Translate this page into:
The genetic relatedness between Salmonella enterica isolated from food samples and outpatient’s clinics using pulsed field gel electrophoresis
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Background and objectives
Salmonella enterica (S. enterica) is an important food-borne pathogen that causes many public health problems, molecular characterizations and fingerprinting of the recovered strain are very important for epidemiological surveillance. Therefore, food of animal origins collected from various marketplaces located in Riyadh, KSA, were tested to evaluate the incidence, molecular characterizations, and fingerprinting of Salmonella species using multiplex-PCR and Pulsed-Field Gel Electrophoresis (PFGE).
Methods
Three hundred and fifty food samples (125 minced poultry meat, 110 ground beef meat, and 115 hamburgers beef meat) were collected from markets placed in Riyadh, KSA, from the period of June 2019 to October 2019. Also, 25 strains of Salmonella species isolated from humans who suffer from food poisoning and gastrointestinal disturbance were collected from hospitalized patients situated in Riyadh from the period of May 2019 to October 2019. Typical bacteriological practices for culturing and identification of Salmonella from the collected food samples, as well as molecular detection and characterizations using multiplex-PCR and fingerprinting of the recovered Salmonellae species using PFGE were carried out.
Results
Thirty-seven strains of S. enterica subspecies (10.57%) were isolated, mostly from chicken samples 19 (15.2%). Identical chromosomal restriction patterns had been observed between the strains of Salmonella Typhimurium recovered from food of animal origins and the strains recovered from outpatients’ clinics.
Conclusions
The study concluded that chickens are the main natural reservoir host of Salmonella infection for human being as the genetic similarity and the degree of relatedness between the recovered strains of S. enterica from food samples and from outpatients clinics using PFGE revealed a high degree of identity (similarity index up to 99%). Therefore the regulations for handling chicken products should be updated to reduce Salmonella infections.
Keywords
S. enterica
Pulsed-field gel electrophoresis
Genetic fingerprinting
Food samples
1 Introduction
S. enterica is a very significant food-borne microorganism that leads to many public health problems for man and animals (Shaw et al., 2018). It is the second bacterial food-borne illness affecting human, especially by consumptions food samples originated from chickens animal (Evans et al., 2015; Persed and Lejeune, 2018). Spreading of S. enterica from domestic and wild animals to human cause food poisoning and severe illnesses, such as diarrhea, fever and general fatigue (Rimet et al., 2019). Salmonella infections had been increased in the Kingdom of Saudi Arabia (KSA), and most of the frequent serotypes recovered from cases of food-borne infections or gastrointestinal tract infections were identified as Salmonella Typhimurium and Salmonella Enteritidis; this finding substantiates the report of the (CDC, 2011, 2014; USDA-FSIS, 2015).
Standard bacteriological techniques for isolation and serotyping of Salmonella Typhimurium and Salmonella Enteritidis required several days for complete identification (Gast, 2013), which is considered problematic, especially for epidemiological surveillance and disease outbreak (Rimet et al., 2019). Molecular diagnosis is more reliable, accurate, and time-consuming for the diagnosis of food-borne pathogens and is recommended especially for epidemiological surveillance than other conventional methods (HuaZou et al., 2016; Imre et al., 2005). The invA gene is more commonly used for diagnosis Salmonella as it’s primarily associated with all Salmonellae except Salmonella Litchfield and Salmonella Senftenberg (Hara-Kudo et al., 2005; Malorny et al., 2003; Soliman et al., 2018). Serotyping of Salmonella Typhimurium and Salmonella Enteritidis had been improved by several multiplex PCR directly from the food samples or after pre-enrichment on Rappaport-Vassiliadis media (Alzwghaibi et al., 2018; Dele Ogunremi et al., 2017). During the outbreak of food poisoning caused by S. enterica, molecular characterizations and fingerprinting are very important to evaluate the genetic similarity between the recovered strains of S. enterica from food samples patients suffering from gastrointestinal tract disturbance such as diarrhea, fever, severe illnesses, and general fatigue is very important for epidemiological surveillance (CDC, 2019) to detect the main source of infection and the degree of food contaminations (USDA-FSIS, 2015).
