Translate this page into:
The effect of up-regulation of NLE1 gene expression on the invasion and migration of colon cancer cells and its mechanism
⁎Corresponding author at: Department of Anorectal Center, Cheeloo College of Medicine, Shandong University, China. xuzhen00616@163.com (Zhen Xu)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Objectives
To explore the regulatory effects of up-regulation of NLE1 gene expression in vitro and the mechanism of colon cancer cell invasion and migration.
Methods
The logarithmic growth phase human colonic fibroblasts CCD-18Co, colon cancer cells HT-29, and NLE1 interference group were transfected with NLE1 siRNA to interfere with expressing viral vectors and liposomes. The NLE1 negative control group was added with liposomes, and NLE1 was overexpressed. The expression changes of RNA interference on the target gene were analyzed.
Results
The expression of NLE1 mRNA in colon cancer cells was significantly higher (P < 0.05). Likewise, the expression of NLE1 protein in colon cancer cells was significantly increased than in control colon cancer cell lines (p < 0.05). Compared with the NLE1 interference group and the NLE1 negative control group, the migration and invasion ability of colon cancer cells in the NLE1 overexpression group were significantly varied (P < 0.05)‘.
Conclusions
Up-regulation of the NLE1 gene in colon cancer cells promotes cell migration and invasion of colon cancer cells and inhibits the apoptosis ability of colon cancer cells.
Keywords
Colon cancer
Cell lines
NLE1 gene
HT-29
Ribosome
Invasion
1 Introduction
Malignant tumour has become one of the major public health problems that seriously threaten the health of people all over the world, and the global cancer incidence and mortality rate are still increasing year by year. The burden of cancer disease has increased in recent times in both developing and developed countries due to various reasons, including socioeconomic development, food habits, ageng, and associated risk factors (Vineis and Wild, 2014). In recent years, the incidence of Global cancer and mortality increased gradually. In China, a malignant tumour caused increased mortality than other countries (Hu et al., 2019). With the increasing population and ageing throughout the world, cancer is involved in premature death and affects the socioeconomic of individuals. In China, the incidence of cancer varied from developed and developing countries and this disease is a ransition stage in China (Feng et al., 2019). Colon cancer is a common malignancy of endothelial origin. The first step in carcinogenesis is the formation of specific types of neoplastic polyps in the colonic mucosa, and polyp histology is critical to determine malignant potential. The two common histological types are proliferative and adenomatous, while most colon cancers originate in adenomas (adenoma-to-carcinoma sequences), and their incidence increases with age. Risk factors include a history of hyperplastic polyps and a family history of colon cancer. Risk factors may vary, but age and family history are the main factors. Central deposits of physical inactivity, overweight, and obesity are consistent risk factors associated with this disease. While some dietary and other factors may have a significant impact on the risk of colon cancer. Moreover, physical inactivity, overweight and obesity are the reported risk factors (Haraldsdottir et al., 2014). Colorectal cancer is a type of cancer that develops in the colon or the rectum of the large intestine (end of the colon) (Ramadan et al., 2022). The colon may be impacted by other cancers. These consist of sarcomas, melanoma, lymphomas, and carcinoid tumours. This research paper analyzes only colorectal cancer. Stage 0 is considered as an early stage of colorectal cancer and stage IV is considered severe case (Giovannucci, 2002). Tumour-infiltrating immune cells are an important source of prognostic information for patients with resectable colon cancer (Zhou et al., 2019). PTBP1 is overexpressed in both colon cancer cell lines and tissues (Li et al., 2018) f-actin regulator LIMK/ Cofilin pathway and SSH1 are associated with CRC progression and chemical resistance, representing a promising tumour biomarker and therapeutic target in CRC (Aggelou et al., 2018; El-Beeh et al., 2022;).
