Translate this page into:
The proximity of Hydrocotyle umbellata L. with araliaceae as evident from plastome and phylotranscriptomic analyses
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The recent massive development in the next-generation sequencing (NGS) platforms and bioinformatics tools including cloud-based NGS data analyses have proven extremely useful in understanding the deeper-level phylogenetic relationships of angiosperms. The family ‘Apiaceae Lindley / Umbelliferae Jussieu’ and Araliaceae Jussieu resemble each other in the structure of their gynoecia, were placed in the order Apiales Nakai, and are closely related. The family Araliaceae with the subfamily 1. Hydrocotyloideae Link, 2. Harmsiopanax Harms and 3. Aralioideae Eaton accepted as a monophyletic branch within the Apiales, an order within the Asterids. Of these, Hydrocotyle L. from Hydrocotyloideae Link of Apiaceae transferred to Araliaceae based on molecular phylogenetic studies. The present study evaluates the proximity of H. umbellata with Araliaceae based on plastome and phylotranscriptomic analyses using the minimum evolution method. The analyses revealed the nesting of the H. umbellata under the family Araliaceae in the MPT (Maximum Parsimony Tree), and the proximity of H. umbellata under the family Araliaceae further supported by the evolutionary divergence between the sequences and plastome alignment.
Keywords
Hydrocotyle L.
Hydrocotyle umbellata L. Apiales
Araliaceae
Apiaceae
Plastome
Phylotranscriptome
1 Introduction
The order Apiales [(Nakai, Hisi-Shokubutsu 58 (1930)] which includes seven families {[i.e. 1. Apiaceae [(Lindl., Intr. Nat. Syst. Bot. (ed. 2) 21, (1836)], 2. Araliaceae [(Juss., Gen. Pl. 217, (1789)], 3. Griseliniaceae [(Akht., Sist. Magnol. 209 (1987)], 4. Myodocarpaceae [(Doweld, Prosyllab. Tracheophyt. Lii (2001)], 5. Pennantiaceae [(J. Agardh, Theoria Syst. Pl. 301 (1858)], 6. Pittosporaceae [(R. Br., Voy. Terra Austral. 2: 542 (1814)], and 7. Torricelliaceae [(H.H. Hu, Bull. Fan Mem. Inst. Biol., Bot. 5: 311, 1934)]} have been placed within the Asterid group of Eudicots as circumscribed by group Campanulids (APG IV, 2016). The family ‘Apiaceae Lindley / Umbelliferae Jussieu’ (with approx. c. 434 genera and c. 3700 species) and Araliaceae Jussieu (with approx. c. 43 genera, c.1450 species; APG IV, 2016; https://www.mobot.org/mobot/research/apweb/) resemble each other in the structure of their gynoecia, and were placed in the order Ariales, superorder Araliiflorae (Dahlgren, 1980), and are closely related (Plunkett et al., 2004). The family Araliaceae with the subfamily 1. Hydrocotyloideae Link, 2. Harmsiopanax Harms and 3. Aralioideae Eaton accepted as a monophyletic branch within the Apiales (Kim et al., 2017), an order within the Asterids (APG IV, 2016). Of these, the genus Hydrocotyle L. which comprises c.130 species (Hiroe, 1979; Pimenov and Leonov, 1993; Du and Ren, 2010; APG IV, 2016, https://www.mobot.org/mobot/research/apweb/) from Hydrocotyloideae Link of the family Apiaceae transferred to the family Araliaceae based on molecular phylogenetic studies (Lowry et al., 2004; Plunkett et al., 2004). The recent massive development in the next-generation sequencing platforms and bioinformatics tools including cloud-based bioinformatic analyses has proven extremely useful in understanding the deeper-level phylogenetic relationships of angiosperms (Ali, 2021). The present study evaluates the proximity of Hydrocotyle umbellata with Araliaceae based on plastome and phylotranscriptomic analyses.
