Studies on Oman elite date palm varieties and preliminary establishment of identity through SSR marker
⁎Corresponding author. hemadrisvu2020@gmail.com (Hemadri Reddy)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The Date palm (Phoenix dactylifera L.) is perennial, diploid plant found abundantly in the Middle East region. It plays a significant role in traditional as well socioeconomic growth in the Sultanate of Oman. The main focus of the current work is to identify different methods of DNA extractions for date palm leaf and further to study the genetic diversity among the seven different cultivar varieties, within the Sultanate of Oman, using SSR marker. Seven cultivars were obtained randomly from different places in Oman namely Khasab, Naghal, Barni, Madloki, Khenezi, Nasho and Fard. Genomic DNA was extracted using Edwards’s method and Maxi kit method (QIAGEN). Fingerprint is established by PCR amplification of DNA samples with five sets of SSR primers to identify the diversity between individual date palm from seven different locations. This study will help the researchers as a tool and the markers for the similarity and differences analysis as well as mapping of date palm in Sultanate of Oman. The findings of the study indicated that five SSR primers tested were able to produce amplicons for date palms from different locations. Percentage of similarity between seven date palm cultivars from different locations was found to be having 74 % significance.
Keywords
Dates
DNA isolation
Fingerprinting
Genetic diversity
Molecular markers
1 Introduction
Phoenix dactylifera L. is commonly known as Date palm. It is a fruit bearing monocotyledon, native to the Aceraceae (Coryphoideae) (Barrow, 1998; Barrow, 1999). The plant is found to have a genome size of approximately 658-M base pairs in length (Al-Dous et al., 2011). This plant is specifically desert growing important fruit crop in Oman. Date palm varieties can be differentiated using morphological markers like ‘’shape, size, weight, color, fruit skin texture’’ etc. during the time of harvest (Salem et al., 2008).
When it comes to the method of propagation, date palms are propagated mainly clonally, by their offshoots, however only on a very few occasions, the process of sexual propagation is done with the pollens of male population. Most commonly, each variety is collected from the individual paternal variety is cloned using vegetative multiplication. Which helps in maintaining a proper identity of the grown crops or cultivars, however when there is an intra-cultivar variation, it could lead to a potential risk in cultivar identification process. An important part of the quality assurance is to have the original nature of the plants and this requires specific markers for effectively distinguishing between different varieties of cultivars. Isoenzyme markers and morphological traits are extensively studied to identify as well describe the varieties of date palm in North Africa. During identification of date palm varieties, the important points to consider is nature of needles, character of fruit and the morphology of the leaves. It has however been noticed that one cannot exclusively depend on the morphological characters as they are often variable and not very precise and, they can be influenced by different environmental conditions (Elhoumaizi et al., 2002a,b). Besides this, the biochemistry based approaches comprising isozyme pattern, is used to study the date palms in North African states. However, these markers have limited advantages due to very low polymorphism (Al-Jibouri and Adham, 1990). The differentiation of varieties between related clones by using morphometric descriptors is complex method. Later on DNA based molecular markers, are extensively used, and are found to be highly useful in detecting the genetic diversity. DNA based markers like AFLP, RFLP and RAPD are used to fingerprint date palm genotypes. There are many date palm growing countries are using RAPD techniques to detect its genotypes (Al-Khalifah and Askari, 2003; Soliman et al., 2003; Adawy et al., 2005; Trifi et al., 2000; Sedra My et al., 1998), though the RAPD is also not successful in identifying date palm genotypes. Where as in Egypt and California the AFLP method was used to detect polymorphisms among date palm cultivars (Cao and Chao, 2002; El-Assar et al., 2003; El-Assar et al., 2005). To explore in detailed genomic organization in date palm varieties the latest and more comprehensive molecular markers like SSR or STR are repeating sequences of 2 to 6 nucleotides in noncoding regions will be useful (Hancock, 1997). SSRs are di-, tri- or tetra-nucleotide repeats shown to be abundant, dispersed throughout the genome and move highly polymorphic than other genetic markers in eukaryotic genomes. SSR resources are useful for pedigree analysis, cultivar identification, genetic mapping studies and characterization of germplasm diversity (Billotte et al., 2004). Microsatellites as markers are found to be highly useful tool in plant genome testing since it is codominant, highly polymorphic, gene specific and shown to be reliable. The SSR markers are used to assess the genomic fingerprinting and relationship between varieties (Billotte et al., 2004; Elshibi and Korpelainen, 2008; Elshibli and Korpelainen, 2010). In the current study, 7 date palm cultivars grown in different areas of Oman were taken for the analysis to find out SSR fingerprinting.
