy outcomes. J. Dent. Res.. 2014;93:49-54.
- [CrossRef] [Google Scholar]
- Epidemiologic relationship between periodontitis and type 2 diabetes mellitus. BMC Oral Health. 2020;20:204.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2023.102707.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







