Translate this page into:
The functionality of plant-microbe interactions in disease suppression
⁎Corresponding author. Olubukola.babalola@nwu.ac.za (Olubukola Oluranti Babalola)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University
Abstract
The plant microbiome can enhance disease suppression by providing life-supporting functions to their host, including stress resilience, health, and growth. However, our understanding of the core mechanisms of microbiome assembly and activity is still emerging. This article explores the role of plant-associated microbes in enhancing host resistance against pathogen infection through disease suppression. We discuss the factors that influence the community assembly and functioning of the plant microbiome, along with an overview of the mechanisms of disease suppression by the plant microbiota. Additionally, we highlight plant characteristics and mechanisms that recruit and stimulate microbial allies for disease suppression. By uncovering the power of plant-microbe interactions, we can create sustainable disease management strategies in agriculture and beyond.
Keywords
Community assembly
Plant defense mechanisms
Plant-microbe interactions
Disease suppression
1 Introduction
A change in thinking has arisen, and microbes are now seen as functional drivers of their eukaryotic hosts. Healthy and asymptomatic plants live together with microbes unanimously known as plant microbiota, forming complex interactions that can provide life-supporting functions comprising the acquisition of nutrients, stress resilience, support of growth, and health of a plant. Specific plant microbiota is connected to plant attributes such as disease suppression. Even though awareness of the immense functional abilities of the plant microbiome has increased substantially in recent years, a fundamental understanding of the core mechanisms of microbiome assembly and action is still emerging. This knowledge will enhance harnessing of the genomic perspective of plants, thereby improving stress resilience of imminent improved crop development under a shifting climate (Hassani et al., 2018; Cordovez et al., 2019).
The communications between plants and microbes occur in various ways and levels. At specific points in their lives, almost all plant organs interact with microbes and those plants profit either directly or indirectly from the microbes. Microbes linked to plants secrete compounds that could make the plants become resistant to biotic and abiotic stress or could protect them against harmful microbes, thereby promoting their growth (Amoo and Babalola, 2017; Olanrewaju et al., 2017). The way plants exploit the valuable functions delivered by microbial communities while tackling microbial pathogens has been a cynosure to researchers for several years (Cheng et al., 2019). The extremely complex microbial assemblages connected to various plant species, plant growth stages, and certain plant organs have been uncovered by several studies (Ajilogba et al., 2022; Meyer et al., 2022). The plant microbiome stands as one of the primary factors influencing plant health and productivity. Microbes perform crucial roles in disease suppression and unfavorable agroecological affairs (Meena et al., 2017; Olanrewaju and Babalola, 2022). Therefore, uncovering the functionality of plant–microbe interactions aids in comprehending how plants can benefit from the microbial communities they associate with.
In this review, we discussed how microbes connected to plants augment host resistance against pathogen infection. We outline the factors that contribute to the assembly and functioning of the plant microbial community. The plant microbial community linked to disease suppression and the mechanisms of beneficial microbe-mediated disease suppression are highlighted. Plant features and mechanisms concerned with the recruitment and stimulation of microbial associates for disease suppression are also compiled.
2 Role of plant-associated microbes in enhancement of host resistance against pathogen infection
Recently, more research projects have shown that plants depend on the innate immunity of their cells for the resistance of pathogenic invasion. These were conducted by sensing and recognizing patterns produced by pathogens (molecules or structures) that help in triggering defense against such infections to block pathogen proliferation. To colonize and infect plants, pathogenic microbes secrete effector proteins and kill host tissues via enzymes and toxins. In response, beneficial microbes (rhizosphere, endosphere, or phyllosphere) help in controlling pathogenic microbes and improve plant health through various direct and indirect mechanisms such as production of ACC deaminase, siderophore, phytohormones (including auxin, gibberellin, cytokinin, ethylene, salicylic acid, jasmonic acid), phosphate solubilization, and induction of systemic resistance (Olanrewaju and Babalola, 2022; Chepsergon and Moleleki, 2023).
2.1 Plant-associated microbes
Plants interact with their environments, including the different microbiota, which confers diverse useful traits to the plants which include stress tolerance, enhancement of nutrient uptake, and disease resistance. In contrast, host plants provide carbon to the associated microbes. Microbes associated with plants are either beneficial, like the plant growth-promoting rhizobacteria, biocontrol rhizobacteria, or detrimental, causing diseases in plants by destroying tissues, wilting, lesions, drying and/or eventual death. These plant-associated microorganisms can also be found in the rhizosphere, where they form association with plants as biocontrol agents and increase plant growth. Some of them are found on the phyllosphere, in plant tissue (endosphere), and sometimes on the stems. Plant-associated microbes have been known to work symbiotically with plants where they impact the development and functionality of plants (Hassani et al., 2018). These impacts include the release of metabolites and hormones from bacteria that improve crop growth and production (Kumar et al., 2017; Olanrewaju et al., 2019) and protection of host plants against pathogen infection by disease suppression, parasitism, induced systemic resistance (ISR) and metabolite production (Adeleke et al., 2019). Bacteria from the phyllosphere can affect plants’ species distribution over time and ecosystem function by influencing plant production under various environmental factors (Hassani et al., 2018).
2.1.1 Pathogenic plant-associated microbes and pathogenic infection in plants
Plant-associated pathogens are involved with causing diseases and infections in plants. With over 7100 classified bacterial species globally, only about 150 of them are disease-causing bacteria (Kannan et al., 2015). These pathogenic microbes normally thrive where environmental conditions are favorable such as warm and humid conditions in the tropical and subtropical regions of the world. Pathogenic infections in plants could originate from the phyllosphere, rhizosphere and/or endosphere. They cause diseases ranging from rots, blights, cancerous wilts, blights to spots, etc. The mechanisms involve penetration of openings such as stomata, stigma, etc. or through wounds on the plant parts such as the leaves, stems and/or the roots. The microbe inside the plant could attack the plant by extracting nutrients from the living plants but keeping them alive (biotrophy), or it could also kill the plant before extracting nutrients from its dead cells (necrotrophy). This interaction can be incompatible with the host plant (cultivar-specific resistance or host-resistance) or non-host plant in which in either case, a hypertensive response (HR). In a compatible case, the microbe causes disease symptoms in host plants.
These infections are a threat to crop production and food security. The use of chemicals, resistance gene engineering, and genetic modification of plant defense metabolites have helped to reduce this challenge. Nevertheless, widespread disease resilience and pathogenic emergence, along with the proliferation of the host variety and host jump, lead to severe disease spreads, particularly in the context of modern farming practices (Vannier et al., 2019).
