Translate this page into:
Study the epigenetic down-regulation of Bim on colorectal cancer chemotherapy response
⁎Corresponding author. nizarmhaidat@yahoo.com (Nizar M. Mhaidat)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Colorectal cancer (CRC) is the third most common malignancy in developed countries. It has been shown that tumor suppressor gene methylation is associated with cancer development and chemoresistance. One of such genes is Bim, a BH3 only proapoptotic member of the Bcl-2 family that was first identified as an essential mediator of apoptosis induced by microtubules damaging agents. In the present study, the association between Bim gene methylation and CRC risk and chemotherapy responsiveness was investigated using CRC and normal colon tissues. Gene methylation assessment was performed using the methylation specific PCR and quantification of gene methylation was performed using the combined bisulfate restriction analysis (COBRA) method. A significant increase in Bim gene methylation among CRC patient compared to healthy controls (P < 0.05). Moreover, a significant correlation was found between patient demographics including cancer recurrence and tumor metastasis and gene methylation (P < 0.05). In addition, a significant inverse correlation was obtained between Bim gene methylation and chemosensitivity (P < 0.05). Collectively, current results indicate that Bim gene methylation is associated with CRC development, metastasis, and chemosensitivity. Thus, prior evaluation of Bim gene methylation might aid in the diagnosis and treatment of CRC patients.
Keywords
BIM
Colorectal cancer
Methylation
Chemosensitivity
Epigenetics
1 Introduction
Colorectal cancer (CRC) is one of the most prevalent malignant diseases (Giatromanolaki et al., 2006). The risk increases with many factors such as family history of CRC, high intake of red meat, early smoking history, and alcohol intake (Chen et al., 2008). Rectal bleeding, change in bowel habits, abdominal pain, and sometimes anemia are the most common symptoms of CRC (Majumdar et al., 1999). CRC is the third most common malignancy in the developed Western countries and represents the third leading cause of death from cancer in the United States, after lung and prostate cancer (Jemal et al., 2011).
Epigenetic regulation is one of the mechanisms by which gene activation and silencing are controlled. Previous studies have shown that perturbation of this process might lead to cancer initiation, progression, and chemo-resistance (Kondo and Issa, 2004; Laird, 2005). This might occur since gene methylation can affect gene expression by interference directly with binding of transcription factors or indirectly by inducing changes in chromatin structure (Bigl et al., 2008). Alterations seen in methylation process, particularly in cancer are produced by either hyper-methylation of tumor suppressors’ gene or hypo-methylation of oncogenes (Feinberg and Tycko, 2004; Hatada, 2006).
Bim, a member of the Bcl-2 family and BH3 only proapoptotic group, acts as a critical mediator of programed cell death induced by several anticancer agents, particularly microtubules damaging agents (Zhang et al., 2013). Bim has also been shown to play an important role in regulation and control the apoptosis in several cell types, such as osteoblasts, endothelial cells, lymphocytes, neuron, osteoclasts and mast cells (Hengartner, 2000; Kawamura et al., 2007; Youle and Strasser, 2008). Bim apoptotic activities are exerted through interactions with other proapoptotic and antiapoptotic Bcl-2 family proteins that result in activation of the proapoptotic proteins Bax and BakB. A suppressor role for Bim in tumorigenesis and tumor metastasis has been suggested. For example, studies in young mouse kidney epithelial (BMK) tumor indicated that the growth and the development of the tumor is associated with the Bim deficiency, which suggested that lack of Bim play a role in formation of tumors (Tan et al., 2005).
Several studies have demonstrated that BIM can be the target of epigenetic silencing in specific types of cancer as leukemia, lymphomas, and solid tumors. Therefore, in this study, we aimed to investigate the role of pro-apoptotic Bim gene methylation in colorectal cancer (CRC) development, progression, metastasis, and sensitivity to chemotherapy. Results might have implications in the diagnosis and managements of CRC.
2 Materials and methods
This experiment was done as a cross-sectional retrospective correlation study. It was approved by the IRB committee of both King Abdullah University Hospital and Jordan University of Science and Technology.