2 Materials and methods
2.1 Materials
Date palm leaf from Khasab (Sinaw, Bahla and Wadibanikhaled) Naghal (Sinaw, Samail and Bahla), Barni (Sinaw, Tawa and Alkamel), Khenezi (Bidiya, Tawa and Alkamil), Madloki (Ibra, Tawa and Bidiya), Nasho (Ibra, Tawa and Alkamil) and Fard (Nizwa, Samail and Sinaw), primers, DNA extraction kit, PCR kit, electrophoresis apparatus, PCR, spectrophotometer. The Date palm cultivar varieties used in the current study fruit morphology, location and specifications were specified in Table 2 and the information is from Oman encyclopedia.
3 Method
3.1 Isolation of genomic DNA
Two methods were used to extract genomic DNA from seven different date palm by following the Edwards method (Edwards et al., 1991) and QIAGEN Maxi Kit method (DNEasy Plant Maxi Kit, Thermo Scientific).
3.2 Edwards’s method
DNA extraction was carried out from date palm leaf sample by following Edwards’s method by taking 20 g of plant tissue in eppendorf tubes. The plant tissue is grounded with pellet pestle by adding little sterile sand and 400 μL of extraction buffer (200 mM Tris-HCl pH 7.5, 250 mM NaCl, 25 mM EDTA, 0.5 % SDS). Grind on ice with pellet pestle until completely ground up. The mixture is centrifuged in cooling centrifuge at 4 °C for 2 min. The supernatant was collected to new Eppendorf tube (about 250 μL). One volume of ice-cold isopropyl alcohol was added and incubated in icebath for 30 min. Further Centrifuged at room temperature for 5 min. Remove the upper layer and pellet was dried. Resuspended the lower portion in 100 μL of TE buffer and stored at −20 °C.
3.3 QIAGEN Maxi Kit method
DNA extraction was carried out from date leaf sample using Maxi protocol. From each of the test sample 1 gm of leaf was weighed and grounded in liquid nitrogen using mortar and pestle. Pinch of sterilized sand was added to it. After that, total mixture is transferred in centrifuge tube, 5 ml of AP1 buffer and 10 µL of RNase is added to it. Then, the Eppendorf tubes were placed in the hot water bath for 30 min at 65 °C. After that, the sample was incubated in the ice for 5 min. 1.8 ml of AP2 was added to hold and keep DNA in upper layer. Then, it was transferred to the refrigerator for 5 min. Next, the sample was centrifuged for 5 min and the supernatant was transferred to the filter tube. Then, it was centrifuged for 5 min. Then, the supernatant was transferred to Eppendorf tube. The volume of supernatant was measured and the amount of AP3 was calculated by using this equation (volume of supernatant *1.5) and added to the Eppendorf. The sample was centrifuged for 5 min. Then, the liquid was removed and 12 ml of AW buffer was added to the filter with supernatant. After that, it was centrifuged for 10 min. Then, it was transferred to new filter tube, and it was kept open to allow the ethanol to evaporate at room temperature for 20 min. Next, 1000 μl of AE buffer it was added to the filter tube and then it was centrifuged for 5 min. Finally, the sample was stored in the refrigerator. Quantity of sample was determined by the Nano drop device and the standard of the sample was determined by gel electrophoresis.