Diet management is one of the alternative steps for the early prevention of colon cancer (Roslan et al., 2019). Surgery is the recommended treatments, but chemotherapy also plays an important role. It is important to clearly distinguish treatment options for colorectal cancer because there are different treatment options. Radiation therapy is a key component of the treatment of colorectal cancer, but its application in colorectal cancer is limited. A wide range of diseases is mainly treated with chemotherapy, although surgery is sometimes used to treat patients with liver or lung metastases. Personalized medicine enables patients to maximize benefits while minimizing side effects, thus improving survival and quality of life. In stage III colon cancer, data from the IDEA trial led to more refined risk stratification, allowing some patients to opt for adjuvant therapy of a shorter duration (Roslan et al., 2019; Krzyszczyk et al., 2018).
Notchless homolog 1(NLE1) is a ribosomal progen protein that is an important factor in ribosome large subunit maturation and is essential for myogenic cell activation and proliferation (Gayraud-Morel et al., 2018). It encodes a protein containing WD40 repeats that is highly conserved in eukaryotes. The absence of NLE leads to deficiency in the synthesis of large ribosome subunits in recess cells and leads to rapid elimination of intestinal stem and progenitor cells through different cell types, including apoptosis, cell cycle arrest, and bias toward the cup cell lineage. The role of NLE in the maturation of large ribosome subunits is conserved in mice and is necessary for the maintenance of hematopoietic stem cells, and the gut is also sensitive to NLE. It has been found that in the absence of p53, the absence of NLE-deficient ISC and progenitor cells, is the basis for the existence of p53-independent cell reactions after ribosomal defects. NLE is a key factor in intestinal homeostasis and provides new insights into ribosome biogenesis on intestinal epithelial cell fate determination (Stedman et al., 2015). The mechanism of NLE1′s involvement in cancer promotion remains unclear, and some recent works revealed that Notchless (Nle; Nle1) is up-regulated through BAX/ Bcl-XL /FAS pathway to promote carcinogenesis in early colorectal cancer (Liu et al., 2019). The main objective of the study is to analyze the relationship between colorectal cancer, metastasis mechanism and NLE1 upregulation of colorectal cancer cells.
2 Materials and methods
2.1 Materials
Colon cancer cell line HT-29 and human colon fibroblast CCD-18CO (Shanghai Cell Bank, Chinese Academy of Sciences) were obtained. Fetal bovine Serum (Changsha Aijia Biotechnology Co., LTD.); Lipofectamine2000 (Invitrogen, USA); TRIzol reagent (Shanghai Jikai Biotechnology Co., LTD.); TaqManPCR Kit (Applied Biosystem, USA); Caspase3/7 Test Kit (G8091, Promega, USA); High-speed refrigerated centrifuge (Hitachi); JY200C Electrophoresis apparatus (Beijing Junyi Oriental electrophoresis Equipment Co., LTD.); Inverted phase contrast microscope (Olympus, Japan); L-420 Low-speed centrifuge (Xiangyi Experimental Instrument Development Co., LTD.) were used in this study.
2.2 Colon cell line and cell culture
After cell resuscitation, human colon fibroblast CCD-18CO cells were transferred to DMEM medium containing 10% FBS, and HT-29 cells were transferred to RPMIA-1640 medium containing 10% FBS. The cells were cultured at 37 °C in a 5% CO2 incubator, and the medium was changed every 2 to 3 days. When the cells were fused to about 80% it was confirmed using an inverted phase contrast microscope. The cultured cells were digested using trypsin digestion and the third generation of logarithmic growth cells was taken for subsequent experiments. CCD-18CO and HT-29 cells in the logarithmic growth phase were added with trypsin for digestion. The cells were centrifuged at 2000 rpm/min for 10 min to collect cells. The total RNA was extracted by the Trizol method. The RNA purity was calculated with the OD260/OD280 value between 1.6 and 1.8 was considered good quality. The RNA was reverse transcribed using the reverse transcription kit and cDNA was obtained. The real-time fluorescence quantitative PCR reaction was performed using the Green PCR quantitative fluorescence kit. The 20 µL reaction system was set according to the kit: Premix Ex Taq 11.5 µL, 10 µmol/L upstream and downstream primer 0.5 µL each, cDNA 2 µL, ddH2O 5.5 µL. PCR reaction conditions: 95 ℃ for 60 s (predenaturation), denaturation at 95 ℃ for 15 s, annealing at 57 ℃ for 30 s, extension at 72℃ for 45 s, 42 cycles were repeated for a final extension. With U6 as the reference gene, the 2-△△CT method was used to calculate the relative expression of the target gene. The primer sequence was synthesized by Shanghai Shenggong Bioengineering Technology Service Co., LTD.