Paraffin-embedded patient samples and controls were kindly provided by the department of pathology at King Abdullah University Hospital, Irbid, Jordan. The patient was diagnosed and confirmed by a clinical and pathological test of having a CRC disease. CRC patients (n = 40) samples and 10 healthy control samples were applicable for DNA extraction due to high chemical DNA degradation ratio caused by Xylene and Bisulfate modification, which are necessary for DNA extraction and conversion.
2.1 DNA extraction from tissue samples
Extraction of DNA from paraffin-embedded colorectal tissue samples was done using QIAGEN QIAamp®DNA FFPE Tissue Kit according to the manufacture instruction.
Three small suctions of sample tissue were used for this purpose and the quality of the DNA was checked by using agarose gel analysis.
2.2 Bisulfate modification protocol
Bisulfate conversion was done on 20–50 ng of the isolated DNA using commercially available kit (MethylEdge™ BisulfiteConversion System, Promega, USA), following the manufacture’s instruction.
2.3 Methylation specific PCR
Methylation specific PCR amplification was used to determine the methylation status of the promoter of Bim gene, which is considered as a qualitative technique. However, two PCR reactions were run as a separate. One reaction is done to detect the methylated parts in the promoters. The other reaction is for detection of the unmethylated parts of the promoter. A positive and negative control was used for both reactions. The positive control was prepared from enzymatically methylated Hela-derived genomic DNA with CpG methylase (EpiScope™Hela Genomic DNA, TAKARA BIO, Japan). The negative control was a genomic unmethylated DNA extracted from DNMT DKO HCT116 cell (EpiScope® Unmethylated HCT116DKO Genomic, TAKARA BIO, Japan).
Primers (IDT, USA) were prepared in stock solution of 100 pmol/µl and working solution of 10 pmol/µl. The sequence for the primers, product size and its corresponding annealing temperature are summarized in Table 1.
Gene
Primer (5′–3′)
Product size
Annealing temperature
BIM (MS-PCR)
Unmethylated Primers (U)
F: GTATTTTTGGTAAATAATGGGGTTG
R: CAAATAAATCAAAAACTCCCAACA139 bp
60 °C
Methylated Primers (M)
F: AGTATTTTCGGTAAATAATGGGGTC
R: GAATAAATCAAAAACTCCCAACG139 bp
60 °C
BIM (COBRA)
F: TGTATTTTGTTTGGGGGTATTT
R: CAATTATCTACCTTCTCAATCACACTC283 bp
58.3 °C
Each 0.2 ml PCR reaction tube included 1–2 μl of bisulfite treated DNA, 7.5 pmol of each of sense and antisense primers, 12.5 μl PCR hot start master mix (Kapa CG Robost, USA), and nuclease free water was added to attain a total volume of 25 μl. The PCR product were amplified in the thermal cycler (iCycler, BIO-RAD, USA; Mygenie, Bioneer, Germany).
PCR cycling was performed as follows: Initial denaturation at 95 °C for 5 min, followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 72 °C for 15 s and extension at 72 °C for 15 s. Finally, 10-min extension step were performed at 72 °C. The same technique was used for unmethylated reaction and for control DNA samples. PCR products were analyzed by 2–3% agarose gel stained with ethidium bromide and visualized under UV-light.
2.4 Combined bisulfate restriction analysis (COBRA)
Combined bisulfite restriction analysis (COBRA) of the promoter was used to study the methylation quantity of the Bim gene promoter. Primers were designed to amplify regions do not have any CpG islands close to the transcription start sites from bisulfite modified DNA. Ten microliters of PCR product was incubated with 2 U Taq1 restriction enzyme (CGCG) for 4 h at 65 °C before visualization on a 2% metaphore agarose gel. The density of the band was detected by using Quantity One software (Bio-Rad, USA). The Methylation quantity was calculated as a percentage using the equation described previously (Adapted from Xiong and Laird, 1997).
2.5 Bioinformatics analysis of cis-acting element and promoter region of Bim gene
Among Bim promoter region; A 700 bp have been scanned via TFSEARCH website (www.cbrc.jp/research/db/TFSEARCH.html) and many transcriptional factors binding sites have been found. The data from TFSEARCH website indicated the transcriptional factors, their binding sites on the promoter and their position according to the other transcriptional factors. Detection of methylation for the transcriptional factor binding site was accomplished by bisulfate modification followed by using methyl-specific PCR.