2 Materials and methods
2.1 Phylotranscriptomic analyses of the selected taxon
The RNA transcriptome SRA data of Hydrocotyle umbellata L. [(Sp. Pl. 1: 234 (1753)] available (https://zenodo.org/record/3255100#.X9QFGtgza70) from the previous study (Leebens-Mack et al., 2019) was retrieved, and analyzed together with (1) ingroup taxon: Angelica archangelica L. [(Sp. Pl. 1: 250–251 (1753)], Centella asiatica (L.) Urb. [(Fl. Bras. 11(1): 287 (1879)], Hedera helix L. [(Sp. Pl. 1: 202 (1753)], Griselinia littoralis (Raoul) Raoul [(Choix Pl. Nouv.-Zél. 22 (1846)], Griselinia racemosa (Phil.) Taub. [(Bot. Jahrb. Syst. 16: 390 (1892)], Pennantia corymbosa J.R. Forst. & G. Forst. [(Char. Gen. Pl. 67 (1775)], Pittosporum sahnianum Gowda [(J. Arnold Arbor. 32(4): 305–307 (1951)], Pittosporum resiniferum Hemsl. [(Bull. Misc. Inform. Kew 1894: 344 (1894)], Dipsacus asper Wall. ex DC. [(Prodr. 4: 646 (1830)], and (2) outgroup: Dipsacus asper [(Prodr. 4: 646 (1830)] (Table 1). The selected aligned data set were imported to MEGA X (Kumar et al., 2018) and converted into .mega format, and the evolutionary analyses were performed using Maximum Parsimony (MP) bootstrap methods (Felsenstein, 1985).
S. No.
Taxon
Family
SRA accession
Ingroup
1.
Angelica archangelica L.
Apiaceae
ERS1829701
2.
Centella asiatica (L.) Urb.
Apiaceae
ERS1829708
3.
Hydrocotyle umbellata L.
Araliaceae
ERS1829705
4.
Hedera helix L.
Araliaceae
ERS1829702
5.
Griselinia littoralis (Raoul) Raoul
Griseliniaceae
ERS1829706
6.
Griselinia racemose (Phil.) Taub.
Griseliniaceae
ERS1829707
7.
Pennantia corymbosa J.R. Forst. & G. Forst.
Pennantiaceae
ERS1829709
8.
Pittosporum sahnianum Gowda
Pittosporaceae
ERS3670337
9.
Pittosporum resiniferum Hemsl.
Pittosporaceae
ERS1829710
Outgroup
10.
Dipsacus asper Wall. ex DC.
Caprifoliaceae
ERS1829762
2.2 Analyses of plastome data
The plastome (chloroplast genome) of the family Apiaceae and Araliaceae e.g. 1. Angelica dahurica (Fisch.) Benth. & Hook. f. {[Enum. Pl. Jap. 1(1): 187 (1873)]}, 2. Eleutherococcus senticosus {(Rupr. ex Maxim.) Maxim. [Mém. Acad. Imp. Sci. St.-Pétersbourg Divers Savans 9: 132 (1859)], 3. Hydrocotyle sibthorpioides Lam. {[Encycl. 3(1): 153 (1789)]}, 4. Kalopanax septemlobus (Thunb.) Koidz. {[Bot. Mag. (Tokyo) 39(468): 306 (1925)]}, and 5. Petroselinum crispum (Mill.) Mansf. {[Repert. Spec. Nov. Regni Veg. 46 (1168–1170): 307 (1939)]} were retrieved from NCBI GenBank (Table 2), and the genomic rearrangements and the relationships among the selected taxon were detected using MAUVE (Darling et al., 2004).
S. No.
Taxon
Family
NCBI GenBank accession number
1.
Angelica dahurica (Fisch.) Benth. & Hook. f.
Apiaceae
NC_029392.1/KT963037.1
2.
Petroselinum crispum (Mill.) Mansf.
Apiaceae
NC_015821.1/HM596073.1
3.
Eleutherococcus senticosus (Rupr. ex Maxim.) Maxim.
Araliaceae
NC_016430.1/JN637765.1
4.
Hydrocotyle sibthorpioides Lam.
Araliaceae
NC_035502.1/KT589392.1
5.
Kalopanax septemlobus (Thunb.) Koidz.
Araliaceae
NC_022814.1/KC456167.1
3 Results
3.1 Plastome and phylotranscriptome dataset characteristics
The present phylotranscriptomic analyses is from 11,495 parsimony informative sites out of a total of 1,42,796 positions in the aligned (the aligned data set contains 85,153 conserved, 48,037 variable and 35,240 singleton sites, Fig. 1) transcriptome dataset of A. archangelica (Apiaceae), C. asiatica (Apiaceae), H. umbellate (Araliaceae), H. helix (Araliaceae), G. littoralis (Griseliniaceae), G. racemose (Griseliniaceae), P. corymbosa (Pennantiaceae), P. sahnianum (Pittosporaceae), P. resiniferum (Pittosporaceae) and D. asper (Caprifoliaceae).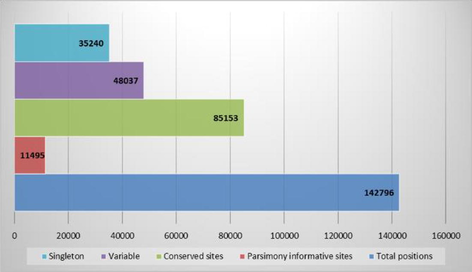
The aligned transcriptome data set showing several different sites. A total number of 11,495 parsimony informative sites out of a total number of 14,27,95 sites were used in phylogenetic analysis.