2.3 Knockout of NLE1 mRNA
Ht-29 colon cancer cells were cultured in a 10% fetal bovine serum medium supplemented with 100 U/mL penicillin and 100 mg/mL streptomycin and incubated at 37 °C in an incubator containing 5% CO2. The experiment was carried out when the cells were in a logarithmic growth phase and digested with trypsin. The experiment was divided into three groups: the NLE1 interference group, the NLE1 negative control group, and the NLE1 overexpression group. The NLE1 interference group was transfected with NLE1 siRNA interference expression virus vector and liposomes, the NLE1 negative control group was added with liposomes, and the NLE1 overexpression group was transfected with NLE1 virus vector and liposomes. After successful transfection, the culture was continued for 48 h for subsequent experimental detection. Three pairs of siRNA sequences targeting the Knockout of NLE1 mRNA (siNLE1) were synthesized by Suzhou Jima Biotechnology Co., LTD. (Shanghai, China). Ht-29 cells were digested with trypsin and inoculated into 24-well plates with a cell density of 105 cells/well, and an antibiotic-free medium of 500 µL was added to each well until 70% to 80% of the cells confluent during transfection. Cells were transfected using Lipofectamine2000 (Invitrogen, USA), and the transfection efficiency was evaluated.
2.4 qRT - PCR detection
After 48 h transfection, RNA was extracted from the cells by Trizol, and then reverse transcription was performed using real-time PCR (rt-qPCR) to detect the expression of NLE1 mRNA and screen siNLE1 with the maximum inhibitory efficiency. qPCR was performed using the SYBR®PremixExTaqTM (Takara) kit. The primer sequence of the NLE1 gene used in this study is as follows: (F:5′ -CCAGGACCGCaccatCAaAG-3 '; R:5′-AGCTTGGGGCTGGTAGAT GA-3′); GAPDH(F:5′-TGACTTCAACAGCGACACCCA-3′; R: 5 '- the CACCCTGTTGCTGT AGCCAAA − 3′). PCR cycles were 95℃ 30 s, 39 cycles, 95℃ 5 s and 60℃ 30 s. All samples were repeated 3 times. β-catenin was used as an internal reference, and 2- δδ Ct was used to calculate the expression level and the amplified length was 203 bp.
2.5 Colony formation assay
To determine colony formation, the cells (1 0 4) were gently seeded onto a sterile dish (60 mm) in duplicate analysis. After fifteen days of culture, the developed colonies were fixed with methanol and further stained with Giemsa stain. The developed colonies were further counted using a microscope.
2.6 Transwell detection of cell migration and invasion
The gel cell suspension was placed in the upper compartment (100 µL/compartment) with a final concentration of 8 × 104 cells/compartment, and 750 µL serum-containing medium was added into the lower compartment by Transwell assay. After 48 h of culture, Transwell chambers were taken out and stained with crystal violet. Five visual fields were randomly selected under the microscope to take photos and collect experimental data for analysis. The transwell cell migration assay quantifies the capacity of cells to chemotactically move toward a chemoattractant. But the transwell cell invasion assay, which is frequently seen in cancer metastasis or embryonic development, assesses both cell chemotaxis and the invasion of cells via the extracellular matrix.