2.1.2 Plant-associated beneficial microbes
Plant-associated beneficial microbes, also known as plant-growth-promoting rhizobacteria/bacteria (PGPR/B), have been studied extensively for their plant growth promoting abilities (Olanrewaju et al., 2017; Etesami and Glick, 2020). These microbes can directly or indirectly affect plant growth by producing plant growth-promoting substances, such as phytohormones, and by improving nutrient uptake and disease resistance (Olanrewaju and Babalola, 2022). The rhizosphere, the soil surrounding plant roots, is a crucial environment for microbial growth and activities (Olanrewaju et al., 2019). The use of microbial inoculants for pathogen biocontrol and biofertilizers has been shown to increase crop productivity and sustainability (Olanrewaju and Babalola, 2019; Tanveer et al., 2023).
One study found that a synthetic community of eight PGPR strains isolated from wheat rhizosphere effectively colonized and interacted with indigenous soil microbiomes, promoting plant growth, and altering soil microbial communities (Liu et al., 2022). Another study reported that Trichoderma strains, a type of beneficial microbe, are effective biocontrol agents against phytopathogens and can improve plant growth (Geng et al., 2022).
Rhizodeposits, root exudates, and root border cells are vital components of the rhizosphere that significantly affect root colonization capacity and multiplication of rhizosphere microbes, as well as secretion of organic bioactive compounds (Olanrewaju et al., 2019). The presence of beneficial microbes in the rhizosphere minimizes the susceptibility to crop diseases and enhances plant growth (Kadiri et al., 2023).
2.2 Plant defense mechanisms
In nature, plants have two main mechanisms of defense against pathogens which are resistance and tolerance. Tolerance is achieved because of a pathogen establishment in the system of the host and the damage it causes. Resistance, usually of chemical nature is observed when the host suppresses or reduces the growth of the pathogen (Burdon et al., 2016; Olanrewaju et al., 2019). Resistance lowers the risk of infection and pathogen multiplication, while tolerance does not. If plants develop resistance, pathogen prevalence will decrease, whereas tolerance has the opposite effect (Montes et al., 2020).
The plant's ability to recognize pathogens and protect itself from infection is essential. Plants have developed many ways to protect themselves, such as being able to recognize chemicals or molecular patterns that are unique to pathogens. When these patterns or chemicals are sensed, the plant's innate immunity is activated (Olanrewaju et al., 2019; Gourion and Ratet, 2021). Furthermore, the presence of foreign molecules sets off several defense mechanisms, such as structural hardening, biochemical deterrents, and the production of pathogenesis-related (PR) proteins (Pathak et al., 2022). The main way a plant protects itself from pathogens is by having an impenetrable shield made of the bark and a hydrophobic cuticle. However, mechanical damage can weaken this defense, making plants easier to invade. The production of antimicrobial chemicals, the release of signaling molecules to attract helpful microbes, and the creation of systemic acquired resistance (SAR) are other ways that plants fight pathogens. The basic mechanisms of innate immunity plants use are microbe-associated molecular pattern (MAMPs) or pathogen-associated molecular pattern (PAMPs)-triggered immunity (MTI or PTI) using pattern recognition receptors (PRR) and effector-triggered immunity (ETI) (Samad et al., 2019; Olanrewaju and Babalola, 2022). Other molecular patterns recognized are the Nematode-Associated Molecular Patterns (NAMPs) which is a group of molecules that are secreted by nematodes (Choi and Klessig, 2016) and the Damage-Associated Molecular Pattern (DAMP) (Olanrewaju and Babalola, 2022). These molecular patterns are highly conserved molecules in microbes, such as flagellin, that are only detected when pathogens interact directly with host plant receptors. (Olanrewaju and Babalola, 2022; Pathak et al., 2022). Examples of MAMPs include bacterial lipopolysaccharide, flagellin which is recognized by the FLS2 receptor in plant cells, lipoproteins, peptidoglycans that interacts with NOD-like receptors, and fungal chitin (Wang et al., 2022). The binding of PRR to MAMP helps plants to recognize beneficial microbes and pathogens.
The detection of MAMPs/PAMPs by plants results in SAR/ISR pathways, which boost host resistance through the activation of PTI or ETI. Pathogens can sometimes evade the plant MTI and still cause diseases; in such cases, ETI using leucine-rich-repeat receptors is triggered by plants against pathogens. Khoshru et al., (2023) and Wang et al., (2022) have written very good reviews on this subject.
To make a plant and microbe partnership work, the plant must avoid setting off its immune response, ETI. Reportedly, certain genes and proteins control beneficial effects of plant microbes (Zhang et al., 2021). If the microbe and plant are not compatible i.e., pathogenic microbes, the plant will not allow the pathogenic microbe in, but beneficial microbes can still enter (Gourion and Ratet, 2021). This can help the plant resist diseases. It is important for a plant to balance its protection and allowing beneficial microbes in for a successful partnership.
2.2.1 Induced systemic resistance
Induced systemic resistance (ISR) is a process in which beneficial or non-pathogenic microbes, including plant growth-promoting microbes (PGPM), help to suppress and/or reduce the harmful effects of plant pathogens by triggering resistance mechanism in plants. When plants are infected by a disease, they respond with a signal that is salicylic acid dependent. This normally leads to them expressing a resistance that is broad-spectrum and long-lasting which is also efficient against bacteria, fungi, and viruses. This is because the endogenous levels of salicylic acid (SA) normally increase in the phloem locally and systemically after infection before the occurrence of ISR. Nevertheless, there are occasions when SA is produced in non-infected regions, consequently inducing a systemic resistance (Nie et al., 2017). In a natural situation where the pathogen for a particular infection is more than one, the level of the basal resistance from the host is increased because of elicited resistance (Sattiraju et al., 2019). In greenhouse or field trials, some strains of Bacillus spp have been found to induce systemic resistance on Arabidopsis spp, bell pepper, cucumber, loblolly pine, muskmelon, sugar beet, watermelon, tobacco, and tomato (Shafi et al., 2017).
Several PGPM have been observed and shown to resist and control plant diseases (Shafi et al., 2017; Sattiraju et al., 2019). Using PGPM to treat plants before planting helps to prepare the plant to act faster against pathogens by inducing its own self-defense mechanism. Seedlings of finger millets dipped in formulation of Pseudomonas fluorescens increased the activity of phenylalanine ammonia-lyase against blast disease of millet (Sattiraju et al., 2019). Lactic acid bacterial strains Lactobacillus plantarum CC100, PM411 and TC92, and Leuconostoc mesenteroides CM160 and CM209 were used as biocontrol agents against Pseudomonas syringae pv. actinidiae in kiwifruit, Xanthomonas arboricola pv. pruni in Prunus and Xanthomonas fragariae in strawberry whose growths were severely inhibited (Daranas et al., 2019).