The genetic relatedness between S. enterica isolates recovered from a different sample using pulsed field gel electrophoresis (PFGE) remains the furthermost reliable, accurate molecular methods for epidemiological studies during the outbreaks (Gatto et al., 2006; Long et al., 2010). Through the PFGE, the whole genome of the recovered strain was cut into several bands of characteristic molecular weight for each strains generating specific chromosomal patterns for each strain. The degree of similarity of such chromosomal patterns determines the genetic relatedness between the strains (Long et al., 2010).
Therefore, this study aimed to evaluate the incidence of Salmonella Enteritidis and Salmonella Typhimurium in food samples collected from KSA, using standard microbiological techniques, molecular serotyping of the recovered strains, and fingerprinting of the recovered strains of Salmonella using PFGE to evaluate the degree of similarity between the strains recovered from foodstuff and outpatients suffering from diarrhea, fever, food poisoning, and general fatigue.
2 Materials and methods
2.1 Samples collection
Three hundred and fifty food samples (125 minced poultry meat, 110 ground beef meat, and 115 hamburgers beef meat) were collected from markets located in Riyadh, KSA, from June 2019 to October 2019. Also, 25 strains of Salmonella species recovered from human suffering from food poisoning and gastrointestinal disturbance were collected from different outpatient clinics located in Riyadh from the period of May 2019 to October 2019.
2.2 Standard bacteriological and serotyping identification of S. enterica
Standard bacteriological and serotyping identification of S. enterica from food samples using ISO/TR6579 -3 (2014) were carried out using a laboratory blender (Stomacher 400) for homogenization of food samples in buffered peptone water and incubated 25 g of the examined specimens were mixed in 225 ml of the peptone water and incubated for16 hours at 37 °C. Then 0.1 ml was injected in 10 ml of Rappaport-Vassiliadis and selenite cystine broth as a selective pre-enrichment broth. One ml from each pre-enrichment broth was sub-cultured into xylose lysine deoxycholate and MacConkey agar media. Non-lactose fermenting colonies were identified by using specific monovalent and polyvalent antisera against somatic, flagella, and virulence antigens. Positive colonies, as well as one milliliter of a selective broth culture of Rappaport-Vassiliadis were used for molecular identification using multiplex-PCR.
2.3 Susceptibility testing
All Salmonella strains isolated from both food specimens and outpatient clinic were tested by Kirby Bauer method to evaluate the sensitivity of the recovered isolates against a wide range of antibiotics (penicillin G P10, ampicillin AMP10, amoxicillin-clavulanic acid AMC 30, ciprofloxacin CIP5, gentamicin CN10, ceftazidime CAZ30, chloramphenicol C 30, erythromycin E15, sulphamethoxazole-trimethoprim SXT 25, nalidixic acid NA30, oxytetracycline OT30, vancomycin VA 30, norfloxacin NOR 10). The result was interpreted according to CLSI (CLSI, 2015).
2.4 Molecular characterization of Salmonella
2.4.1 DNA extraction
The DNA of the suspected colonies was extracted by boiling methods in Tris-EDTA buffer using heat block at100°C/20 min after washing off the suspected colonies with phosphate buffer saline (Alzwghaibi et al., 2018). After centrifugation at 14000 rpm/10 min, 5 μl from the supernatant can be applied as a template for multiplex PCR. The DNA from the pre-enriched broth and the suspected Salmonella isolates collected from outpatient clinics were extracted by DNA extraction Kits from Thermo Fisher Scientific.
-
Molecular identification and serotyping of the isolated S. enterica using Multiplex-PCR.
The genomic DNA of the suspected colonies were being tested by three primer pairs for molecular identification and molecular serotyping, the first primer pair targeting a specific sequence for all species of Salmonella (Dele Ogunremi et al., 2017; Moussa et al., 2012; Soumet et al., 1999) using ST11&ST15. The second primer pair amplifies a specific sequence within the sefA gene-specific for Salmonella Enteritidis (Soumet et al., 1999) using Sef167-Sef478 primers. The third primer pair amplify specific sequence within the flic gene-specific for Salmonella Typhimurium (Moussa et al., 2012) using (Fli15&Typ04) primers.
