Translate this page into:
Survey sequencing and in-silico development and validation of genomic SSR markers in Indian dill seed
⁎Corresponding author. sushil254386@yahoo.com (Sushil Kumar)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Indian dill seed (Anethum sowa Roxb. ex Fleming) is an important member of family Apiaceae. It is a significant seed spice having many medicinal properties. But, this aromatic herb is still orphan from breeding and crop improvement perspective as no serious attention has been given by breeders. To swift the breeding program, molecular markers play the main role. But due to the paucity of markers, breeding is slower in dill seed. Therefore, an attempt was made to develop the genomic SSR markers in dill seed through next-generation sequencing. A total of 2119.51 Mbp of raw data were generated on the Miseq NGS platform during survey sequencing. In 2,25,956 contigs created by Spades assembler, a total 48,951 repeat motifs were identified. A set of 20,294 primer pairs (dimer to hexamer repeats) were produced. Among detected repeat motifs, 48.89% was mononucleotides. Of 12 primers, 10 (83%) primers could be successfully amplified in dill seed and produced 11 amplicons. During cross-genera amplification of markers, 9 out of 10 primers could be successfully amplified in related genera. Developed markers can also be used to initiate the molecular breeding program, association mapping and to assess the evolutionary relationship among seed spices.
Keywords
Anethum
Seed spices
SSR
Transferability
1 Introduction
Globally, seed spices are an integral part of food as it plays a pivotal role in human diets by providing pleasant flavour to culinary preparations. Seed spices are annual crops whose seeds are consumed. For Ages, spices are well known for their effective therapeutic effects. Secondary metabolites and many biologically active phytochemicals make them effective therapeutic food. Seed spices are also a vital group of agri-commodities and play a significant role in the world economy (Rathore et al., 2013). Among various seed spices, the members of family Apiaceae (Umbelliferae) viz. cumin, coriander, fennel, ajwain, dill, etc. are more prominent.
Except for carrot, most of the members in family Apiaceae are less studied. Therefore breeding and crop improvement programs in Apiaceae is inadequate (Alison and Caswell, 2016). The crop improvement of seed spices of Apiaceae is mainly focused on cumin and coriander. A substantial conventional breeding work especially phenotypic based variability assessment has been carried out in most of the spices of Apiaceae. It is well established that morphological traits especially quantitative traits are highly influenced by the environment. This genotype by environment interaction cause deviation in phenotyping value; consequently the information on variability may not be true (Kumar et al., 2015). To mitigate this problem, molecular markers may be deployed (Rukhsar et al., 2017). Though, despite their economic importance, most of the seed spices of Apiaceae have relatively underdeveloped molecular resources as result inaccessibility of modern breeding methods for breeders.
Limited work on diversity analysis using molecular markers like Random Amplified Polymorphic DNA (RAPD) and Inter-Simple Sequence Repeat (ISSR) has been carried out in seed spices. But dominant nature and low reproducibility of such markers hinder the molecular breeding program in Apiaceae. Underdeveloped molecular resources especially Simple Sequence Repeat (SSR) markers, enforced the researcher to use SSR markers of carrot and other non-spices plants of Apiaceae to examine the variability as genomic levels (Kumar et al., 2014). Though cross-amplification is a cost-effective method to identify molecular resources in orphan crops. But in most of the cases, cross-amplification results in monomorphic marker due to genome conservation. Recently, gSSR has been reported in cumin (Bharti et al., 2018).
Among seed spices crops of Apiaceae, Indian dill (Anethum sowa Roxb. ex Fleming) is a popular annual spice which is under cultivation in both irrigated as well as rainfed condition. This glabrous crop is widely grown during the cold season in India, Myanmar, Pakistan and Bangladesh (Husain et al., 1988). In India, Gujarat, Rajasthan, Madhya Pradesh are the leading states concerning area and production (Patel et al., 2017).