2.6 Statistical analysis
Data was analyzed using SPSS version 16 package (SPSS Inc, Chicago, USA) for windows. Chi-square test was used to test statistical differences between groups within the population. Fisher exact test was used for subgroups studied with a small sample size. Kaplan-Meier survival curves were used to assess the survival rates for overall samples. A significant difference is considered to exist with a P value of less than 0.05. All values of results were presented as frequency and percentage.
3 Results
To qualify Bim Promoter methylation in colorectal malignant and normal tissues, methyl-specific PCR was used. Results were visualized using agarose gel and UV light (see Fig. 1 as an example). Among 40 of CRC paraffin embedded tissues, a total of 26 were found methylated with a methylation percentage of 65%. In the control tissues, 2 were found methylated and this represents 20%. The difference between the two groups (CRC and controls) is statistically significant (P < 0.05).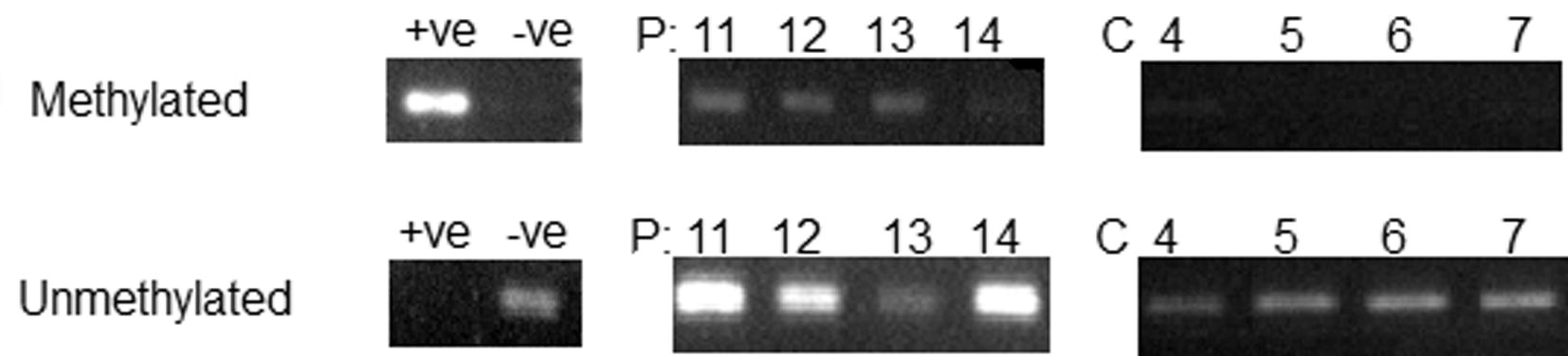
Promoter specific polymerase chain reaction (PS-PCR) of Bim gene-representative image: Examples of PCR products using methylated and unmethylated primers. P indicates patient samples, C indicate control samples. Numbers indicate subjects ID, +ve: positive control, −ve indicates negative control.