3.2 Phylotranscriptomic analyses
The phylotranscriptomic analyses recovered the MPT with tree length of 0.628 [consistency index 0.689, retention index 0.404, composite index 0.362]. The MPT revealed the proximity (bootstrap support 99%) of H. umbellata (ex Apiaceae, Araliaceae) with H. helix (Araliaceae), while A. archangelica (Apiaceae), C. asiatica (Apiaceae) clade (supported by 99% bootstrap support)- P. sahnianum (Pittosporaceae)- G. littoralis (Griseliniaceae)- P. corymbosa (Pennantiaceae) shows proximity of Araliaceae clade (70% bootstrap support) (Fig. 2), which is also evident from the estimates of evolutionary divergence between the sequences (Table 3) and the alignment of plastome of A. dahurica (Apiaceae), P. crispum (Apiaceae), E. senticosus (Araliaceae), H. sibthorpioides (Araliaceae) and K. septemlobus (Araliaceae). The Araliaceae clade [constitute with E. senticosus (Araliaceae) - H. sibthorpioides (Araliaceae) with branch length 0.08] clade with K. septemlobus (Araliaceae) (branch length 0.15), while A. dahurica (Apiaceae) (branch length 0.15) clade together with P. crispum (Apiaceae) (branch length 0.17) (Fig. 3). In the maximum likelihood analyses, the tree topology and the proximity of H. umbellata with the taxon included in the analyses found similar to MPT.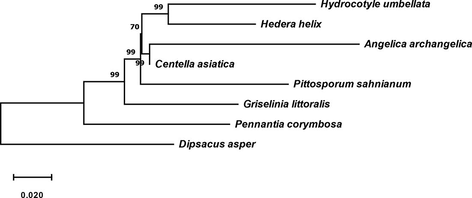
The evolutionary tree to evaluates the proximity of the genus Hydrocotyle with Araliaceae based on phylotranscriptomic analyses.
G. littoralis
P. sahnianum
H. umbellata
P. corymbosa
H. helix
A. archangelica
C. asiatica
D. asper
G. littoralis
P. sahnianum
0.145
H. umbellata
0.164
0.171
P. corymbosa
0.150
0.182
0.202
H. helix
0.118
0.134
0.123
0.168
A. archangelica
0.187
0.193
0.201
0.220
0.180
C. asiatica
0.158
0.167
0.179
0.198
0.146
0.195
D. asper
0.223
0.243
0.248
0.211
0.231
0.269
0.252
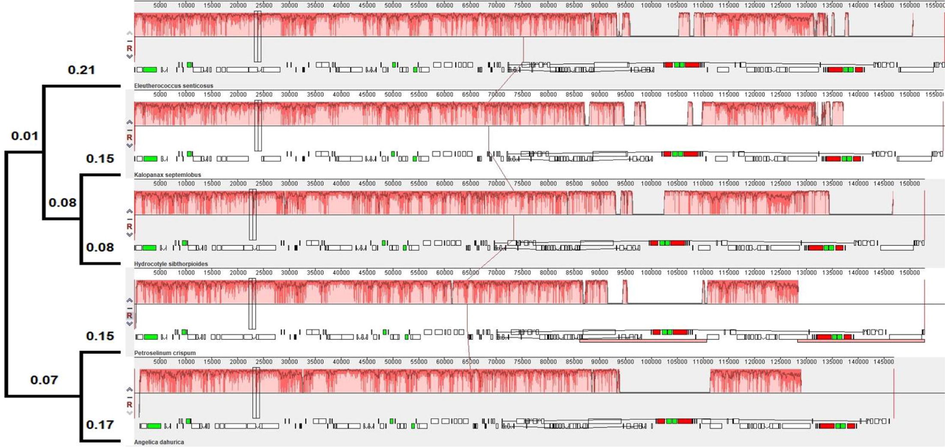
The plastome alignment of the representative of the family Apiaceae and Araliaceae using MAUVE (Darling et al., 2004). Alignment Lane1: Eleutherococcus senticosus, 2. Kalopanax septemlobus, 3. Hydrocotyle sibthorpioides, 4. Petroselinum crispum, 5. Angelica dahurica.