2.4.2 Pulsed field gel electrophoresis
Tryptic soy agar plates were applied for growing the isolates at 37 °C for one successive day. Two and a half ml of PIV buffer were thoroughly mixed with the bacterial cells. Five hundred µl of these suspensions were mixed with 500 µl of 1.6% low melting point agarose. This agar-cell suspension mixture was directly disseminated into the bores of the plug molds and allowed to solidify at 4 °C for 30 min. The plugs were moved into tubes comprising 1 ml of 1X Lysis buffer (6 mM Tris-HCl [pH 7.4]; 1 M NaCl; 10 mM EDTA [pH 7.5]; 0.5% Brij 58; 0.2% deoxycholate; 0.5% sodium lauroyl sarcosine; 0.5 mg/ml lysozyme; 10 mg/ml RNase A) and incubated at 37 °C in a water bath. The lysis buffer was replaced with 1 ml ESP buffer (10 mM Tris-HCl (pH 7.4); 1 mM EDTA; 100 µg/ml Proteinase K; 1% SDS), the combination was incubated at 50 °C/overnight in a water bath. Bacterial plugs were washed 4 times with 5 ml TE for 30 min each wash at room temperature (Long et al., 2010). The plug slices of 3 × 5 mm-wide were placed in a 1.5 ml microcentrifuge tube containing 200 µl of 1X restriction buffer (NE Buffer with 50 U of XbaI-digested restriction enzyme, and incubated overnight at room temperature. Plug slices were washed for 30 min with 0.5X TBE and subsequently inserted on the well of 1% agarose gel, and the wells were overlaid with 1% LMP agarose dissolved in 0.5X TBE. After solidification, the gel was run.
3 Results
3.1 Results of standard bacteriological and serotyping identification of Salmonella from food samples
Thirty-seven strains of Salmonella species (10.57%) out of 350 examined food samples (minced poultry meat 125, ground beef meat 110, and hamburger 115 samples) were isolated (Table 1). Most of the strains recovered from chicken samples, 19 (15.2%), serotyping of such strains revealed, 8 strains (6.4%) Salmonella Enteritidis, 4 strains (3.2%) Salmonella Typhimurium, 4 strains (3.2%) Salmonella Gallinarum, and 3 strains (2.4%) Salmonella Pullorum (Table 1). Thirteen strains (11.31%) of S. enterica were isolated from hamburger, typing of such strains revealed 5 strains (4.35%) Salmonella Typhimurium, 3 strains (2.61%) Salmonella Newport, 3 strains (2.61%) Salmonella Agona, and two strains (1.74%) Salmonella Dublin Dublin as shown in Table 1.
Types of Food samples
Food samples
Salmonella enterica subspecies
Total number
Positive for Salmonella
Salmonella enterica subspecies
Number of isolates
Minced poultry meat
125
19 (15.2%)
Salmonella Enteritidis
8 (6.4%)
Salmonella Typhimurium
4 (3.2%)
Salmonella. Gallinarum
4 (3.2%)
Salmonella. Pullorum
3 (2.4%)
Ground beef meat
110
5 (4.55%)
Salmonella Typhimurium
3 (2.73%)
Salmonella Dublin
2 (1.82%)
Hamburger
115
13 (11.31%)
Salmonella Typhimurium
5 (4.35%)
Salmonella Dublin
2 (1.74%)
Salmonella Newport
3 (2.61%)
Salmonella Agona
3 (2.61%)
Total
350
37 (10.57%)
Salmonella enterica
37 (10.57%)
Five strains of S. enterica (4.55%) were isolated from ground minced meat; serotyping of such strains revealed 3 strains (2.73%) Salmonella Typhimurium and 2 strains (1.82%) Salmonella Dublin as shown in Table 1.
3.2 Antibiotic susceptibility test of the recovered S. enterica
The recovered strains showed 95% tolerance to penicillin and 85% resistance to both colistin sulphate and norfloxacin. While the degree of resistance to gentamicin, nalidixic acid, and flumequine were 75%, 70%, and 60% correspondingly, as revealed in Table 2. Molecular serotyping of All the strains of Salmonella (25 strains) collected from outpatients by multiplex-PCR were identified as Salmonella Typhimurium.