Combined bisulfate restriction analysis (COBRA) was used to quantify the methylation level of Bim promoter in the CRC and control groups (see Fig. 2 as an example). The results (Table 2) showed that the average of the methylation percentage was 83% in RCR and was 61% in controls (P < 0.05).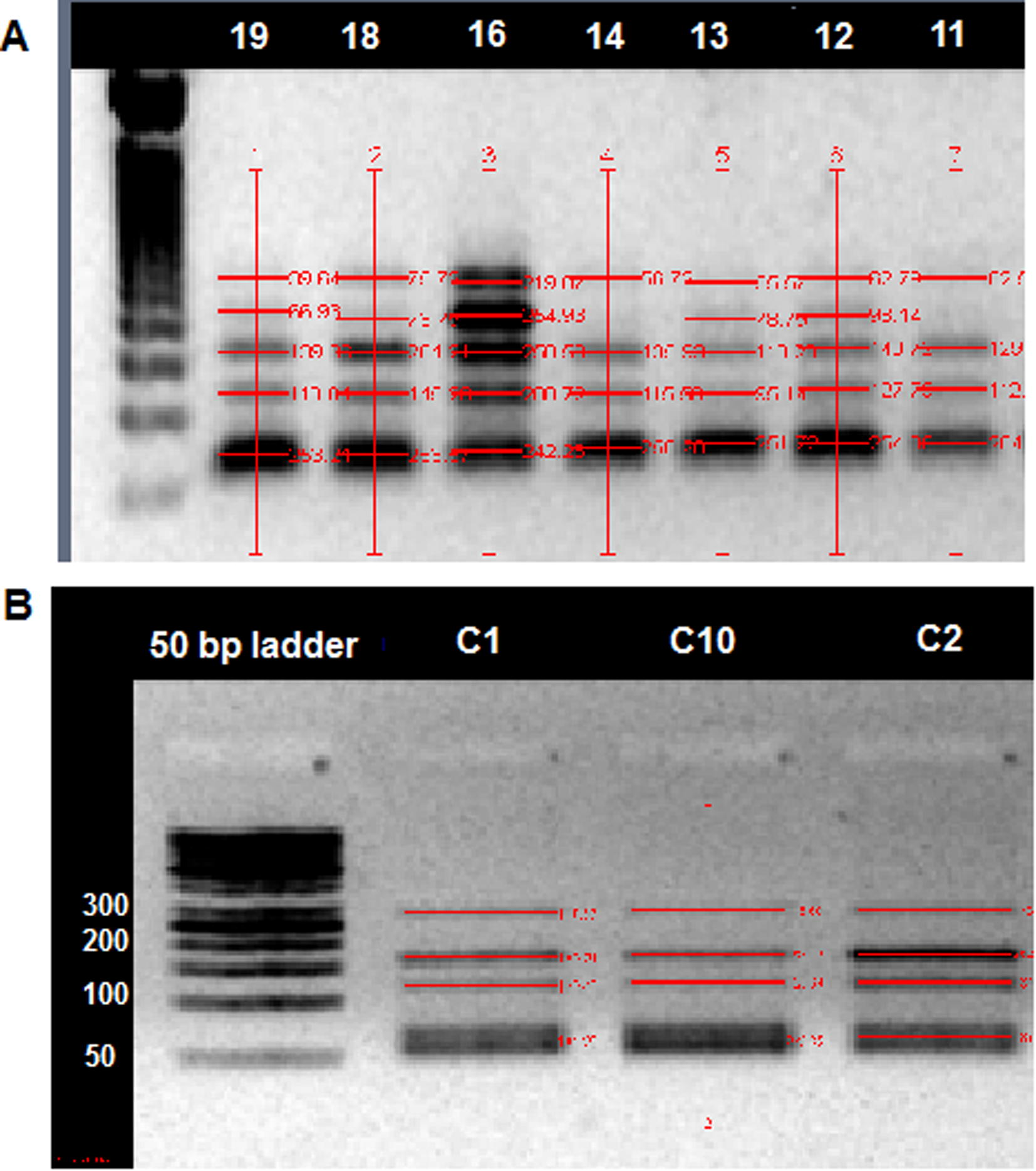
Methylated quantity analysis for Bim gene promoter: Quantitation was performed using combined bisulfate restriction analysis (COBRA) method in CRC patient (A) and controls (B). Numbers indicate subjects ID.