2.2.2 Local induced plant resistance
The multifaceted process of induced resistance in plants is crucial for their survival against pathogens. Alongside SA mentioned earlier, other chemical inducers such as ethylene, jasmonic acid (JA), 2,6-dichloroisonicotic acid, and DL-3 aminobutyric acid can increase plant resistance to a wide variety of pathogens (Li et al., 2020). Moreover, ISR triggered by plant-beneficial microbes is a well-studied phenomenon that can boost plant immunity (Salwan et al., 2023). Upon activation of ISR, plants engage in long-distance systemic signaling that protects distal tissue, eliciting rapid and robust immune responses against pathogen invasions, which are typically mediated by JA and ethylene.
The complex nature of the interaction of multiple signaling pathways in the induction of plant resistance must also be emphasized. Pathogen-induced activation of the plant defensin gene PDF1.2 in Arabidopsis requires simultaneous activation of the JA and ethylene signal pathways (Guo et al., 2020; Ederli et al., 2021). Moreover, there are reports indicating that ETHYLENE RESPONSE FACTOR1 assimilates cues originating from the ethylene and JA pathways to bolster plant defense (Zhou et al., 2022). This underscores the necessity for an enhanced comprehension of the intricate interplay among numerous signaling pathways to induce pathogen resistance in plants.
The multifaceted nature of induced resistance in plants exemplifies the intricacy of plant defense against pathogens. Chemical inducers and ISR, initiated by beneficial microbes, can enhance plant immunity. However, the involvement of multiple signaling pathways underscores the necessity for further research to explore this domain.
2.2.3 Host resistance in plants
Diversity in genetic composition observed in cultivar and crop genotype influences the pattern of interaction between plant hosts and beneficial microbes. Some of these variations have been found in the way plants interact with a variety of microbes including rhizobia, mycorrhizal fungi, and microbial biocontrol agents. Host resistance is regarded as one of the important and effective strategies for controlling and preventing plant diseases (Huzar-Novakowiski et al., 2017). Host resistance (R-genes) has been applied through backcrossing or transformation in crops. This is one of the main ways through which plants defend themselves from pathogenic infection attacks. The mechanism used is the effector-triggered immunity (ETI) in which case, the plant host effector R proteins recognizes the effector protein from the pathogen leading to a hypertensive response that ends up in eventual death of the pathogen or suppression of disease development. It has worked effectively in some plants such as Arabidopsis thaliana and tomato (Lopez et al., 2019). In some other plants such as wheat (using the pyramiding multiple stem rust R-genes), Puccinia graminis Pers. f. sp. Tritici, causative agent of wheat stem rust from a new aggressive race Ug99 caused about 70% loss in wheat yield worldwide. This is because some plant pathogens can adapt in such a way that they can suppress host resistance over time (Lee et al., 2016).
3 Plant microbiota associated with disease suppression and their mechanisms of action
Plant-associated microbiota interactions influence the physiological, functional, and metabolic health of their plant hosts. This is however seen as a contribution by both partners to the overall wellbeing and fitness of the system. Therefore, the association between the plant host and its symbiotic microbes is seen as a unified entity that is termed holobiont (Berg et al., 2020).
The vulnerability of plants to phytopathogens, causing various diseases, can be attributed to multiple factors, including the population of disease-causing microbes, favorable conditions for pathogen growth, and the host's susceptibility to pathogens, among others. These different factors are the determinants of whether the plant associated microbiota can suppress/resist phytopathogens or are susceptible to the phytopathogens (Brader et al., 2017).
3.1 Plants and disease challenges
Massive losses of planted crops are incurred in agricultural outputs globally due to pathogenic microbes and plant pests. This reduction in productivity leads to huge losses economically and affects food security globally, nationally and at household levels (De Silva et al., 2019; Savary et al., 2019).
One prevalent approach to addressing this issue involves the application of synthetic agrochemicals. Nonetheless, this method proves costly and has detrimental effects on the environment, as well as the health of animals and humans. Moreover, the persistent use of these chemicals fosters the development of pathogen resistance and the emergence of new pathogens and pests. Additionally, the use of synthetic agrochemicals can disrupt the balance of existing beneficial soil microbiota (French et al., 2021), leading to increase in the susceptibility of plants to pathogens in the soil. A sustainable and eco-friendly alternative is the deployment of antagonistic microbes that can suppress disease-causing pathogens. These diseases and pests suppressing organisms are generally called biological control agents (BCA). These biocontrol agents, depending on their beneficial applications, could be termed biopesticides or biofertilizers.
The benefits of BCAs over synthetic agrochemicals applications include no need for re-application at every cropping season as they could persist in the ecological environment by establishing, replicating, and colonizing the above and below ground plant regions (Zeilinger et al., 2016) unlike synthetic fungicides.
3.2 Plant microbiota allied with disease suppression
Plant microbiota play a pivotal role as key suppressors of plant diseases, while the holobiont ensures fitness through the maintenance of health and the suppression of microbial dysbiosis (Berg et al., 2017). They achieve this feat through enhancing plant vigor by mobilizing nutrients, directly antagonizing phytopathogens through parasitism or antibiosis, and competing for available resources through niche colonization. Another mechanism employed by these microbes is the induction of immune activity (Teixeira et al., 2019).
Numerous microbiota are actively involved in disease-suppressive soils. Newer high throughput sequencing techniques and omics technologies have exposed researchers to insights that communities of diverse microbes are involved in interactions between microbial biocontrol agents and the suppression of phytopathogens (rather multi-species/ consortia of beneficial microbial control agents against multi-species pathogens). Insights from disease-suppressive soils have revealed that pathogens encounter an unsuitable environment for growth due to the presence and influence of beneficial microbiome communities. These communities exert both specific and general suppressiveness on disease-causing pathogens (Mazzola and Freilich, 2017). Mousa and Raizada (2016) reported that Gaeumannomyces graminis var. tritici disease infection in wheat plant was suppressed with consortia of Pseudomonas and Fusarium species acting synergistically in producing metabolites and inducing physiological changes in the soil that led to disease suppressive attributes.