Antibiotics
resistance %
Ampicillin (A10, 10 µg)
25%
Chloramphenicol (C30, 30 µg)
35%
Cefotaxime (CTX30, 30 µg)
20%
Colistine Sulphate (CL10, 10 µg)
85%
Ciprofloxacin (CF5, 5 µg),
50%
Flumequine (UB30, 30 µg)
60%
Gentamicin (G10, 10µ g)
75%
Nalidixic acid (NA30, 30 µg)
70%
Norfloxacin (Nor10, 10 µg)
85%
Neomycin (N30, 30 µg)
45%
Penicillin-G (P10, 10 I.U)
95%
Oxytetracycline (O30, 30 µg)
5%
Streptomycin (S10, 10 µg)
50%
3.3 Molecular identification and serotyping of the recovered Salmonella isolates by multiplex-PCR
Magnification of 429 bp fragments specific for all members of Genus Salmonella were observed with all strains previously identified as Salmonella by standard microbiological techniques. Amplification of 559 bp fragments sequence of the flic gene were observed with the strains of Salmonella Typhimurium. In contrast, the strains of Salmonella Enteritidis showed amplification of 559 bp fragments specific fore sefA gene as presented in Fig 1. Molecular serotyping of All the strains of Salmonella (25 strains) collected from outpatients by multiplex-PCR revealed amplification of 429 bp and 559 bp fragments of Salmonella Typhimurium (Fig. 1).
-
Genetic fingerprinting of the recovered S. enterica subspecies by Chromosomal DNA analysis using PFGE.
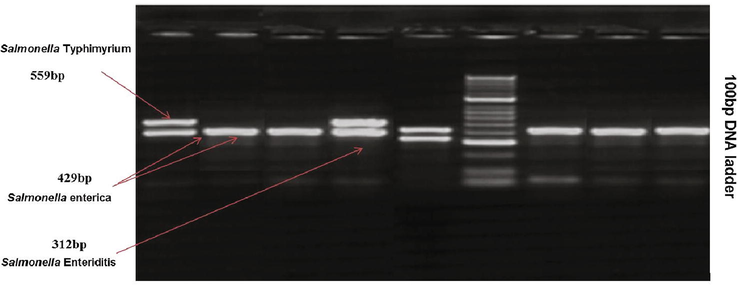
- Multiplex-PCR showing amplification of 429 base pair fragments of Salmonella species, 312 base pair fragments of Salmonella Enteriditis and 559 bp fragments of Salmonella Typhimyrium.
Restriction digestion of the recovered strains by PFGE using XbaI- restriction enzyme revealed 24 chromosomal digestion patterns as shown in Figs. 2 and 3. The XbaI- restriction enzyme digests the DNA of the studied S. enterica into 11 to 12 fragments of different molecular weight. Meanwhile, Salmonella Typhimurium strains isolated from food samples and from outpatients showed identical chromosomal restriction patterns.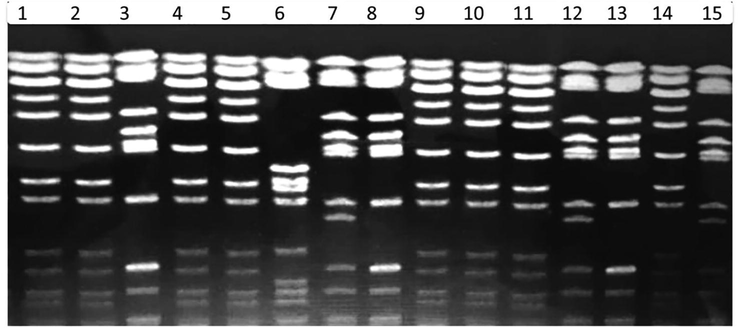
PFGE separation of restriction fragments of the Salmonella enterica digested genome from food of animal origin and from human samples collected from outpatient clinics with Xbal-digested restriction enzyme.
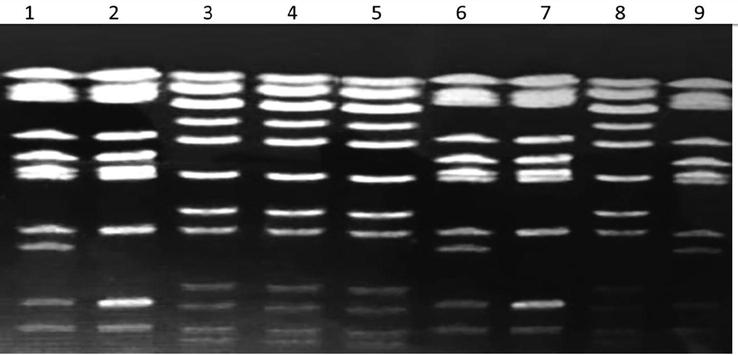
PFGE separation of restriction fragments of the Salmonella enterica digested genome from food of animal origin and from human samples collected from outpatient clinics with Xbal-digested restriction enzyme.