3.3 Plastome genome size and CDS
The comparison of the plastome size and coding sequence (CDS) of 29 species of the family Apiaceae (e.g. Anethum graveolens L. (KR011055.1), Angelica acutiloba (Siebold & Zucc.) Kitag. (KT963036.1), Angelica dahurica (Fisch.) Benth. & Hook. f. (KT963037.1), Angelica gigas Nakai (KT963038.1), Anthriscus cerefolium (L.) Hoffm. (GU456628.1), Arracacia xanthorrhiza Bancr. (KY117235.1), Bupleurum boissieuanum H. Wolff (MF663725.1), Bupleurum falcatum L. (KM207676.1), Bupleurum latissimum Nakai (KT983258.1), Carum carvi L. (KR048286.1), Cicuta virosa L. (KX352466.1), Coriandrum sativum L. (KR002656.1), Crithmum maritimum L. (HM596072.1), Daucus carota L. (DQ898156.1), Foeniculum vulgare Mill. (KR011054.1), Glehnia littoralis F. Schmidt ex Miq. (KU866532.1), Hansenia forbesii (H.Boissieu) Pimenov & Kljuykov (KX808492.1), Hansenia forrestii (H.Wolff) Pimenov & Kljuykov (KX808494.1), Hansenia oviformis (R.H.Shan) Pimenov & Kljuykov (KX808493.1), Hansenia weberbaueriana (Fedde ex H.Wolff) Pimenov & Kljuykov (KX808491.1), Ledebouriella seseloides (Hoffm.) H.Wolff (KU866529.1), Ligusticum tenuissimum (Nakai) Kitag. (KT963039.1), Ostericum grosseserratum (Maxim.) Kitag. (KT852844.1), Petroselinum crispum (Mill.) Fuss (HM596073.1), Peucedanum insolens Kitag. (KU041143.1), Peucedanum japonicum Thunb. (KU866530.1), Pleurospermum camtschaticum Hoffm. (KU041142.1), Prangos trifida (Mill.) Herrnst. & Heyn (MG386251.1), Pterygopleurum neurophyllum (Maxim.) Kitag. (KT983257.1), and 19 species of the family Araliaeae (e.g. Aralia undulata Hand.-Mazz. (KC456163.1), Brassaiopsis hainla (Buch.-Ham.) Seem. (KC456164.1), Dendropanax dentiger (Harms) Merr. (KP271241.1), Dendropanax morbiferus H.Lév. (KR136270.1), Eleutherococcus senticosus (Rupr. & Maxim.) Maxim. (JN637765.1), Fatsia japonica (Thunb.) Decne. & Planch. (KR021045.1), Hydrocotyle sibthorpioides Lam. (KT589392.1), Hydrocotyle verticillata Thunb. (HM596070.1), Kalopanax septemlobus (Thunb.) Koidz. (KC456167.1), Metapanax delavayi (Franch.) J.Wen & Frodin (KC456165.1), Panax ginseng C.A.Mey. (AY582139.1), Panax japonicus (T.Nees) C.A.Mey. (KP036469.1), Panax notoginseng (Burkill) F.H.Chen (KJ566590.1), Panax quinquefolius L. (KM088018.1), Panax stipuleanatus H.T.Tsai & K.M.Feng (KX247147.1), Panax trifolius L. (MF100782.1), Panax vietnamensis Ha & Grushv. (KP036470.1), Schefflera delavayi (Franch.) Harms (KC456166.1), Schefflera heptaphylla (L.) Frodin (KT748629.1), revealed that the plastome size ranges between 140,000 to 160,000 nucleotides base-pairs in both Apiaceae and Araliaceae, but the CDS varies e.g. from 71 to 99 CDS in Apiaceae, and while narrowly varies (CDS 85 to 87) in Araliaceae, indicating similar content and pattern of CDS in the genus Hydrocotyle with Araliaceae.