4 Microbial interactions and disease suppression
Microbial interactions in the plant’s rhizosphere contribute to disease suppression through various forms. One such form involves rhizobacteria in plant roots acting as BCAs against other bacteria that are pathogenic to the plant, known as bacterial-bacterial pathogen interactions. Additionally, different forms of interactions include bacteria-fungal pathogen interactions, fungal-bacterial pathogen interactions, fungal-fungal pathogen interactions, and multiple microbial interactions. This text explores the diverse ways in which microbes interact in plant’s rhizosphere, along with their specific methods of interaction.
4.1 Bacterial – Assisted disease suppression
As earlier said, this is a kind of interaction that is seen between bacteria (biocontrol agent) and a bacterial pathogen. Here, the bacteria having a biocontrol capability is applied to plant roots to contain bacterial-causing diseases in plants. An example is the involvement of Bacillus subtilis MBI 600 in the control of gray mold in cucumber plants caused by Botrytis cinerea (Samaras et al., 2021). This interaction involves biocontrol, where microbes in the rhizosphere compete for space and nutrients, exhibit induced systemic resistance (ISR), or engage in antibiosis. A recent study highlighted the utilization of beneficial induced systemic resistant bacteria in the rhizosphere for controlling leaf infections caused by the biotrophic bacterial pathogen, Pseudomonas syringae (Berendsen et al., 2018). In another study, Trong et al., (2022) observed a significant reduction in the incidence of tumors, commonly referred to as crown gall disease, in grapevine crops caused by the pathogenic Allorhizobium vitis using Paraburkholderia phytofirmans PsJN. However, the process of bacterial-based biocontrol may also encompass other mechanisms, such as the development of antibiotics, contributing to this phenomenon. Samaras et al., (2021) examined whether treating plants with the biocontrol agent B. subtilis MBI 600 can induce defense responses and enhance control efficacy against B. cinerea. The authors analyzed the transcription patterns of five marker genes at different time points and found that all tested genes were highly induced in B. subtilis MBI 600 inoculated plants compared to mock-inoculated plants. The findings showed that the bacterial strain studied had the ability to activate the plant’s basal immune responses. Furthermore, this result suggests that plant treatment with B. subtilis MBI 600 can not only control B. cinerea effectively but also stimulate the plant's immune system. This is a significant finding, as it indicates that the use of biocontrol agents like B. subtilis MBI 600 can be an effective strategy to manage plant diseases sustainably. The induction of basal immune responses also suggests that the plants are better equipped to defend themselves against future pathogen attacks. Other studies have proven that plants under pathogenic attack engage microbes in their rhizospheres for protection (Perea-Molina et al., 2022; Yang et al., 2023). The recruitment of beneficial microbes in the rhizospheres of attacked plants have also been demonstrated in wheat monocultures which developed disease suppressiveness because of the production of 2,4-dicetylphoroglucinol by Pseudomonads, after an outbreak of take-all disease (Weller et al., 2002).
4.2 Bacterial antagonists: Effective management of soil-borne fungal infections in plants
There are several pathogenic fungi that infect plants, and these pathogens are capable of infecting different plant parts including the leaves, fruits, roots, and stems. However, numerous studies have also exhibited the management of soil borne fungal infections using bacterial communities and this is due to the rising simplicity with the use of molecular approaches that can be applied to determine the occurrence, distribution, parallel significance, and the specific ways in which various bacterial antagonists act (Madkour et al., 2019; Gogoi et al., 2020). Various bacteria can control the emergence and spread of soil-borne fungal diseases by applying them in the soil, and in plant seeds (Ueki et al., 2018). Certain species of bacteria such as Pseudomonas putida, P. aeruginosa, P. aureofaciens, Bacillus subtilis, B. cereus, B. Polymyxa, and Burkholderia cepacia have shown outstanding results in the control of several soil-borne diseases of fungi (Shafi et al., 2017; Murugan, 2019). The application of these bacterial antagonists to most agricultural plants like rice, sugar beet, wheat, cotton and different vegetables can control the growth of fungal pathogens including Fusarium oxysporum, Verticillium dahlia, F. solani, Rhizoctonia solani, Gaeumanannomuces graminis that are capable of causing soil-borne diseases of plants, for instance, the take all, damping off, root rot, vascular wilt, and seed rot (Majeed et al., 2018; Vurukonda et al., 2018; Abbasi et al., 2019). Streptomyces species such as S. lividans, S. griseus, S. coelicolor have also been found to protect plants against fungal pathogens (Vurukonda et al., 2018). Anderson et al., (2004) in their study, indicated the possibility of using biological materials to control the growth of fungal pathogens in the phyllosphere. Therefore, they proposed that the emergence and development of various diseases can be reduced with the use of bacterial antagonists. A taxonomic analysis of sugar beet root microbiome planted in a soil which suppresses the root associated fungal pathogen R. solani showed a consistency in the association of many bacterial genera with the disease suppressive state. Hence, the upregulation of genes associated with stress in bacterial families was more exuberant on the roots of plants grown in the suppressive soil (Cordovez et al., 2019). The authors of this study hypothesized that the fungal pathogen activates responses in the root microbiome, either directly or indirectly, which causes shifts in composition and triggering of special antagonist characteristics that prevent the growth of pathogens (Cordovez et al., 2019). Similarly, some species of Pseudomonas obtained from Greenland soil repressed potato scab infection triggered by R. solani (Gómez Expósito et al., 2017; Cordovez et al., 2019). A study reported an enhanced resistance against Erysiphe pisi, a plant pathogen that causes powdery mildew by rhizobia species in Medicago truncatula (Smigielski et al., 2019).
4.3 Fungal-assisted disease suppression
Numerous researchers have focused on the interactions between fungal biocontrol agents and plant diseases caused by fungi on an equal footing with studies involving bacterial-fungal pathogen interactions that were earlier discussed. Nevertheless, fungi-fungi interactions are a special type of interactions because fungi can grow better than bacteria in the surrounding soils because of hyphal growth. The biocontrol capacity of several fungi has been investigated in recent research works, although majority of the studies seem to be on Trichoderma species which could reflect their easy growth and ability to grow within a wide range of host. Fungal species of particular interest in biocontrol studies include those of P. oligandrum Drechsler (Baturo-Cieśniewska et al., 2018) and Trichoderma species (Degani and Dor, 2021), among others. Plant pathogens that are mostly targeted by these biocontrol fungi are: Rhizoctonia solani, Pythium, and Fusarium species. These fungal pathogens are of immense agricultural and world-wide importance and are easily controlled under protected cropping systems.