Moreover, the digested fragments of the chromosomal DNA of all strains of Salmonella generated by XbaI- restriction enzyme revealed a number of identical bands similar in their molecular weight (shared bands) in all strains indicating the remarkable similarity of the tested strains. However, these sharing bands differed in their intensity as the concentration of the DNA templates differs in each sample (Figs. 2 and 3).
The phylogenetic analysis of the examined S. enterica subspecies with pulsed field gel electrophoresis revealed five major clusters according to the degree of similarity between the strains.
The first cluster contains two groups of each of 5 strains, all of them belonging to Salmonella Enteritidis; the strains of this cluster were isolated from food samples collected from chickens. The second clusters contain two groups (12 strains), all of the previously identified as Salmonella Typhimurium and all of them isolated from food samples, and it’s of value to say that some of such strains showed identical chromosomal patterns to each other with similarity index up to 100% as shown in Fig. 4. The third clusters contain two groups (5 strains of them are Salmonella Typhimurium) of identical chromosomal patterns to each other with a similarity index up to 99%, all of them isolated from outpatients clinics. The strains of the second and third cluster previously identified as Salmonella Typhimurium and isolated from food samples and human showed identical chromosomal patterns to each other with a similarity index up to 98%, indicating. The fourth showed one group of one strain (salmonella Dublin) isolated from food and the fifth clusters also showed one group of one strain (Salmonella Newport) isolated from food (Fig. 4).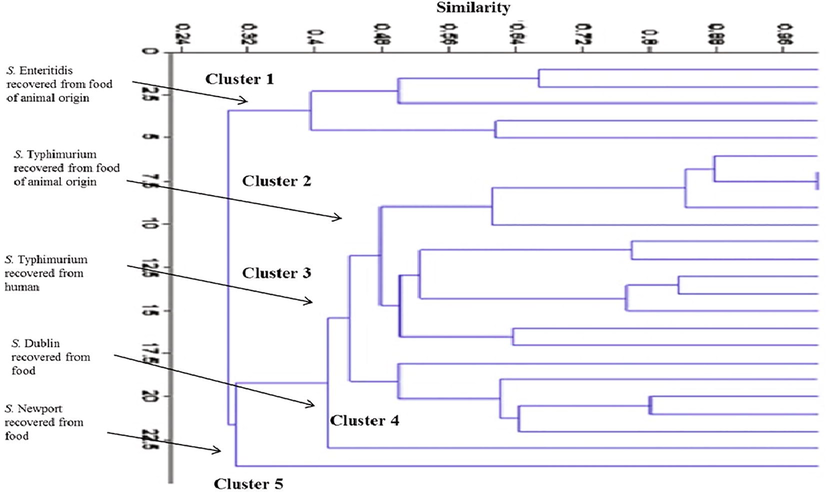
A dendrogram showing the degree of similarity between Salmonella enterica recovered from food of animal origin and clinical isolates recovered from outpatient clinics.
4 Discussion
Salmonella enterica is an essential food-borne pathogen that causes many public health problems. Molecular diagnosis is more reliable, accurate, and time-consuming for the diagnosis of food-borne pathogens and is recommended especially for epidemiological surveillance than other conventional methods (HuaZou et al., 2016; Imre et al., 2005). The study was aimed to evaluate the incidence of Salmonella Enteritidis and Salmonella Typhimurium in food samples collected from KSA, using standard microbiological techniques. Therefore, three hundred and fifty food samples (125 minced poultry meat, 110 ground beef meat, and 115 hamburgers beef meat) were tested. Thirty-seven strains of S. enterica (10.57%) were recovered from the tested food specimens, and most of the isolates recovered from chickens 19 (15.2%) strain out of 125 examined minced poultry meat as shown in Table 1, the same result had been reported by Cortez et al., (2008) and Velusamy et al., (2010), they isolated 10% Salmonella species from foot samples. However, the incidence of Salmonella in the examined food samples collected from Riyadh, KSA were lower than the prevalence rate (18%) of Salmonella isolated from food samples reported in the USA (USDA-FSIS, 2015).