4 Discussion
The phylogenetic reconstruction of evolutionary histories in plants based on molecular data has been based largely on nuclear sequences or chloroplast/plastid DNA markers (Ali et al., 2014). The recent massive development in the next-generation sequencing platforms has brought cost-effective sequencing of genome or organelle genome e.g. chloroplast and mitochondria, which have proven extremely useful in understanding the deeper-level phylogenetic relationships of angiosperms (Ali, 2021). The transcriptome/ RNA-Seq (Wang et al., 2009) have attracted the attention for the reconstruction of evolutionary histories in plants because it allows massively parallel sequencing of expressed genes within a single genome which offers a powerful means of investigating the signals for evolutionary implications in higher taxonomic level such as Charophytes, Caryophyllales, Vitaceae, Brassicaceae, Betulaceae, Asteraceae, Hydnoraceae, Asclepias, Ferns; at a lower taxonomic level such as Camelina sativa, Artemisia tridentata, Ranunculus, Flaveria, or at angiosperms level; and have also been proven very useful in comparative transcriptomics, character evolution, domestication of crops and genome evolution (Wen et al., 2014).
The present phylotranscriptomic analyses revealed the proximity of H. umbellata (ex Apiaceae, Araliaceae) with H. helix (Araliaceae). The family Araliaceae (-Ginseng family) have distributed mostly in tropical regions, consists of c. 41 plus genera under Subfamily Aralioideae (1. Anakasia Philipson, 2. Aralia L., 3. Astrotricha DC., 4. Brassaiopsis Decne. & Planch., 5. Cephalaralia Harms, 7. Cheirodendron Nutt. ex Seem., 8. Cussonia Thunb., 9. Dendropanax Decne. & Planch., 10. Eleutherococcus Maxim., 11. Fatsia Decne. & Planch., 12. Gamblea C.B. Clarke, 13. Harmsiopanax Warb., 14. Hedera L., 15. Heteropanax Seem., 16. Hunaniopanax C.J. Qi & T.R. Cao, 17. Kalopanax Miq., 18. Macropanax Miq., 19. Megalopanax Ekman ex Harms, 20. Merrilliopanax H.L. Li, 21. Meryta J.R. Forst. & G. Forst., 22. Metapanax J. Wen & Frodin, 23. Motherwellia F. Muell., 24. Oplopanax (Torr. & A. Gray) Miq., 25. Oreopanax Decne. & Planch., 26. Osmoxylon Miq., 27. Panax L., 28. Plerandra A. Gray, 29. Polyscias J.R. Forst. & G. Forst., 30. Pseudopanax C. Koch, 31. Raukaua Seem., 32. Schefflera J.R. Forst. & G. Forst., 33. Sciadodendron Griseb., 34. Seemannaralia R. Vig., 35. Sinopanax H.L. Li, 36. Stilbocarpa (Hook. f.) Decne. & Planch., 37. Tetrapanax (K. Koch) K. Koch, 38. Trevesia Vis., 39. Woodburnia Prain) and Subfamily Hydrocotyloideae: 40. Hydrocotyle, 41. Trachymene (APG, 2016); out of these, some possess immense medicinal impotence such as Hedera sp., Panax sp. (Ginseng), and Eleutherococcus senticosus (Wen et al., 2000; Plunkett et al., 2004), are distinctive in being palmate or pinnate leaves, heads, reduced calyx, apopetalous to sympetalous corolla, and a 1–∞-carpellate inferior ovary, apical-axile placentation, fruit a berry, drupe, or schizocarp (Plunkett et al. 2004). The family Araliaceae show considerable floral variation. The family Apiaceae and Araliaceae resemble each other in the structure of their gynoecia, and are closely related (Plunkett et al., 2004), the family Araliaceae with the subfamily Hydrocotyloideae, Harmsiopanax and Aralioideae accepted as a monophyletic branch within the Apiales, an order within the Asterids; of these, Hydrocotyle from Hydrocotyloideae of Apiaceae transferred to Araliaceae based on molecular phylogenetic studies (Lowry et al., 2004; Plunkett et al., 2004). The Hydrocotyloideae are sister to the rest of the family (Chandler and Plunkett, 2004; Plunkett et al., 2004; Nicolas and Plunkett, 2009), the species of the genus Hydrocotyle are highly polyploidy (Yi et al., 2004), show early corolla tube initiation (Leins and Erbar, 1997; Erbar and Leins, 2004), calyx remains absent (Tseng, 1967), and possess has trilacunar nodes, laterally flattened fruits with a sclerified endocarp and stipules are cauline or borne on the leaf base (Sinnott and Bailey 1914).