4.4 Mechanisms of microbial disease suppression: An overview
Research indicates that various enzymes and bacteria contribute to disease suppression and plant growth promotion (Olanrewaju and Babalola, 2019; Mir et al., 2022). Pseudomonas, Bacillus, Streptomyces, and other beneficial microbes play a role in plant disease suppression through siderophore-mediated competition for iron, antibiosis, production of lytic enzymes, and ISR (Ankati et al., 2021; Agbodjato et al., 2022; Zeng et al., 2023). Additionally, bacteria that produce the enzyme 1-aminocyclopropane-1-carboxylate (ACC) deaminase contribute to plant growth, particularly under environmental stress conditions (Glick and Nascimento, 2021). ACC deaminase breaks down ACC, the precursor of ethylene, into α-ketobutyrate and ammonia. By reducing ethylene production in plants, this enzyme helps sustain plant growth and development during biotic and abiotic stress conditions (Fig. 1).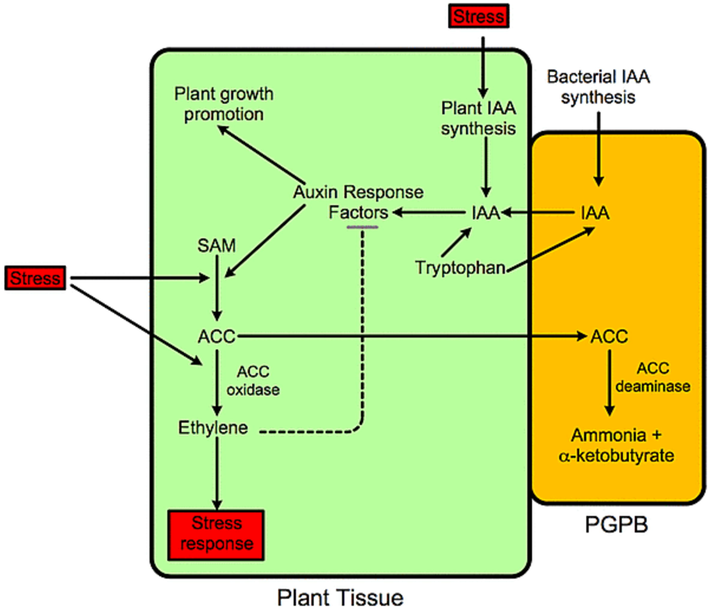
Schematic diagram of ACC deaminase-containing beneficial microbes stimulating plant development. Stress boosts IAA and ethylene production, reducing plant growth. ACC deaminases reduce ethylene levels which keeps bacterial IAA thereby promoting plant development. Thus, beneficial microbe that produce both IAA and ACC deaminase reduce plant growth inhibition from many environmental stresses. Beneficial microbes protect plants from ethylene-producing conditions like fungal and bacterial phytopathogens (Olanrewaju et al., 2017).
Bacillus spp. has been reported to produce indole-3-acetic acid, gibberellic acid, and ACC deaminase that helps in regulating the intracellular phytohormone, initiating the antioxidant and defense systems, and increasing plant stress tolerance (Narayanasamy et al., 2020; Patel et al., 2023).
Siderophores are small molecules that are produced by bacteria and fungi to scavenge iron from the environment (Olanrewaju et al., 2017). Iron is an essential nutrient for the growth and survival of microbes, and siderophores play a key role in high-affinity iron acquisition in both pathogenic and non-pathogenic bacteria and fungi (Fig. 2) (Olanrewaju and Babalola, 2022). Siderophores are also involved in the biocontrol mechanism of beneficial microbes by depriving the pathogen of iron nutrition, thus resulting in increased yields of crops (Nithyapriya et al., 2021). The role of siderophores in natural conditions is still not fully understood. In addition, siderophores can function as virulent factors in pathogenic organisms by facilitating iron uptake from the host. Therefore, the mechanism of siderophore-mediated disease suppression involves the production of siderophores by microbes to scavenge iron from the environment, which deprives pathogens of iron nutrition and reduces their virulence.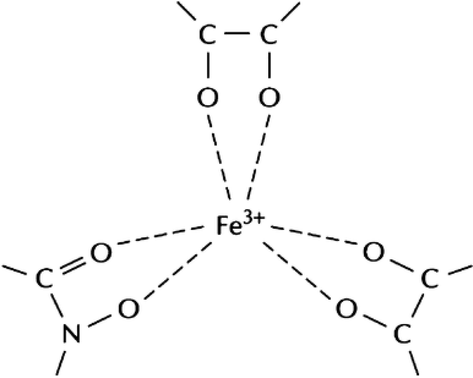
Schematic representation of three bidentate groups of a siderophore molecule binding to iron (Olanrewaju et al., 2017).
5 Plant traits and mechanisms implicated in the recruitment and stimulation of microbial partners for disease suppression
Plant growth and development are hampered by disease causing microbes invading the plants. On pathogen invasion, plants respond through myriads of processes. Such processes include secretion of exudates, ISR, and SAR. These three are the major responses shown by plants. Among these responses, ISR and SAR are elicited within the plant tissue as a direct response to pathogen attack while root exudates act through the recruitment of microbial partners against infection. Our focus will therefore be on root exudates and how they help in shaping the rhizosphere microbial community against plant pathogens. Upon the release of root exudates, microbes are attracted to the plant rhizosphere. The type of exudate determines the type of microbes that will be attracted to the rhizosphere meaning that the type of exudate determines the rhizosphere microbial community.