Botanically, the seed of dill is a fruit which can be split into two halves, each containing one true seed. With miraculous medicinal benefits, Indian dill is also referred to as a variant of European dill (A. graveolens) (Singh, 2012). But compared to European dill, the fruit of Indian dill is longer but is less in thickness and having low fragrance. Indian dill is rich in apiol while carvone content is higher in European dill (Saleh-e-In et al., 2010). The essential oil in seed (1.5–4.5%) is responsible for its medicinal properties (Singh, 2012). It is mainly used to cure hiccup, intestinal spasms and griping and also to improve appetite and digestion (Jana and Shekhawat, 2010). Essential oil of fruit also has insecticidal and ovicidal activity (Tripathi et al., 2001; Walia et al., 2017). Compared to A. graveolens, no marker-based study has been carried out in Indian dill which hampers the genetic diversity assessment during its improvement program. In the current study, attempts have been made to develop genomic SSR markers to support the breeders working for the improvement of seed spices.
2 Materials and methods
2.1 DNA isolation and DNA sequencing
To isolate microbial genome free DNA of dill seed, the cultivar GAVD-1 was gown in the controlled condition in seed germinator (Percival). For high-quality DNA isolation, leaves from aseptically grown 15 days old seedlings were collected. The CTAB protocol of Doyle and Doyle (1990) was used for genomic DNA isolation. Isolated was assessed on 0.8% agarose gel and spectrometer to confirm the intactness and quantity of extracted genomic DNA.
PCR-Free TruSeq (LT) DNA sample Prep kit (FC-121–3001) was used to generate the library following the protocol recommended by Illumina. Paired-end sequencing was performed in the departmental laboratory on MiSeq platform (Illumina) with MiSeq Reagent Kit version 2 (2 × 250 bp). The details on up- and down-stream procedure for DNA sequencing is given in Bharti et al. (2018).
2.2 Bioinformatic analysis for SSR marker development
During downstream processing of raw sequences, quality checking was carried out using FastQC tool (Andrews, 2010) for confirmation of high-quality reads. For removing adaptor sequences and trimming of low-quality bases Trimmomatic (Bolger et al., 2014) was used. During raw reads processing, reads were filtered with the custom setting (Phred 33, Sliding window:4:15, Leading: 10, Trailing: 10, Minlen: 35) to remove adaptor sequence or low-quality reads. After trimming, FastQC analysis was again performed to check the high quality of trimmed reads. Subsequently, high-quality clean reads were utilized for de-novo assembly using SPAdes-3.11.1 (Bankevich et al., 2012), Megahit -1 (Li et al., 2015) and SOAPdenovo 2.04 (Luo et al., 2012) software. SPAdes was performed with the read error correction program. QUAST was used to evaluate the results of each assembly (Gurevich et al., 2013). Eventually, the best assembly was used as an input file for MISA to detect microsatellite using default parameters and primer3 was used to generate SSR markers.
2.3 Marker validation and cross-amplification
For validation of primers, a total of 12 SSR primers were selected randomly. The DNA of dill seed cultivar GAVD 1 was used for primer amplification. The DNA amplification was achieved through PCR in a final volume of 15 μl with 1× PCR buffer, 1 μlMgCl2, 0.5 μl dNTPs, 1 μM of primers (10 pmol), 0.3 μl of Taq DNA polymerase (5 U/μl), and 20 ng of template DNA. PCR reactions were completed with initial denaturation at 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, primer-specific annealing temperature (Ta) for 40 s, and 72 °C for 60 s with a final extension of 10 min at 72 °C. The amplified products were separated in agarose gel (1.5%) and photographed in a gel documentation system (G-box, Genei). For cross-amplification of SSR primers, DNA six seed spices (Ajwain, Celery, Coriander, Cumin, Fennel and Parsley) belonging to the Apiaceae family was extracted followed by PCR as described above. The amplification products were scored in base pair on a visual basis. For evaluation of SSR marker transferability, the microsatellite products were classified into two classes based on the amplicon intensity and scoring ease: (++) strong amplicon and easy to score and (+) weak amplicon and hard to score. The amplicons were declared as non-specific if the product is of dissimilar size (within +100 bp) to that of dill seed.
3 Results and discussion
3.1 Filtering of reads and de novo genome assembly
Survey sequencing of the library generated 2119.51 Mbp of raw data containing 7,102,148 paired-end reads. The average read length was ∼298 bp. After cleaning and filtering the raw reads, 6,516,297 reads were obtained from high-quality 1541.62 Mbp. The mean read length of clean data was ∼207 bp which is similar to Shiwani (2017) where it was 203 bp. The GC content of filtered reads was 34%.