2.7 Statistical analysis
SPSS 20.0 statistical software was used for statistical analysis. The expression level of NLE1 mRNA, the number of invasive cells, and the number of migrating cells in colon cancer ht-29 cells in each group were expressed by x ± s. The comparison between groups was performed by t-test, and the comparison within groups was performed by snk-q test. The p-value < 0.05 was considered statistically significant.
3 Results
3.1 Expression of NLE1 in colon cancer cells
Compared with normal colon cells, the expression of NLE1 mRNA in colon cancer cells was significantly higher (P < 0.05) (Table 1) (Fig. 1). Likewise, the expression of NLE1 protein in colon cancer cells was significantly increased than control colon cancer cell lines (p < 0.05) (Fig. 2). Note: *P < 0.05 compared with normal colon cells.
Group
Normal colon cells
Colon cancer cells
NLE1 mRNA
1.295 ± 0.15
6.541 ± 0.523*
NLE1 protein
1.434 ± 0.23
6.817 ± 0.816*
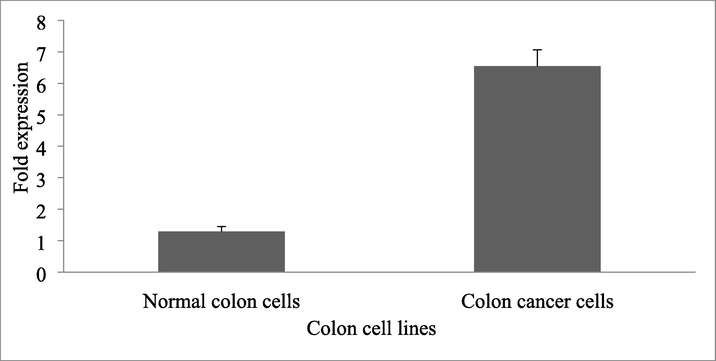
Expression of NLE1 mRNA in colon cancer cells and normal colon cells. The result was expressed as fold changes.
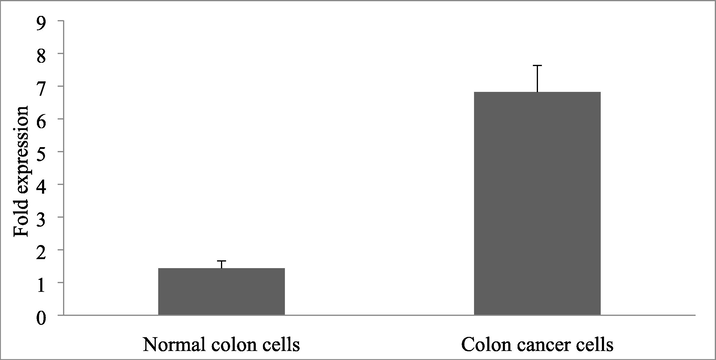
Expression of NLE1 protein in colon cancer cells and normal colon cells. The result was expressed as fold changes.
3.2 Hyperexpression of NLE1 mRNA and protein in NLE1 mRNA in vitro
Compared with the NLE1 interference group, the mRNA and protein in the NLE1 negative control group and the NLE1 overexpression group were significantly increased (P < 0. 05). The present finding revealed that the mRNA and protein in the NLE1 overexpression group were significantly increased compared with the NLE1 negative control group (P < 0. 05) (Table 2). Note: aP < 0.05 compared with the NLE1 interference group; Compared with the NLE1 negative control group, bP < 0.05.
Group
NLE1 interferential group
NLE1 negative control group
NLE1 overexpression group
NLE1 mRNA
0.24 ± 0.09
0.95 ± 0.07a
15 ± 6.72ab
NLE1 protein
0.36 ± 0.13
1.41 ± 0.06a
18 ± 9.37ab
3.3 Overexpression of NLE1 on the migration and invasion ability of colon cancer cells
Compared with the NLE1 interference group and the NLE1 negative control group, the migration and invasion ability of colon cancer cells in the NLE1 overexpression group were significantly varied (P < 0.05) (Table 3). These results indicated that the overexpression of NLE1 could promote the invasion of HT29 cells (Fig. 3) (Fig. 4). Note: aP < 0.05 compared with NLE1 interference group; Compared with NLE1 negative control group,bP < 0.05.