Clinical Characteristic
Category
Overall (N = 40)
%
Unmethylated
Methylated
P value
N
%
N
%
Age
≤50 y
17
43
5
12
12
28
0.944
>50
23
57
7
12
13
17
Gender
Male
15
38
7
47
20
13
0.075
Female
25
62
5
20
8
32
Cancer Location
Colon
31
78
11
35
21
68
0.868
Rectum
9
23
3
33
5
55
T category
T 2
7
18
2
29
5
71
0.686
T3
22
55
9
41
13
59
T4
11
28
3
27
8
73
N category
NO
17
43
6
35
11
65
0.948
N 1
13
33
4
30
9
70
N2
10
25
3
30
7
70
Surgery
Yes
31
78
9
29
22
71
0.804
No
9
23
3
33
6
67
Recurrence
Yes
17
43
3
18
14
82
0.048*
No
23
58
11
48
12
52
Metastasis
Yes
20
50
3
15
17
85
0.008*
No
20
50
11
55
9
45
Chemotherapy
Received
22
55
7
32
15
68
0.641
Not received
18
45
7
39
11
78
Among CRC patients, twenty cases were investigated. As shown in Fig. 3 and using Kaplan-Meier survival curve, no significant association was found between survival period and Bim promoter methylation (P = 0.267).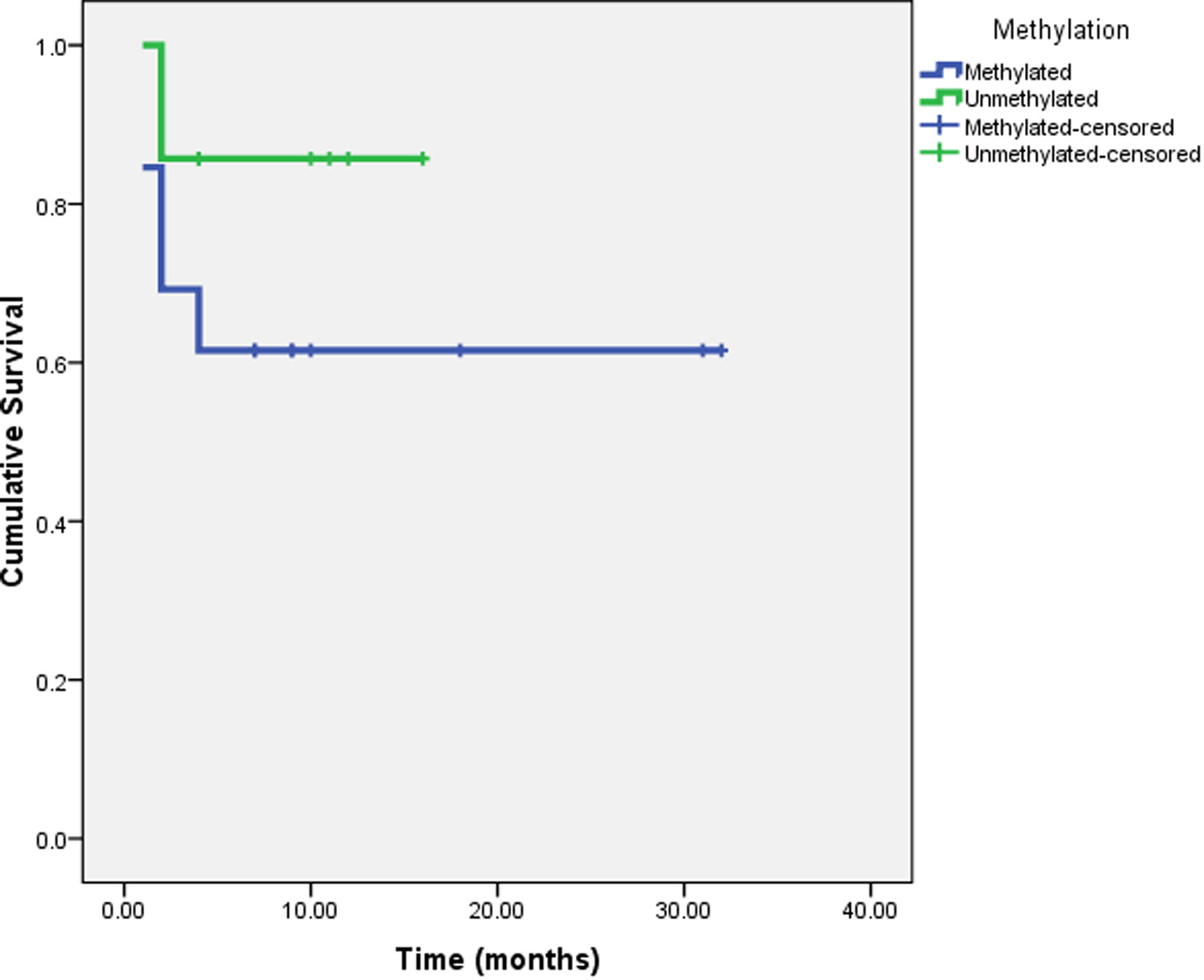
Kaplan-Meier Survival curve of CRC patients vs. Bim gene methylation.