Evaluation of all three assemblies through QUAST suggested that assembly generated by SPADES was best. Using high-quality reads, SPADES generated 2,25,956 contigs (183.54 million bases). A set of 156,521 contigs were of ≥500 bp long. The maximum and minimum contig length was 92,006 and 128 bp, respectively. The N50 was 604 and 832 with SOAP denovo and Megahit, respectively. However, the N50 value of this study was 1020 bp in case of SPADES assembly. Compared to reported of Shiwani (2017), where N50 in dill seed was 661 bp, N50 was 1.5X higher. The GC content (32.46%) in the current study was closer to carrot (34.8%) and coriander (34.65%) as reported by Iorizzo et al. (2011) and Tulsani (2013).
3.2 Characteristics of genomic SSRs
A total of 37,863 (16.75%) contigs, out of 2,25,956 were containing microsatellite motifs. Earlier, in cumin, SSRs motif in survey sequences was 12% of the total contings (Bharti et al., 2018). A total of 48,951 repeat motifs were detected suggesting one SSR/3.75 kb considering 183.54 million bases. Moreover, 8185 contigs were found to be containing more than 1 SSR, and 5114 contigs were identified having SSRs present in compound formation (Table 1). Previously, in fennel, 954 (18%) transcripts exhibited more than one microsatellite region (Palumbo et al., 2018).
Features
Values
Total number of sequences examined
2,25,956
Total size of examined sequences (bp)
18,35,45,799
Total number of identified SSRs
48,951
Number of SSR containing sequences
37,863 (16.75%)
Number of sequences containing more than one SSR
8185
Number of SSRs present in compound formation
5114
Among detected repeat motifs, the 48.89% (23936) was mononucleotides. Compared to cumin (15%) and celery (21.66%) genome, the mononucleotide repeats was very high (Li et al., 2014a; Bharti et al., 2018). The frequency of A/T SSRs was more in mononucleotide repeats, almost 27x more than C/G SSRs which is in congruence with Li et al. (2014a). After mononucleotide, the majority (49.02%) of SSRs were detected in the di- and tri-nucleotide categories (37.34% and 11.67%, respectively). Penta- and hexa-mers were least common motifs (0.24% and 0.11%, respectively) (Table 2). This is in agreement with the previous report in various seed spices crops where di- and tri-mers were frequent while penta- and hexa-mers are exceptional (Li et al., 2014a; Bharti et al., 2018; Palumbo et al., 2018). Similar to Li et al. (2014a) in celery, the repeat-motif between 5 and 8 were dominant, accounting for 56% of all the SSRs identified. AT/AT was the greatly frequent (64.91%) dimer repeat whereas AC/GT and AG/CT were present in equal frequency (17% each) (Table 3). The similar pattern was also recorded in cumin (Bharti et al., 2018). Many researchers have also recorded richness of AT/AT repeat motif in the genome of different crops (Kalia et al., 2011; Xu et al., 2013). The less existence of CG/CG motif also demonstrated depicted in different Apiaceae crops high (Li et al., 2014a; Bharti et al., 2018).