Experiment
NLE1 interferential group
NLE1 negative control group
NLE1 overexpression group
Migration
73.44 ± 5.36
30.84 ± 2.89a
186.17 ± 16.34ab
Invasion
86.32 ± 5.42
42.56 ± 5.63a
291.53 ± 20.11ab
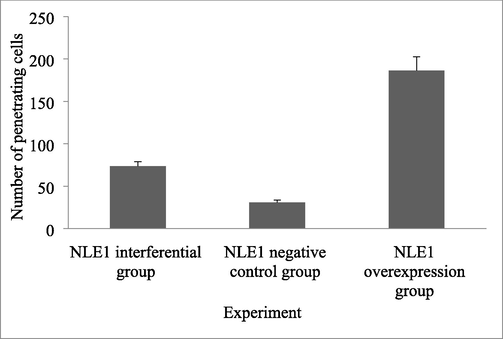
Migration ability of colon cancer cells between Transwell cell migration groups.
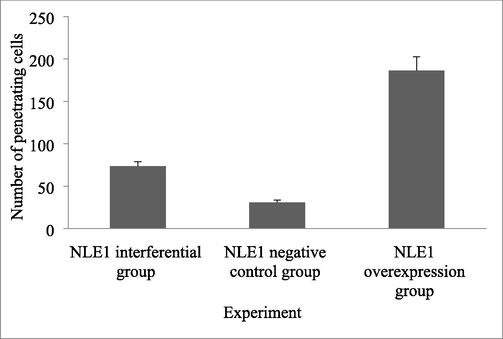
Invasion ability of colon cancer cells between Transwell cell migration groups.
3.4 In vitro knockdown of NLE1 affects proliferation and generation of colon cancer cells
The expression of NLE1 in colon cancer cell lines was evaluated using quantitative real-time PCR analysis. The present finding revealed that NLE1 was highly expressed in Ht-29 colon cancer cells and was statistically significant (p < 0.05). To analyze the impacts of NLE1 in colon cancer siRNAs targeting NLE1 were employed for silencing NLE1. The transfection potential was analyzed to be > 85% and the knockdown potential was further analyzed by qRT-PCR analysis (p < 0.05).
3.5 Effect of NLE1 on colon cancer cell colony development in vitro
The influence of NLE1silencing in Ht-29 colon cancer cells was analyzed. The colony formation assay revealed the number of colonies in cells, in which NLE1 was downregulated. The expression level was significantly varied than CCD-18CO cells (p < 0.05).
4 Discussion
Notchless Homolog 1 plays a potent role in signal transduction, transcription and cell proliferation. It has been previously reported that the development of colorectal cancer (CRC), among which Mir-506-3p affects CRC progression by affecting EZH2 expression, which shows a molecular target for the treatment of CRC (Ai et al., 2021). NLE1 participates in the assembly of 60S ribosome (Gayraud-Morel et al., 2018). Loss of Nle function leads to dysfunction of the ribosome that significantly reduced WnT-driven tumour initiation. Moreover, ribosome biogenesis significantly reduces the development of cancer cells in the intestinal epithelium (Raveux et al., 2020). Tumour-associated calcium signal transduction protein (TROP)–2 has been reported to be a potential function with tumour necrosis factor (TNF)–α levels in colon cancer. TROP–2 protein levels have been analyzed in HCT–116 human colon cancer cells cultured with various concentrations of TNF–α and analyzed using western blot analysis (Belo et al., 2011).