The relationship between Bim promoter methylation and Clinicopathological Characteristics of the patients was studied using Chi-square Test and Fisher “Exact Test (Table 2). Result revealed no significant association between methylation status and patient’s demographics including age, gender. Similarly, no significant association was detected between the gene methylation and disease demographics except for recurrence and metastasis.
With respect to the association between Bim methylation and clinical outcomes, result (Table 3) indicate significant association was detected among CRC patients receiving chemotherapy between Bim gene promoter methylation and disease recurrence and tumor metastasis (P < 0.05). Moreover, a significant association was found between Bim gene promoter methylation and the tumor respond for chemotherapy treatment (P < 0.05).
Category
Chemotherapy
Received
Not Received
M
N(%)U
N(%)M
N(%)U
N(%)
Recurrence
Yes
10(71)
1(17)
5(33)
1(17)
No
4(29)
5(83)
10(67)
5(83)
P value
0.024*
0.445
Metastasis
Yes
13(93)
1(17)
5(39)
2(29)
No
1(7)
5(83)
8(61)
5(71)
P value
0.001*
0.658
Chemotherapy Responds
Respond
4(29)
6(100)
No Respond
10(71)
0(0)
P value
0.005*
Analysis of the promoter region that examined in this study showed that the AP-1 binding site is overlapped with the sequence of methyl-specific PCR reverse primer at the Bim promoter. This indicates the aberrant regulation of AP-1 pathway in the studied samples of RCR.
4 Discussion
Previous studies have shown that the development and progression of CRC are associated with epigenetic mutations affecting the phenotypes of the gene expression rather than changes in DNA sequence (Baylin and Ohm, 2006). For regulation of gene expression pattern, in mammalian cells DNA methylation play a critical role (McCabe et al., 2009). Aberrant methylation is widespread in several gene promoters including hypermethylation of tumor suppressor genes and hypomethylation of oncogens (Daura-Oller et al., 2009). Hypomethylation has a critical role in tumor progress and carcinogenesis. Reports indicated DNA hypomethylation as major reason behind genomic instability and high level of the transposon insertion mutation. Moreover, a whole genome hypomethylation can activate transcription of oncogene, which results in carcinogenesis and tumor development. Furthermore, recent studies suggested that also hypomethylation plays an important role in tumor metastasis and invasion by activating certain genes. Some anticancer agents such as S-Adenosylmethionine might be used as methyl donors in the methylation reaction and used in breast and colon cancer regimens (Guruswamy et al., 2008; Pakneshan et al., 2004).
Results of the present study revealed that Bim gene promoter methylation was significantly higher in the CRC patients group compared to the healthy group indicating that Bim gene promoter methylation might lead to the development of CRC. This finding is an extension of other studies in which hypermethylation of the proapoptotic genes might favor and accelerate disease progression (Yokoyama et al., 2003). In contrast, studies on the epigenetic down-regulation of proapoptotic gene Bax in CRC did not influence disease progression (Alipour et al., 2013).
Furthermore, results in here indicated that CRC cell sensitivity to chemotherapy is inversely associated with Bim gene promoter methylation. Similarly, studies on other cancers including breast cancer, anaplastic large cell lymphoma, B-cell lymphomas and Epstein–Barr virus related tumors, revealed that Bim gene promoter methylation has led to reduced levels of the gene expression and cancer cell response to anticancer remedies (Fernandez et al., 2012; Mestre-Escorihuela et al., 2007; Paschos et al., 2009; Piazza et al., 2013; San Jose-Eneriz et al., 2009). However, this finding was contradicted in another study on CML that concluded that Bim or Bid promoter methylation did not associate with cancer cell response to imatinib (Bozkurt et al., 2013).
Moreover, results indicated, for the first time, a significant association between Bim gene promoter methylation and CRC recurrence and metastasis (P = 0.048, P = 0.008, respectively). This indicates that early detection of Bim gene promoter methylation might be regarded as a bio-indicator for CRC progression. In addition, a significant association was detected among CRC patients receiving chemotherapy between Bim gene promoter methylation and disease recurrence and tumor metastasis indicating non-functionality of Bim as a sensor for chemotherapy-induced apoptosis in CRC patients. Although Bim methylation was found to play a role in CRC metastasis, progression and chemotherapy response, it has no effect on survival rate.