3.4 PCR amplification
For PCR amplification the total volume of 20 μL was prepared by mixing: 12.9 μL of H2O, 50 ng of total cellular DNA (2 μL) as template, 2 μL/PCR buffer, 1 μL of 0.2 mM dNTP PCR mix, 0.1 μL (0.625 U) of Taq DNA polymerase and 1 μL of 0.2 mM of each selected primer. The PCR procedure as follows: “Denaturation step was carried out at 95 °C for 5 min followed by 35 cycles of 30 s, 1 min at 48–56 °C (Primer annealing temperature) and 1 min at 72 °C, and a final extension step at 72 °C for 7 min”. Amplified products were separated by AGE (1.2 %) based to their MW and visualized by staining with EtBr as described by (Sambrook et al., 1989). The amplified DNA was visualized using ultraviolet trans illuminator and documented by using a gel doc system. Amplifications were performed for twice and only reproducible products were considered for further data analysis.
3.5 Data analysis
For genome lab fragment analysis, Sample was prepared as following steps: 21.5 μl of sample loading solution, 2 μl standard, 1ul PCR products and drop of mineral oil were mixed together and centrifuged for few seconds. Then, it was placed in the plate and applied to the genome lab fragment analysis. Finally, data was analyzed by using the software GenAlEX to identify the percentage of dissimilarity and the “phylogenetic relationship among the genotypes on the basis of the allele’s size”.
4 Results and discussion
4.1 Genetic diversity analysis
The morphological descriptors are used traditionally to detect the genetic variations in date palm germplasm. However, these descriptors are often not satisfactory and ambiguous due to various environmental factors involved in plant developmental stages (Elhoumaizi et al., 2002a,b). Further, the traditional method of detecting germplasm is also a time taking process. The advanced SSRs and SNPs, are very effective than other methods therefore they become a tool of primary consideration with many advantages with most of the population (Brumfield et al., 2003). In addition, the SNPs and SSR markers are quickly and easily used through bioinformatics tools and can be used in the detection of alleles involved in disease resistance, genomic imprinting, genetic diversity analysis, paternal assessment, forensic studies and influence of population history (Gong et al., 2011; Ashkani et al., 2012; Katti et al., 2001). SSRs are dominant molecular markers, because of their genetic codominance, large volume, present in the whole genome, multiallelic dissimilarity, high robustness and level of polymorphism (Tóth et al., 2000; Schlötterer, 2000). The high level of polymorphism is because of mutations that affects repeating unit’s numbers. Using SSR markers as tool has many advantages than other types like more number of SSR alleles can be identified by a simple PCR based screening at a single locus, minute amount of DNA is enough, and analysis is manageable in automated allele detection (Mortimer et al., 2005). There are evidences strongly recommends that some SSRs in exon sites are also can be used in analysis (Jubrael et al., 2005). In the present analysis, the binary data was prepared using excel work sheet based on SSR banding pattern and was further analysed using geneAlex is used to evaluate variance in coefficient data. The variance between seven samples is identified to be 26 %. Also, it indicates the percentage of variance between Omani cultivars and variance within tested individuals is 74 %. The Date palm varieties tested were showed highly diverse in their Genome sequence. The present study shows that, the similarity of date palm individuals tested ranged between 0.00 and 0.74. The remaining cultivars showed differences in their dissimilarity but still they were grouped with each other's. A variance grid between tested cultivars (Table 1) were found a mean variance range from 0.098 to 0.423. The lowest dissimilarity number was found in between Chair and Nashi cultivars (0.098). Based on this finding it is clear that these two varieties can be close to each other therefore they can be regrouped. The variance 0.423 was obtained between Barny and Far. All the other cultivars displayed low level of variance but still they have been grouped with each other (Table 3).