5.1 Root exudation to recruit microbial partners: Plant’s rescue call
The first study to report root-microbe relationship was carried out on ultrathin sections of wheat rhizosphere microbiome by Foster and Rovira (1976). Many processes occurring in the rhizosphere microbiome are not passive, they are induced by external influences. These external factors serve as mediators linking the various processes (Olanrewaju et al., 2019). They are said to therefore act as signals in the activation or regulation of different processes in the rhizosphere. This is one of the key functions of root exudates in rhizosphere microbial interplay. They help in recruiting beneficial microbes to the rhizosphere thereby connecting communications that occur in the rhizosphere (Fig. 3). During this process, they exert a significant effect on the plant’s health.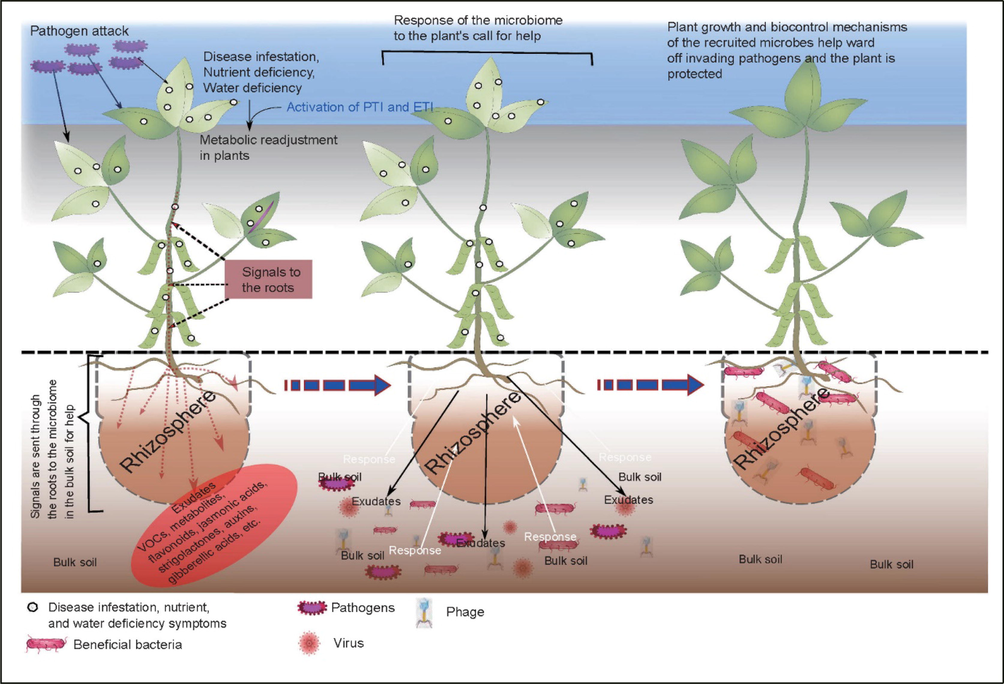
Soil microbiome response to plant signals. Left panel, plant signals to the soil microbiome arising from pathogen attack, nutrient, or water deficiency; center panel, the response of the various microbes to the exudates released by the plant; right panel, plant growth response to biocontrol mechanisms provided by recruited microbes. There is a positive response as pathogens do not respond to the signals (Olanrewaju and Babalola, 2022).
Plants secrete root exudates in response to various external factors. We are going to deal only with the release of exudates in response to pathogen attack on plants. The infestation of plants by diseases induces multiple biological, biochemical, and genetic changes and modifications to plant processes and development. These changes invariably induce the exudation of various metabolites majorly through the roots into the rhizosphere. These exudates serve as signal molecules to the plant environment to call for help and support because they attract beneficial microbes to the roots of the plants. These microbes, through various mechanisms, would then help the plants to fight against various diseases. This process of interactions of the microbes with the plants as an effect of the root exudates is still not adequately fathomed (Hayat et al., 2017). However, there have been recent developments in this regard for this purpose. However, common elements of signal pathways in the rhizosphere because of root exudate impact induces a high level of interactions between plants and microbes thus, regulating responses in the rhizosphere (Olanrewaju et al., 2019). Exudates such as monosaccharide sugars (fructose, mannose, and glucose), disaccharide sugars such as maltose, five-carbon sugars such as arabinose, and oligosaccharide sugars. There are also amino acids such as glutamine, aspartate, arginine, cysteine, and asparagine; organic acids (acetic, malic, ferulic, and benzoic acids); phenolics like coumarin. Others include high-molecular-weight compounds like auxins, tannins, alkaloids, terpenoids, flavonoids, fatty acids, vitamins, enzymes, and polyacetylenes (Hayat et al., 2017).
6 Perspective
The highly diverse interactions of the plant microbiota are part of the key determining factors of the health and productivity of plants. Although acuity of the numerous functional capacities of the plant microbiome has considerably improved recently, perception of the core mechanisms of the assembly and activity of the microbiome is still developing. This knowledge is necessary for exploiting the genomic potential of plants thus advancing stress resilience of imminent crop production under a shifting climate. Various factors influence the community assembly and function of the plant microbiome and are necessary for critically comprehending these associations. A fundamental understanding of the mechanisms involved in the recruitment of microbial allies for plants is still needed. Even though root exudates can be stated to be emissaries that call for rescue when plants are distressed, a comprehension of how they recruit microbes for this purpose is crucial.
Declaration of Competing Interest
Work in OOB lab is supported by the National Research Foundation of South Africa (Grant Ref: UID123634; UID132595).
References
- Soil incorporation of buckwheat as a pre-plant amendment provides control of Rhizoctonia damping-off and root rot of radish and Pythium damping-off and root rot of cucumber. Can. J. Plant Pathol.. 2019;41(1):24-34.
- [CrossRef] [Google Scholar]
- Applications of plant-microbe interactions in agro-ecosystems. In: Microbiome in Plant Health and Disease. Springer; 2019. p. :1-34.
- [CrossRef] [Google Scholar]
- Formulation of biostimulants based on arbuscular mycorrhizal fungi for maize growth and yield. Front. Agron.. 2022;4:894489
- [CrossRef] [Google Scholar]
- Plant growth stage drives the temporal and spatial dynamics of the bacterial microbiome in the rhizosphere of Vigna subterranea. Front. Microbiol.. 2022;13:825377
- [CrossRef] [Google Scholar]
- Ammonia-oxidizing microorganisms: key players in the promotion of plant growth. J. Soil Sci. Plant Nutr.. 2017;17(4):935-947.
- [CrossRef] [Google Scholar]
- Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell. 2004;16(12):3460-3479.
- [CrossRef] [Google Scholar]
- Streptomyces consortia-mediated plant defense against Fusarium wilt and plant growth-promotion in chickpea. Microb. Pathog.. 2021;157:104961
- [CrossRef] [Google Scholar]
- Development of Sclerotinia sclerotiorum (Lib.) de Bary on stored carrot treated with Pythium oligandrum Drechsler determined by qPCR assay. Acta Sci. Polonorum. Hortorum Cultus.. 2018;17(5):111-121.
- [CrossRef] [Google Scholar]
- Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J.. 2018;12(6):1496-1507.
- [CrossRef] [Google Scholar]
- Plant microbial diversity is suggested as the key to future biocontrol and health trends. Microbiol. Ecol.. 2017;93(5):fix050.
- [CrossRef] [Google Scholar]
- Microbiome definition re-visited: old concepts and new challenges. Microbiome.. 2020;8:1-22.
- [CrossRef] [Google Scholar]
- Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu. Rev. Phytopathol.. 2017;55:61-83.