Chickens samples showed a high incidence of Salmonella isolation 19 strains (15.2%) if it compared with hamburger 13 strains (11.31%) and ground beef meet 5 strain (4.55%), these explain the variation between the incidence according to the sample types as chickens considered as a natural reservoir host for Salmonella (CDC, 2011, 2014; Rimet et al., 2019; USDA-FSIS, 2015). The methods of isolation and previous enrichment play an essential role in the isolation of Salmonellae (Moussa et al., 2012). Serotyping of the strains isolated from chickens revealed 8 strains (6.4%) Salmonella Enteritidis, 4 strains (3.2%) Salmonella Typhimurium, 4 strains (3.2%) Salmonella Gallinarum, and 3 strains (2.4%) Salmonella Pullorum as presented in Table 1 the results confirm that chickens and chicken product are the main sources of Salmonellae that causing severe gastroenteritis and food-borne infection of human worldwide (Abd El Halim et al., 2017; CDC, 2014; Rimet et al., 2019).
Ground beef meat, minced meat, hamburger, and all meat products are responsible for Salmonella food poisoning by the ingestion of contaminated foods (CDC, 2019). The incidence of S. enterica from hamburger showed a high degree of Salmonella isolation and 13 strains (11.31%) were isolated, while only five strains of S. enterica (4.55%) were isolated from the minced meat.
Serotyping of the strains isolated from hamburger revealed 5 strains (4.35%) Salmonella Typhimurium, 3 strains (2.61%) Salmonella Newport, 3 strains (2.61%) Salmonella Agona and two strains (1.74%) Salmonella Dublin Dublin while, serotyping of such strains revealed 3 strains (2.73%) Salmonella Typhimurium, and 2 strains (1.82%) Salmonella Dublin as shown in Table 1.
Fingerprinting of the Salmonella strains isolated from food and from outpatient clinics by PFGE using XbaI- restriction enzyme revealed 24 chromosomal digestion patterns as shown in Figs. 2 and 3. The phylogenetic analysis of the recovered 24 chromosomal digestion patterns of the examined S. enterica subspecies with pulsed field gel electrophoresis revealed five major clusters according to the degree of identity between the strains. Identical chromosomal patterns had been observed between the strains of Salmonella Typhimurium isolated from food samples and the strains isolated from outpatient clinics. These findings are confirmed by the identity matrix where the similarity between the strains recovered from human and ground meat beef was 99.1%, while with poultry meat was 98.7–97.9, as shown in Fig. 4.
5 Conclusions
The study concluded that chickens are the main natural reservoir host of Salmonella infection for human being as the genetic similarity and the degree of relatedness between the recovered strains of S. enterica from food samples and outpatients clinics using PFGE revealed a high degree of identity (similarity index up to 99%). Therefore the regulations for handling chicken products should be updated to reduce Salmonella infections.
6 Disclosure of funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Alzwghaibi, A.B., Yahyaraeyat, R., Nayeri Fasaei, B., Ghalyanchi Langeroudi, A., Zahraei Salehi, T., 2018. Rapid molecular identification and differentiation of common Salmonella Serovars isolated from poultry, domestic animals and foodstuff using multiplex PCR assay. Arch. Microbiol. 200(7), 1009–1016. doi: 10.1007/s00203-018-1501-7. Epub 2018 Apr 7.
- Center for Disease Control and Prevention (CDC), 2011. Multistate Outbreak of Human Salmonella Heidelberg Infections Linked to Ground Turkey. Atlanta, GA. Available online at: http://www.cdc.gov/Salmonella/2011/ground-turkey-11-10-2011.html (accessed June 21, 2016).
- Center for Disease Control and Prevention (CDC), 2014. Salmonella. Atlanta, GA. Available online at: http://www.cdc.gov/Salmonella/ (accessed June 21, 2016).