Motif length
Repeats number
5
6
7
8
9
10
11
12
>12 (13–128)
Total
%
Dinucleotide
–
4522
2759
2049
1559
1084
864
741
4704
18,282
73.08
Trinucleotide
2090
898
451
268
227
179
158
121
1324
5716
22.85
Tetranucleotide
473
156
88
36
26
21
10
8
22
840
3.36
Pentanucleotide
86
24
4
3
3
–
–
1
1
122
0.49
Hexanucleotide
37
9
4
1
2
–
1
–
1
55
0.22
Total
2686
5609
3306
2357
1817
1284
1033
871
6052
25,015
–
%
10.74
22.42
13.22
9.42
7.26
5.13
4.13
3.48
24.19
–
–
Repeat motif
Repeats number
5
6
7
8
9
10
11
12
>12 (13–128)
Total
%
AC/GT
–
1257
700
452
291
177
112
81
183
3253
17.79
AG/CT
–
766
529
390
278
179
162
127
682
3113
17.03
AT/AT
–
2467
1522
1202
987
727
590
533
3839
11,867
64.91
CG/CG
–
32
8
5
3
1
–
–
–
49
0.27
Total
0
4522
2759
2049
1559
1084
864
741
4704
18,282
–
AAC/GTT
102
51
27
13
15
6
6
3
14
237
4.15
AAG/CTT
227
91
31
18
9
10
5
3
29
423
7.40
AAT/ATT
1351
609
321
197
189
150
139
112
1257
4325
75.66
ACC/GGT
55
16
3
3
3
–
–
–
–
80
1.40
ACG/CTG
8
6
1
1
–
–
–
–
–
16
0.28
ACT/ATG
65
18
7
7
3
–
3
1
5
109
1.91
AGC/CGT
81
29
14
4
–
2
–
–
2
132
2.31
AGG/CCT
42
10
14
7
3
–
–
1
–
77
1.35
AGT/ATC
145
63
33
18
5
11
5
1
17
298
5.21
CCG/CGG
14
5
–
–
–
–
–
–
–
19
0.33
Total
2090
898
451
268
227
179
158
121
1324
5716
–
An overabundance of both di- and tri-nucleotide repeats in cumin, celery and parsley -members of Apiaceae- has been recorded (Li et al., 2014a,b; Bharti et al., 2018). The overabundance of di- and tri-mer repeats disagree with SSRs in Triticum aestivum and Saccharum spp, wherein tetranucleotide repeats were dominant (Cordeiro et al., 2001; Gupta et al., 2003). In the current study, dinucleotide repeats were higher frequent that trinucleotide repeats. This is in agreement with findings in citrus (Biswas et al. 2014) and sesame (Wei et al., 2014).
3.3 Validation of the SSR markers and their transferability
A small set of 12 SSR primers were used for amplification to validate the primers. Of 12 primers, 10 (83%) primers could successfully amplify in dill seed. Representative amplification profiles of few primers are shown in Fig. 1. High amplification rate suggested that assembly was perfect and primers have a proper binding site in the genome. The amplification rate has also been reported by Bharti et al. (2018) in cumin for genomic SSR markers. Amplicon size of validated primers ranged from 120 to 260 bp. A total of 11 alleles were amplified by ten primers in dill seed (Table 4). ++strong band and easy to score; +weak band and difficult to score; #amplified product of not similar size (within 100 bp) to that of dill seed.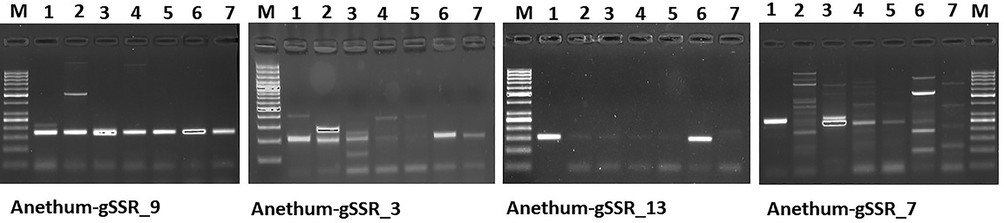
Amplification profile of genomic SSR markers in members of family Apiaceae; M = 50 bp ladder, 1 = Dill seed; 2 = Ajwain; 3 = Celery; 4 = Coriander; 5 = Cumin; 6 = Fennel; 7 = Parsley.