NLE1 is only one reported gene that is involved in the development and regulation of the brain regions. It has been earlier reported that the expression was highly limited to the embryo-fetal metaphase and late development (Al-Mubarak et al., 2020). NLE1 knockdown by shRNA has inhibited cell proliferation, improved the apoptotic sensitivity and affected the migration of melanoma cells in vitro conditions. Mouse xenograft experimental trial revealed that NLE1 knockdown could prevent the development of tumour melanoma in vivo. Also, the induction of apoptosis of melanoma by NLE1 knockdown involved the participation of various apoptosis-related proteins. NLE1 can stimulate the activation of PI3K/AKT signalling pathway (Ren et al., 2021).
Notchless homolog 1 is a molecular regulator of Notch and was upregulated in colonic HGIN adenoma tissues and T1-T2 colon tumours. It has been previously reported that the expression of the NLE1 gene could affect migration, invasion, and cell proliferation. In vivo and in vitro knockdown of NLE1 inhibited the development of cancer, and NLE1 may promote early carcinogenesis through the BAX/ Bcl-XL /FAS pathway (Liu et al., 2019). It has been previously reported that BAX/ Bcl-XL /FAS pathway can induce apoptosis and Cloverin reduces B-cell lymphoma 2(Bcl-2) and increases (Bax) protein levels. It effectively induces apoptosis in NCHI-H460 human non-small cell lung cancer (NSCLC) cells (Kim et al., 2016). Karimzadeh et al. (2017) have analyzed the developmental changes in NLE1 and Notch 1 expression in a genetic model of absence epilepsy.
SCU therapy has been recommended and which increased the expression of Fas and ligand (FasL), which induced the cleavage of Caspase-3, Caspase-8, polymerized adenosine diphosphate, whereas the amount of death receptor 4(DR4) considerably decreased. It has been previously stated that BAX/ Bcl-XL /FAS pathway in the cells of Hep3B was mediated apoptosis pathway. KRG extract effectively increase the expression of BAD, BAK and BAX proteins and reduce the expression of anti- apoptotic proteins such as Bcl-XL and induce the apoptosis of McF-7 breast cancer cells McF-10a non-malignant cells (Kim et al., 2021). Lung adenocarcinoma HT-29 cells revealed that overexpression of NLE1 gene promoted migration, invasion and proliferation. This finding revealed that NLE1 is highly related to the development of the progression of adenocarcinoma, which is actively involved in the regulation of Fas gene expression.
It has been previously reported that NLE1 can promote the proliferation and activation of myoblast cells C2C12 in mice (Stedman et al., 2015). NLE1 has the potential for differentiation and proliferation (Eldafrawy et al., 2018). The present study revealed that NLE1 could promote melanoma development through inhibiting cell apoptosis of melanoma cells, migration and promoting cell proliferation. The Notch signaling pathway is one of the important pathways involved in cell growth. During embryonic growth, NLE1 binds to the Notch receptor and specifically regulates the Notch signalling pathway (Karimzadeh et al., 2017). In this study, compared with normal colon cell lines, the expression of NLE1 mRNA and NLE1 protein in colon cancer cell lines was lower than that in normal colon cells. In the present study, we also analyzed the mechanism of gene expression. The overexpression of NLE1 could promote the invasion of HT29 cells in colorectal cancer cells.
5 Conclusions
The mechanism and up-regulation of the NLE1 gene in colon cancer cells and cell cycles were determined. The expression of NLE1 mRNA and NLE1 protein in colon cancer tissue was lower than that in normal colon tissue (P < 0. 05). The mRNA and protein in the NLE1 negative control group and the NLE1 overexpression group were significantly increased. These results indicated that the overexpression of NLE1 could promote the invasion of HT29 cells in colorectal cancer. The NLE1 gene is expressed highly in colon cancer cells. To conclude, NLE1 is a useful target for the treatment of colon cancer cells.
Authors contribution
All the authors contributed equally to this work.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- LIMK/cofilin pathway and Slingshot are implicated in human colorectal cancer progression and chemoresistance. Virchows Archiv.. 2018;472:27-737.
- [Google Scholar]
- MicroRNA-506-3p inhibits colorectal cancer cell proliferation through targeting enhancer of zeste homologue 2. Bioengineered.. 2021;12(1):4044-4053.