- Center for Disease Control and Prevention (CDC), 2019. Surveillance [Internet]. [Updated 2019 Feb 8; cited 2018 May 29]. Available from: https://www.cdc.gov/Salmonella/reportspubs/surveillance.html.
- Identification of Salmonella spp. isolates from chicken abattoirs by multiplex-PCR. Res. Vet. Sci.. 2008;81:340-344.
- [Google Scholar]
- Dele Ogunremi, Susan Nadin-Davis, Andrée Ann Dupras, Imelda Gálvan Márquez, Katayoun Omidi, Louise Pope, John Devenish, Teresa Burke, Ray Allain, Daniel Leclair, 2017. Evaluation of a multiplex PCR assay for the identification of Salmonella serovars enteritidis and typhimurium using retail and abattoir samples. J. Food Prot. 80(2), 295–301.
- Preharvest Salmonella detection for evaluation of fresh ground poultry product contamination. J. Food Prot.. 2015;78:1266-1271.
- [Google Scholar]
- Claire-Sophie Rimet, John J. Maurer, Larissa Pickler, Lisa Stabler, Kasey K. Johnson, Roy D. Berghaus, Ana M. Villegas, Margie Lee and Monique França 2019. Frontiers Sustainable Food System, 04 February 2019, Salmonella Harborage Sites in Infected Poultry That May Contribute to Contamination of Ground Meat. Frontiers Sustainable Food System. 04 February 2019. https://doi.org/10.3389/fsufs.2019.00002.
- Gast, R.K., 2013. Paratyphoid infections, in Diseases of Poultry, 13th Edn., eds D. E. Swayne, J. R. Glisson, L. R. Mcdougald, L. K. Nolan, D. L. Suarez, and V. Nair (Ames, IA: Wiley-Blackwell), 693–706.
- Loop mediated isothermal amplification for the rapid detection of Salmonella. FEMS Microbiol. Lett.. 2005;253:155-161.
- [Google Scholar]
- Genotyping of Salmonella with lineage-specific genes: correlation with serotyping. Int. J. Infect. Dis.. 2016;49:134-140.
- [Google Scholar]
- Development of a PCR system for the characterisation of Salmonella flagellin genes. Acta Vet Hung.. 2005;53:163-172.
- [Google Scholar]
- ISO/TR6579 -3, 2014. Microbiology of food chain horizontal method for the detection, enumeration and serotyping of Salmonella –part3 guidelines for serotyping of Salmonella spp. ed , p33
- Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl. Environ. Microbiol.. 2003;69:290-296.
- [Google Scholar]
- Rapid detection and characterization of Salmonella enterica serovars by multiplex polymerase chain reaction assay. Afr. J. Biotechnol.. 2012;11(14):3452-3458.
- [Google Scholar]
- Persed, A.K., LeJeune, J., 2018. A review of current research and knowledge gaps in the epidemiology of shiga toxin-producing Escherichia coli and Salmonella spp. in Trinidad and Tobago. Veterinary Sci. 5(2), 42. doi:10.3390/vetsci5020042.
- Shaw, J.A., Henard, C.A., Liu, L., Dieckman, L., Vázquez-Torres, A., Bourret, T.J, 2018. Salmonella enterica serovars Typhimurium has three transketolase enzymes contributing to the pentose phosphate pathway. J. Biol. Chem., 293(29), 11271–11282. https: //doi.org/ 10.1074/ jbc.RA118.003661.
- Salmonella infection in Broiler flocks in Egypt. Biosci. Res.. 2018;15(3):1925-1930.
- [Google Scholar]
- Identification by a multiplex-PCR-based assay of Salmonella Typhimurium and Salmonella Enteritidis strains from environment swabs poultry houses. Lett. Appl. Microbiol.. 1999;29:1-6.
- [Google Scholar]
- Pulsed-field Gel Electrophoresis for Salmonella Infection Surveillance, Texas, USA, 2007. Emerg. Infect. Dis.. 2010 Jun;16(6):983-985.
- [Google Scholar]
- USDA-FSIS, 2015. Progress Report on Salmonella and Campylobacter Testing of Raw Meat and Poultry Products, CY, 1998–2014.
- An overview of food-borne pathogen detection: in the perspective of biosensors. Biotechnol. Adv.. 2010;28:232-254.
- [CrossRef] [Google Scholar]