Primer
Primer Sequence
Product Size (bp), quality (+/++) and specificity#
Dillseed
Ajwain
Celery
Coriander
Cumin
Fennel
Parsley
Anethum-gSSR_1
cggcccaaaccaataaacta/agagaaggggaaaggaggtg
260
260 (++)
260 (++)
200, 260 (++)
200, 260 (++)
260, 310 (++)
260, 610# (+)
Anethum-gSSR_3
ttcaagaggatgatgatgcaa/agacccctcctgcacattta
200
200, 290 (++)
120, 200 (++)
230, 390# (+)
230, 390# (+)
230 (+)
220 (+)
Anethum-gSSR_5
aaagcaacccctgcataaaa/tgtggttgggtatttttggg
150
140 (+)
150 (+)
–
–
–
–
Anethum-gSSR_12
gcacgatattactcgtaacccc/ttctttgacttgacaatttgcag
120
–
–
–
–
–
–
Anethum-gSSR_7
agcgagaaaggtggaaacac/agacccacaatcaaagctcc
250
–
240, 260 (++)
240 (++)
240 (++)
250, 520# (++)
–
Anethum-gSSR_9
agttccttccccgaacctta/atcctgatgcgactgattcc
160
160, 500 (++)
160 (++)
160 (++)
160 (++)
160 (++)
160 (++)
Anethum-gSSR_20
ttttgttttctgtttggtgtgg/ccctataagggaagcagcct
230
130, 300 (+)
190, 260 (+)
–
–
–
–
Anethum-gSSR_22
agagggaactaaggggcaac/cggatttaacggaaatacgc
200
200, 400# (++)
200, 350# (++)
400# (++)
190, 400# (++)
400# (++)
–
Anethum-gSSR_23
gcagctgatgaaaatgcaag/catttatgttgggggtttgg
190, 230
240, 300# (++)
120, 170 (+)
–
240 (+)
–
300#, 390# (+)
Anethum-gSSR_13
acaagccgtttgtcgatacc/acctcgtggagaatcgaaga
150
150 (+)
150 (+)
150 (+)
150 (+)
150 (++)
180 (+)
Most of the member of family Apiaceae are deficient in genomic resources and molecular markers (Kumar et al., 2014). Most of the (EST)SSR markers are produced through transcriptomic study in fennel, celery, parsley etc. (Li et al., 2014a, b; Palumbo et al., 2018). EST-SSRs are superior to genomic SSRs due to a segment of transcripts. However, EST-SSRs are less polymorphic due to the selection against the variation in the conserved regions of the EST-SSRs (Zhang et al., 2014). This suggests that there is a need to develop genomic SSR markers in the above crops as well as other members of the carrot family. Though SSR development with NGS is cost-effective, but still, it is better to investigate the cross-transferability of SSRs in related genera to reduce the cost especially for underdeveloped labs (Kumar et al., 2014; Bharti et al., 2018). Evolutionary relatedness of the species affects the success of marker transferability. During cross-genera amplification of markers, 9 out of 10 primers could successfully be amplified, and merely one primer could not amplify in any of the related genera. To mitigate the amplification problem caused by genome complexity, PCR was carried out by changing the annealing temperature as suggested by Bharti et al. (2018).
The SSR transferability rate varied between 50% (Parsley) to 90% (celery). The frequency of clear and strong (scored as ++) bands varied from 20% (parsley) to 83.3% (fennel). A varied level of transferability of SSRs has been earlier reported in family Apiaceae (Cavagnaro et al., 2011; Kumar et al., 2014; Bharti et al., 2018). The reason for low to high transferability and amplicon quality (+or ++) may be due to (dis)similarity of dill seed genome with other studied crops in the current study.
4 Conclusion
In this study, we have first developed the genomic SSR for Indian dill seed through next generation survey sequencing. Eventually, it can be concluded that SSRs developed in dill seed can be further exploited to dissect the genetic variability present in dill seed and another member of the family Apiaceae gene pool. Developed markers can also be used to initiate the molecular breeding program, association mapping and to assess the evolutionary relationship among seed spices.
Conflict of interest
The authors declare no conflict no interest.
Acknowledgement
Authors acknowledge Anand Agricultural University, India for providing all necessary facilities. SK acknowledges the INSA, N. Delhi for providing visiting scientist fellowship at ICRISAT, Hyderabad during the analysis of data.
References
- Applications of doubled haploidy for improving industrial oilseeds. In: Hayes T.A., Hildebrand D.G., Weselake R.J., eds. Industrial Oil Crops McKeon. Academic Press; 2016. p. :359-378.
- [Google Scholar]
- Andrews, S., 2010. FastQC: A quality control tool for high throughput sequence data. Available: http://www.bioinformatics.babraham.ac.uk.
- SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol.. 2012;19(5):455-477.
- [Google Scholar]
- Development of genomic simple sequence repeat (gSSR) markers in cumin and their application in diversity analyses and cross-transferability. Ind. Crop Prod.. 2018;111:158-164.
- [Google Scholar]
- Genome wide characterization of short tandem repeat markers in sweet orange (Citrus sinensis) PLoS One. 2014;9(8):e104182
- [Google Scholar]
- Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinfo.. 2014;30:2114-2120.
- [Google Scholar]
- Microsatellite isolation and marker development in carrot - genomic distribution, linkage mapping, genetic diversity analysis and marker transferability across Apiaceae. BMC Genomics. 2011;12:386-405.