In conclusion, the present study evaluates the proximity of Hydrocotyle with Araliaceae based on plastome and phylotranscriptomic analyses using the minimum evolution method. The family ‘Apiaceae/Umbelliferae and Araliaceae resemble each other in the structure of their gynoecia, and were placed in the order Apiales, and are closely related. The family Araliaceae with the subfamily 1. Hydrocotyloideae Link, 2. Harmsiopanax Harms and 3. Aralioideae Eaton accepted as a monophyletic branch within the Apiales, an order within the Asterids. Of these, Hydrocotyle L. from Hydrocotyloideae Link of Apiaceae transferred to Araliaceae based on molecular phylogenetic studies. The present analyses revealed the nesting of the Hydrocotyle under Araliaceae in the MPT, and the proximity is further supported by the evolutionary divergence between the sequences and plastome alignment.
Acknowledgements
The authors thank the Deanship of Scientific Research and RSSU at King Saud University for their technical support.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- The changing epitome of species identification – DNA barcoding. Saudi J. Biol. Sci.. 2014;21(3):204-231.
- [Google Scholar]
- Taxonomic implication of phylotranscriptomic analysis of Dillenia indica L. (Dilleniales, Dilleniaceae). Saudi. J. Biol. Sci. 2021 https://www.sciencedirect.com/science/article/pii/S1319562X21000395
- [Google Scholar]
- An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linnean Soc.. 2016;181:1-20.
- [Google Scholar]
- Evolution in Apiales: Nuclear and chloroplast markers together in (almost) perfect harmony. Bot. J. Linnean Soc.. 2004;2:123-147.
- [Google Scholar]
- A revised system of classification of the angiosperms. Bot. J. Linnean Soc.. 1980;80(2):91-124.
- [Google Scholar]
- Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res.. 2004;14(7):1394-1403.
- [Google Scholar]
- Hydrocotyle changanensi (Araliaceae), a new species from Shaanxi. China. Ann. Bot. Fenn.. 2010;47:403-407.
- [Google Scholar]
- Sympetaly in Apiales (Apiaceae, Araliaceae, Pittosporaceae) South Afr. J. Bot.. 2004;70:458-467.
- [Google Scholar]
- Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39(4):783-791.
- [Google Scholar]
- Recent advances in understanding Apiales and a revised classification Author links open overlay panel. S. Afr. J. Bot.. 2004;70:371-381.
- [Google Scholar]
- Hydrocotyle. In: Umbelliferae of World. Matsuo Biru, Tokyo, Japan: Ariake Book Company; 1979. p. :103-168.
- [Google Scholar]
- Evolution of the Araliaceae family inferred from complete chloroplast genomes and 45S nrDNAs of 10 Panax-related species. Sci. Rep.. 2017;7:4917.
- [Google Scholar]
- MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol.. 2018;35:1547-1549.
- [Google Scholar]
- Floral developmental studies: Some old and new questions. Int. J. Plant Sci.. 1997;158:S3-S12.
- [Google Scholar]
- One thousand plant transcriptomes and the phylogenomics of green plants. Nature. 2019;574:679-685.
- [Google Scholar]
- Generic relationships in Araliaceae: looking into the crystal ball. S. Afr. J. Bot.. 2004;70:382-392.
- [Google Scholar]
- The demise of subfamily Hydrocotyloideae (Apiaceae) and the re-alignment of its genera across the whole order Apiales. Mol. Phyl. Evol.. 2009;53:134-151.
- [Google Scholar]
- The Genera of the Umbelliferae: A nomenclature. Kew: Royal Botanic Gardens; 1993. p. :5-161.
- Investigations on the phylogeny of the angiosperms. 3. Nodal anatomy and the morphology of stipules. Am. J. Bot.. 1914;1:441-453.
- [Google Scholar]
- Anatomical studies of flower and fruit in Hydrocotyloideae (Umbelliferae) Univ. California Publ. Bot.. 1967;42:1-79.
- [Google Scholar]
- RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet.. 2009;10:57-63.
- [Google Scholar]
- The evolution of Araliaceae: a phylogenetic analysis based on ITS sequences of nuclear ribosomal DNA. Syst. Bot.. 2000;26:144-167.
- [Google Scholar]
- Utility of transcriptome sequencing for phylogenetic inference and character evolution. In: Hörandl E., Appelhans M.S., eds. Next-Generation Sequencing in Plant Systematics. IAPT; 2014. p. :1-41. C2
- [Google Scholar]
- Evolutionary divergence and convergence in proteins. In: Bryson V., Vogel H.J., eds. Evolving Genes and Proteins. New York: Academic Press; 1965. p. :97-166.
- [Google Scholar]