Regarding Bim cis-acting element analysis, previous studies indicated that XFD-3, known as FOXA2 and HFH2, also known as FOXD3 (Cheng et al., 2013), can interfere with epigenetic hypermethylation and has a role in CRC progression and tumorigenesis (Cheng et al., 2013; Hasson et al., 2014; Lam et al., 2013). Furthermore, other reports suggested that Clox, also known as CDP protein could play a role in cell cycle progression, which is one of the main functions of Bim gene (Li et al., 2013). In addition, studies indicated that low expression of CDP gene have a role in chemotherapy resistance in gastric cancer (Li et al., 2013). On the other hand, methylation of the binding site for these transcriptional factors could not be detected due to lack of the appropriate detection methods. In contrast, an association was found for the transcriptional factor AP-1, which encompasses the CG dinucleotide methylation in its binding site. The AP-1 is a transcriptional factor associated with several mitogenic signal pathways (Angel and Karin, 1991). This factor play a role in multiple biological function including proliferation, transformation, differentiation, stress response and apoptosis, depending on surrounding environment and cell type (Angel and Karin, 1991). Two types of mammalian AP-1 have been identified as homodimers and heterodimers, mostly composed of the Jun families (c-Jun, JunB and JunD), and Fos families (c-Fos, FosB, Fra-1 and Fra-2) (Angel and Karin, 1991). However, the role of AP-1 in regulation of cell proliferation is not fully understood. AP-1function in both direction, induction and prevention of apoptosis (Ashida et al., 2005), which suggested that AP-1 involved in regulation of growth to adjust the gene expression that allows cells to adapt to the new environments (Ashida et al., 2005; Shaulian and Karin, 2001).
Recent reports suggested two pathways including K-ras mutation pathway and Wnt pathway that can induce the activation of AP-1 transcriptional factor, thus triggering cell proliferation in CRC (Ashida et al., 2005). In contrast, studies on Acute lymphoblastic leukemia (ALL) demonstrated the activation of AP-1 transcription factor can induce apoptosis via activation of Bim transcription (Chen et al., 2012). Furthermore, previous studies indicated that increasing in methylation level at the AP-1 binding site associated with the decrease of its binding capacity (Zhang et al., 2007).
The present study has shown that AP-1 binding site is overlapped with the sequence of methyl-specific PCR reverse primer at the Bim promoter. However, methylation of CG dinucleotide of this site could block the binding of AP-1 transcription factor. Detection of methylation for this site was accomplished by bisulfate modification followed by using methyl-specific PCR.
5 Conclusions
Together, our data could represent a significant association with the methylation of Bim promoter at the AP-1 binding site and CRC recurrence and metastasis. Furthermore, our data might suggest a correlation with chemotherapy drug resistant and suppression of AP-1 binding via methylation of the CG dinucleotide. In summary, these findings add new insights to understand the role of Bim gene promoter methylation and CRC development, progression, and sensitivity to chemotherapy.
Funding
This study was funded by a grant from Deanship of Research, Jordan University of Science and Technology.
References
- The study of DNA methylation of bax gene promoter in breast and colorectal carcinoma cell lines. Iran. J. Cancer Prev.. 2013;6:59-64.
- [Google Scholar]
- The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta. 1991;1072:129-157.
- [Google Scholar]
- Epigenetic gene silencing in cancer – a mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer. 2006;6:107-116.
- [Google Scholar]
- Aberrant methylation of human L- and M-fructose 1,6-bisphosphatase genes in cancer. Biochem. Biophys. Res. Commun.. 2008;377:720-724.
- [Google Scholar]
- The roles of epigenetic modifications of proapoptotic BID and BIM genes in imatinib-resistant chronic myeloid leukemia cells. Hematology. 2013;18:217-223.
- [Google Scholar]
- Modeling the mechanism of GR/c-Jun/Erg crosstalk in apoptosis of acute lymphoblastic leukemia. Front. Physiol.. 2012;3:410.
- [Google Scholar]
- PINCH-1 regulates the ERK-Bim pathway and contributes to apoptosis resistance in cancer cells. J. Biol. Chem.. 2008;283:2508-2517.