- [CrossRef] [Google Scholar]
- Addressing the challenges of pathogen evolution on the world’s arable crops. Phytopathol.. 2016;106(10):1117-1127.
- [CrossRef] [Google Scholar]
- Plant-microbe interactions facing environmental challenge. Cell Host Microbe. 2019;26(2):183-192.
- [CrossRef] [Google Scholar]
- Rhizosphere bacterial interactions and impact on plant health. Curr. Opin. Microbiol.. 2023;73:102297
- [Google Scholar]
- DAMPs, MAMPs, and NAMPs in plant innate immunity. BMC Plant Biol.. 2016;16(1):232.
- [CrossRef] [Google Scholar]
- Ecology and evolution of plant microbiomes. Annu. Rev. Microbiol.. 2019;73:69-88.
- [CrossRef] [Google Scholar]
- Biological control of bacterial plant diseases with Lactobacillus plantarum strains selected for their broad-spectrum activity. Ann. Appl. Biol.. 2019;174(1):92-105.
- [CrossRef] [Google Scholar]
- Use of endophytes as biocontrol agents. Fungal Biol. Rev.. 2019;33(2):133-148.
- [CrossRef] [Google Scholar]
- Trichoderma biological control to protect sensitive maize hybrids against late wilt disease in the field. J. Fungi.. 2021;7(4):315.
- [CrossRef] [Google Scholar]
- In the tripartite combination Botrytis cinerea–Arabidopsis–Eurydema oleracea, the fungal pathogen alters the plant–insect interaction via jasmonic acid signalling activation and inducible plant-emitted volatiles. J. Plant Res.. 2021;134(3):523-533.
- [CrossRef] [Google Scholar]
- Halotolerant plant growth–promoting bacteria: Prospects for alleviating salinity stress in plants. Environ. Exp. Bot.. 2020;178:104124
- [CrossRef] [Google Scholar]
- Emerging strategies for precision microbiome management in diverse agroecosystems. Nat. Plants. 2021;7(3):256-267.
- [CrossRef] [Google Scholar]
- Biocontrol potential of Trichoderma harzianum against Botrytis cinerea in tomato plants. Biol. Control. 2022;174:105019
- [CrossRef] [Google Scholar]
- Pseudomonas 1-Aminocyclopropane-1-carboxylate (ACC) Deaminase and Its Role in Beneficial Plant-Microbe Interactions. Microorganisms.. 2021;9(12):2467.
- [CrossRef] [Google Scholar]
- Plant growth-promoting rhizobacteria in management of soil-borne fungal pathogens. In: Management of Fungal Pathogens in Pulses. Springer; 2020. p. :1-13.
- [CrossRef] [Google Scholar]
- Current insights into the role of rhizosphere bacteria in disease suppressive soils. Front. Microbiol.. 2017;8:2529.
- [CrossRef] [Google Scholar]
- Avoidance of detrimental defense responses in beneficial plant–microbe interactions. Curr. Opin. Biotechnol.. 2021;70:266-272.
- [CrossRef] [Google Scholar]
- Heterologous expression of poplar WRKY18/35 paralogs in Arabidopsis reveals their antagonistic regulation on pathogen resistance and abiotic stress tolerance via variable hormonal pathways. Int. J. Mol. Sci.. 2020;21(15):5440.
- [CrossRef] [Google Scholar]
- Microbial interactions within the plant holobiont. Microbiome.. 2018;6(1):58.
- [CrossRef] [Google Scholar]
- Root exudates: composition and impact on plant-microbe interaction. Biofilms Plant Soil Health. 2017:179-193.
- [CrossRef] [Google Scholar]
- Host resistance and chemical control for management of Sclerotinia stem rot of soybean in Ohio. Phytopathol.. 2017;107(8):937-949.
- [CrossRef] [Google Scholar]
- Pan-genome analysis and molecular docking unveil the biocontrol potential of Bacillus velezensis VB7 against Phytophthora infestans. Microbiol. Res.. 2023;268:127277
- [CrossRef] [Google Scholar]
- Kannan, V.R., Bastas, K.K., Devi, R.S., 2015. Scientific and Economic Impact of Plant Pathogenic Bacteria. Sustainable approaches to controlling plant pathogenic bacteria. V. R. Kannan and K. K. Bastas. CRC Press, Boca Raton, FL, USA, pp. 369–392. https://doi.org/10.1201/b18892-21.
- Decrypting the multi-functional biological activators and inducers of defense responses against biotic stresses in plants. Heliyon.. 2023;9(3):e13825.
- [Google Scholar]
- Endophytic and epiphytic modes of microbial interactions and benefits. In: Plant-Microbe Interactions in Agro-Ecological Perspectives. Springer; 2017. p. :227-253.
- [CrossRef] [Google Scholar]
- Mini review: potential applications of non-host resistance for crop improvement. Front. Plant Sci.. 2016;7(997)
- [CrossRef] [Google Scholar]
- Translocation of PpNPR1 is required for β-aminobutyric acid-triggered resistance against Rhizopus stolonifer in peach fruit. Sci. Hortic.. 2020;272:109556
- [CrossRef] [Google Scholar]
- Effective colonisation by a bacterial synthetic community promotes plant growth and alters soil microbial community. J. Sust. Agri. Environ.. 2022;1(1):30-42.
- [CrossRef] [Google Scholar]
- A bacterial effector mimics a host HSP90 client to undermine immunity. Cell. 2019;179(1):205-218.e221.
- [CrossRef] [Google Scholar]
- biological control of soil-borne fungal pathogens. Arab Uni. J. Agri. Sci.. 2019;27(1):749-760.
- [CrossRef] [Google Scholar]
- Damping off in chilli and its biological management-A review. Int. J. Curr. Microbiol. Appl. Sci.. 2018;7(4):2175-2185.
- [CrossRef] [Google Scholar]
- Prospects for biological soilborne disease control: application of indigenous versus synthetic microbiomes. Phytopathol.. 2017;107(3):256-263.
- [CrossRef] [Google Scholar]
- Beneficial microbes for disease suppression and plant growth promotion. In: Plant-Microbe Interactions in Agro-Ecological Perspectives. Springer; 2017. p. :395-432.
- [CrossRef] [Google Scholar]
- Plant neighborhood shapes diversity and reduces interspecific variation of the phyllosphere microbiome. ISME J.. 2022;16(5):1376-1387.