- [Google Scholar]
- Whole exome sequencing in ADHD trios from single and multi-incident families implicates new candidate genes and highlights polygenic transmission. Eur. J. Hum. Genet.. 2020;28(8):1098-1110.
- [Google Scholar]
- Muscarinic receptor agonists stimulate human colon cancer cell migration and invasion. Am. J. Physiol. Gastrointestin. Liver Physiol.. 2011;300(5):G749-G760.
- [Google Scholar]
- Anti-aging trait of whey protein against brain damage of senile rats. J. Umm Al-Quara Univ. App. Sci.. 2022;8:8-20.
- [Google Scholar]
- Bonding to CAD-CAM composites: an interfacial fracture toughness approach. J. Dent. Res.. 2018;97(1):60-67.
- [Google Scholar]
- Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun.. 2019;39:1-12.
- [Google Scholar]
- Notchless defines a stage-specific requirement for ribosome biogenesis during lineage progression in adult skeletal myogenesis. Development. 2018;145(23):dev162636.
- [Google Scholar]
- Modifiable risk factors for colon cancer. Gastroenterol Clin North Am.. 2002;31(4):925-943.
- [Google Scholar]
- Identification of differently expressed genes associated with prognosis and growth in colon adenocarcinoma based on integrated bioinformatics analysis. Frontiers in Genetics. 2019;10:1245.
- [Google Scholar]
- Developmental changes in Notch1 and NLE1 expression in a genetic model of absence epilepsy. Brain Struct. Funct.. 2017;222:2773-2785.
- [Google Scholar]
- Trifolin induces apoptosis via extrinsic and intrinsic pathways in the NCI-H460 human non-small cell lung-cancer cell line. Phytomedicine.. 2016;23(10):998-1004.
- [Google Scholar]
- Korean red ginseng induces extrinsic and intrinsic apoptotic pathways in MCF-7 breast cancer cells and MCF-10A non-malignant breast cells. J. Obstet. Gynaecol. Res.. 2021;47(8):2758-2766.
- [Google Scholar]
- The growing role of precision and personalized medicine for cancer treatment. Technology. 2018;6(03n04):79-100.
- [Google Scholar]
- PTBP1 promotes tumorigenesis by regulating apoptosis and cell cycle in colon cancer. Bulletin du Cancer. 2018;105(12):1193-1201.
- [Google Scholar]
- Mo1738–Notchless Homolog1 (NLE1), a Stepwisely Upregulated Gene in Early-Stage Colorectal Cancer Promote the Carcinogenesis Via Bax/Bcl-Xl/Fas Pathway. Gastroenterology. 2019;156(6):S-827.
- [Google Scholar]
- Biochemical and histopathological alterations induced by subchronic exposure to zinc oxide nanoparticle in male rats and assessment of its genotoxicicty. J. Umm Al-Quara Univ. App. Sci.. 2022;8:41-49.
- [Google Scholar]
- Compensation between Wnt-driven tumorigenesis and cellular responses to ribosome biogenesis inhibition in the murine intestinal epithelium. Cell Death Differ.. 2020;10:2872-2887.
- [Google Scholar]
- Knockdown of NLE1 inhibits development of malignant melanoma in vitro and in vivo NLE1 promotes development of malignant melanoma. Exp Cell Res.. 2021;404(2):112636
- [Google Scholar]
- A review on dietary intervention in obesity associated colon cancer. Asian Pac J Cancer Prev.. 2019;20(5):1309-1319.
- [Google Scholar]
- Ribosome biogenesis dysfunction leads to p53-mediated apoptosis and goblet cell differentiation of mouse intestinal stem/progenitor cells. Cell Death Differ.. 2015;22(11):1865-1876.
- [Google Scholar]
- Immune cell infiltration as a biomarker for the diagnosis and prognosis of stage I-III colon cancer. Cancer Immunol Immunother. 2019;68:433-442.
- [Google Scholar]