- [Google Scholar]
- Helicobacter pylori causes epigenetic dysregulation of FOXD3 to promote gastric carcinogenesis. Gastroenterology. 2013;144(122–133):e129.
- [Google Scholar]
- Specific gene hypomethylation and cancer: new insights into coding region feature trends. Bioinformation. 2009;3:340-343.
- [Google Scholar]
- Expression and DNA methylation changes in human breast epithelial cells after bisphenol A exposure. Int. J. Oncol.. 2012;41:369-377.
- [Google Scholar]
- Angiogenesis in colorectal cancer: prognostic and therapeutic implications. Am. J. Clin. Oncol.. 2006;29:408-417.
- [Google Scholar]
- S-adenosyl l-methionine inhibits azoxymethane-induced colonic aberrant crypt foci in F344 rats and suppresses human colon cancer Caco-2 cell growth in 3D culture. Int. J. Cancer. 2008;122:25-30.
- [Google Scholar]
- Estrogen receptor alpha or beta loss in the colon of Min/+ mice promotes crypt expansion and impairs TGFbeta and HNF3beta signaling. Carcinogenesis. 2014;35:96-102.
- [Google Scholar]
- Emerging technologies for genome-wide DNA methylation profiling in cancer. Crit. Rev. Oncog.. 2006;12:205-223.
- [Google Scholar]
- Akt1 in osteoblasts and osteoclasts controls bone remodeling. PLoS ONE. 2007;2:e1058.
- [Google Scholar]
- Laird, P.W. 2005. Cancer epigenetics. Hum Mol Genet 14 Spec No 1, R65-76.
- Forkhead box proteins: tuning forks for transcriptional harmony. Nat. Rev. Cancer. 2013;13:482-495.
- [Google Scholar]
- Transcription factor CUTL1 is a negative regulator of drug resistance in gastric cancer. J. Biol. Chem.. 2013;288:4135-4147.
- [Google Scholar]
- How does colorectal cancer present? Symptoms, duration, and clues to location. Am. J. Gastroenterol.. 1999;94:3039-3045.
- [Google Scholar]
- Cancer DNA methylation: molecular mechanisms and clinical implications. Clin. Cancer Res.. 2009;15:3927-3937.
- [Google Scholar]
- Homozygous deletions localize novel tumor suppressor genes in B-cell lymphomas. Blood. 2007;109:271-280.
- [Google Scholar]
- Reversal of the hypomethylation status of urokinase (uPA) promoter blocks breast cancer growth and metastasis. J. Biol. Chem.. 2004;279:31735-31744.
- [Google Scholar]
- Epstein-barr virus latency in B cells leads to epigenetic repression and CpG methylation of the tumour suppressor gene Bim. PLoS Pathog.. 2009;5:e1000492.
- [Google Scholar]
- Epigenetic silencing of the proapoptotic gene BIM in anaplastic large cell lymphoma through an MeCP2/SIN3a deacetylating complex. Neoplasia. 2013;15:511-522.
- [Google Scholar]
- Epigenetic down-regulation of BIM expression is associated with reduced optimal responses to imatinib treatment in chronic myeloid leukaemia. Eur. J. Cancer. 2009;45:1877-1889.
- [Google Scholar]
- Key roles of BIM-driven apoptosis in epithelial tumors and rational chemotherapy. Cancer Cell. 2005;7:227-238.
- [Google Scholar]
- COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res.. 1997;25:2532-2534.
- [Google Scholar]
- Methylation of ASC/TMS1, a proapoptotic gene responsible for activating procaspase-1, in human colorectal cancer. Cancer Lett.. 2003;202:101-108.
- [Google Scholar]
- The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol.. 2008;9:47-59.
- [Google Scholar]
- Maternal cocaine administration causes an epigenetic modification of protein kinase Cepsilon gene expression in fetal rat heart. Mol. Pharmacol.. 2007;71:1319-1328.
- [Google Scholar]
- A review of the role of Puma, Noxa and Bim in the tumorigenesis, therapy and drug resistance of chronic lymphocytic leukemia. Cancer Gene Ther.. 2013;20:1-7.
- [Google Scholar]