- [CrossRef] [Google Scholar]
- Multifarious indigenous diazotrophic rhizobacteria of rice (Oryza sativa L.) rhizosphere and their effect on plant growth promotion. Front. Nutr.. 2022;8:781764
- [CrossRef] [Google Scholar]
- Trade-offs between host tolerances to different pathogens in plant–virus interactions. Virus Evol.. 2020;6(1):veaa019.
- [CrossRef] [Google Scholar]
- Natural disease control in cereal grains. In: Colin W., Harold C., Koushik S., Jon F., eds. Encyclopedia of Food Grains. Elsevier; 2016. p. :257-263.
- [CrossRef] [Google Scholar]
- Biological control of pest using antagonistic microorganism. BIOINFOLET-A Quarterly. J. Life Sci.. 2019;16(1and2):79-89.
- [Google Scholar]
- Plant growth-promoting Bacillus sp. cahoots moisture stress alleviation in rice genotypes by triggering antioxidant defense system. Microbiol. Res. 2020:239. 126518
- [CrossRef] [Google Scholar]
- Induced systemic resistance against Botrytis cinerea by Bacillus cereus AR156 through a JA/ET-and NPR1-dependent signaling pathway and activates PAMP-triggered immunity in Arabidopsis. Front. Plant Sci.. 2017;8:238.
- [CrossRef] [Google Scholar]
- Production, purification, and characterization of bacillibactin siderophore of Bacillus subtilis and its application for improvement in plant growth and oil content in Sesame. Sustainability.. 2021;13(10):5394.
- [CrossRef] [Google Scholar]
- Plant health: feedback effect of root exudates-rhizobiome interactions. Appl. Microbiol. Biotechnol.. 2019;103(3):1155-1166.
- [CrossRef] [Google Scholar]
- Bacterial consortium for improved Maize (Zea mays L.) production. Microorganisms.. 2019;7(11):519.
- [CrossRef] [Google Scholar]
- The rhizosphere microbial complex in plant health: A review of interaction dynamics. J. Integr. Agric.. 2022;21(8):2168-2182.
- [CrossRef] [Google Scholar]
- Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol.. 2017;33:1-16.
- [CrossRef] [Google Scholar]
- Multi-trait halotolerant plant growth-promoting bacteria mitigate induced salt stress and enhance growth of Amaranthus Viridis. J. Soil Sci. Plant Nutr.. 2023;1–24
- [CrossRef] [Google Scholar]
- Plant-endophyte interaction during biotic stress management. Plants.. 2022;11(17):2203.
- [CrossRef] [Google Scholar]
- A biocontrol Bacillus velezensis strain decreases pathogen Burkholderia glumae population and occupies a similar niche in rice plants. Biol. Control. 2022;176:105067
- [CrossRef] [Google Scholar]
- Insights into plant beneficial microorganism-triggered induced systemic resistance. Plant Stress.. 2023;7:100140
- [CrossRef] [Google Scholar]
- Plant-associated bacteria. In: Elsas J.D., Trevors J.T., Rosado A.S., Nannipieri P., eds. Modern Soil Microbiology. CRC Press; 2019. p. :163-179.
- [CrossRef] [Google Scholar]
- Insights into the multitrophic interactions between the biocontrol agent Bacillus subtilis MBI 600, the pathogen Botrytis cinerea and their plant host. Microbiol. Res.. 2021;248:126752.
- [CrossRef] [Google Scholar]
- Plant growth-promoting microbes: contribution to stress management in plant hosts. In: Environmental Biotechnology: For Sustainable Future. Springer; 2019. p. :199-236.
- [CrossRef] [Google Scholar]
- The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol.. 2019;3(3):430-439.
- [CrossRef] [Google Scholar]
- Bacillus species as versatile weapons for plant pathogens: a review. Biotechnol. Biotechnol. Equip.. 2017;31(3):446-459.
- [CrossRef] [Google Scholar]
- Nodulation induces systemic resistance of Medicago truncatula and Pisum sativum against Erysiphe pisi and primes for powdery mildew-triggered salicylic acid accumulation. Mol. Plant Microbe Inter.. 2019;32(9):1243-1255.
- [CrossRef] [Google Scholar]
- Interactive effects of Pseudomonas putida and salicylic acid for mitigating drought tolerance in canola (Brassica napus L.) Heliyon. 2023;9(3):e14193.
- [CrossRef] [Google Scholar]
- Beyond pathogens: microbiota interactions with the plant immune system. Curr. Opin. Microbiol.. 2019;49:7-17.
- [CrossRef] [Google Scholar]
- Biological Control of Grapevine Crown Gall Disease, Caused by Allorhizobium vitis, Using Paraburkholderia phytofirmans PsJN. PhytoFrontiers™.. 2022;2(4):391-403.
- [CrossRef] [Google Scholar]
- Role of anaerobic bacteria in biological soil disinfestation for elimination of soil-borne plant pathogens in agriculture. Appl. Microbiol. Biotechnol.. 2018;102(15):6309-6318.
- [CrossRef] [Google Scholar]
- Microbiota-mediated disease resistance in plants. PLoS Pathog.. 2019;15(6):e1007740-e.
- [CrossRef] [Google Scholar]
- Plant growth promoting and biocontrol activity of Streptomyces spp. as endophytes. Int. J. Mol. Sci.. 2018;19(4):952.
- [CrossRef] [Google Scholar]
- Evasion of plant immunity by microbial pathogens. Nat. Rev. Microbiol.. 2022;20(8):449-464.
- [CrossRef] [Google Scholar]
- Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol.. 2002;40(1):309-348.
- [CrossRef] [Google Scholar]
- Plant disease resistance-related pathways recruit beneficial bacteria by remodeling root exudates upon Bacillus cereus AR156 treatment. Microbiol. Spectr.. 2023;11(2):e03611-03622.
- [CrossRef] [Google Scholar]
- Friends or foes? Emerging insights from fungal interactions with plants. FEMS Microbiol. Ecol. Rev.. 2016;40(2):182-207.
- [CrossRef] [Google Scholar]
- Effects of rice blast biocontrol strain Pseudomonas alcaliphila Ej2 on the endophytic microbiome and proteome of rice under salt stress. Front. Microbiol.. 2023;14:550.
- [CrossRef] [Google Scholar]
- Glycine max NNL1 restricts symbiotic compatibility with widely distributed bradyrhizobia via root hair infection. Nat. Plant.. 2021;7(1):73-86.
- [CrossRef] [Google Scholar]
- Multilayered synergistic regulation of phytoalexin biosynthesis by ethylene, jasmonate, and MAPK signaling pathways in Arabidopsis. Plant Cell. 2022;34(8):3066-3087.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2023.102893.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