- [Google Scholar]
- Microsatellite markers from sugarcane (Saccharum spp.) ESTs cross transferable to erianthus and sorghum. Plant Sci.. 2001;160(6):1115-1123.
- [Google Scholar]
- Transferable EST-SSR markers for the study of polymorphism and genetic diversity in bread wheat. Mol. Genet. Genomic. 2003;270(4):315-323.
- [Google Scholar]
- QUAST: quality assessment tool for genome assemblies. Bioinfo.. 2013;29(8):1072-1075.
- [Google Scholar]
- Major Essential Oil-bearing Plants of India. Lucknow: CIMAP; 1988. p. :87-95.
- De novo assembly and characterization of the carrot transcriptome reveals novel genes, new markers, and genetic diversity. BMC Genomics. 2011;12:389.
- [Google Scholar]
- Anethum graveolens: an Indian traditional medicinal herb and spice. Pharmacogn Rev.. 2010;4(8):179-184.
- [Google Scholar]
- Microsatellite markers: an overview of the recent progress in plants. Euphytica. 2011;177:309-334.
- [Google Scholar]
- Transferability of carrot (Daucus carota) microsatellite markers to cumin (Cuminum cyminum) Int. J. Seed Spices. 2014;4(1):88-90.
- [Google Scholar]
- Understanding Cuminum cyminum: an important seed spice crop of arid and semi arid regions. Int. J. Seed Spices. 2015;5(2):1-19.
- [Google Scholar]
- MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinfo.. 2015;31(10):1674-1676.
- [Google Scholar]
- De novo transcriptome sequence assembly and identification of AP2/ERF transcription factor related to abiotic stress in parsley (Petroselinum crispum) PLoS One. 2014;9(9):e108977
- [Google Scholar]
- Identification of SSRs and differentially expressed genes in two cultivars of celery (Apium graveolens L.) by deep transcriptome sequencing. Horti. Res.. 2014;1:10.
- [Google Scholar]
- SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1(1):18.
- [Google Scholar]
- The leaf transcriptome of fennel (Foeniculum vulgare Mill.) enables characterization of the t-anethole pathway and the discovery of microsatellites and single-nucleotide variants. Sci. Rep.. 2018;8(1):10459.
- [Google Scholar]
- Identification of reference genes for real time PCR analysis in Dill seed (Anethum sowa) Int. J. Seed Spices. 2017;7(1):82-85.
- [Google Scholar]
- Potential health benefits of major seed spices. Int. J. Seed Spices. 2013;3(2):1-12.
- [Google Scholar]
- Morphological and molecular diversity patterns in castor germplasm accessions. Ind. Crops Prod.. 2017;97:316-323.
- [Google Scholar]
- Chemical constituents of essential oil from Anethum sowa herb (leaf and stem) growing in Bangladesh. Bangladesh J. Sci. Ind. Res.. 2010;45(2):173-176.
- [Google Scholar]
- Genome sequencing of dill (Anethum graveolens l.) for development and validation of simple sequence repeat markers. Junagadh, India: Junagadh Agricultural University; 2017. M.Sc. Thesis submitted to
- Chemical constituents of essential oil from Anethum Sowa Kurz. seed. J. Chem. Pharma. Res.. 2012;4(9):4156-4160.
- [Google Scholar]
- Insecticidal and ovicidal activity of the essential oil of Anethum sowa Kurz against Callosobruchus maculatus F. (Coleoptera: Bruchidae) Int. J. Trop Insect Sci.. 2001;21(1):61-66.
- [Google Scholar]
- Genome and transcriptome sequencing of coriander (Coriandrum sativum L.) to reveal its genome architecture. Junagadh, India: Junagadh Agricultural University; 2013. M.Sc. Thesis submitted to
- Phytochemical biopesticides: some recent developments. Phytochem. Rev.. 2017;16(5):989-1007.
- [Google Scholar]
- Development of simple sequence repeat (SSR) markers of sesame (Sesamum indicum) from a genome survey. Molecules. 2014;19:5150-5162.
- [Google Scholar]
- Development and characterization of simple sequence repeat markers providing genome-wide coverage and high resolution in maize. DNA Res.. 2013;20:497-509.
- [Google Scholar]
- Development and characterization of polymorphic EST-SSR and genomic SSR markers for Tibetan annual wild barley. PLoS One. 2014;9(4):e94881
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2019.04.006.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