| Country | Cultivars studied | Location of samples | Coordinates |
|---|---|---|---|
| Sultanate of Oman, Middle East | Khasab | Sinaw (1a) | 22.51 56°N; 58.03 32°E |
| Bahla (2a) | 22.96 80°N;57.29 80°E | ||
| Wadi bani Khaled (3a) | 22.60 20°N;59.08 30°E | ||
| Naghal | Sinaw (1b) | 22.51 56 °N,58.03 32 °E | |
| Samail (2b) | 23.30 00 °N,57.98 33 °E | ||
| Bahla (3b) | 22.96 80 °N,57.29 80 °E | ||
| Barni | Sinaw (1c) | 22.51 56 °N,58.03 32 °E | |
| Tawa (2c) | 22.23 59°N,59.14 55° E | ||
| Al kamil (3c) | 22.13 11°N,59.12 9°E | ||
| Khenezi | Bidiya (1d) | 22.44 00°N,58.80 00°E | |
| Tawa (2d) | 22.23 59°N,59.14 55° E | ||
| Alkamil (3d) | 22.13 11°N,59.12 9°E | ||
| Madloki | Ibra (1e) | 22.68 33°N,58.55 00°E | |
| Tawa (2e) | 22.23 59°N,59.14 55° E | ||
| Nasho | Ibra (1f) | 22.68 33°N,58.55 00°E | |
| Tawa (2f) | 22.23 59°N,59.14 55° E | ||
| Alkamil (3f) | 22.13 11°N,59.12 9°E | ||
| Fard | Nizwa (1g) | 22.93 33°N,57.53 33°E | |
| Samail (2g) | 23.30 00 °N,57.98 33 °E | ||
| Sinaw (3g) | 22.51 56 °N,58.03 32 °E |
| Cultivar | Barni | Naghal | Khasab | Madloki | Khenizi | Fard | Nasho(25) | |
|---|---|---|---|---|---|---|---|---|
| Location | All Oman Region especially in Al Dakhliyah Governorate | All Oman region especially in Al Dakhliyah and Al Dahirah Governorate | One of the basic Palm species in Oman grown in muddy rich region | Anterior and coastal regions | All Oman regions | All Oman regions but good quality present in Wilayat Samael | ||
| Fruit Morphology | Length (cm) | 4 | 4.3 | 3.7 | 3.4 | 3.3 | 3.9 | |
| Weight (g) | 9.8 | 11.2 | 8 | 9.6 | 10.4 | 15.4 | ||
| Diameter (cm) | 1.7 | 2.3 | 2.5 | 2.1 | 2.2 | 2.1 | ||
| Fruit color | Golden to brown | Dark gold | Dark red | Golden | Dark red | Brown | ||
| Time of maturation | Middle of season | Beginning of season | End of season | Middle of season | Middle of season | Middle of season | ||
| Fruit Shape | Cylindrical shape with circular top and convex base | Cylindrical shape with circular top and base | Elliptical shape with circular top and convex base | Small cylindrical shape | Elliptical shape, cylindrical top and convex base | Cylindrical shape with cone top | ||
| Seed Morphology | Length (cm) | 2.7 | 2.7 | 1.9 | 2.2 | 2.3 | 2.2 | |
| Weight (g) | 1 | 0.9 | 0.7 | 0.9 | 0.5 | 0.6 | ||
| Seed color | Brown | Brown | Golden | Brown | Dark brown | Dark yellow to light brown | ||
| % of Fruit Weight to the total weight | 88 % | 88.8 % | 91 % | 90 % | 95 % | 94 % | 95 % | |
| Locus code | Primer sequences (5′–3′) | Repeated motif |
Allelic range (bp) |
Annealing temperature (°C) |
|---|---|---|---|---|
| PDCAT2 | F: GGCCTTCTCTTCCCTAATGGG R: AGTTTCTTGCCCCTGTTCTTTC |
(GA)29 | 166–194 | 53 |
| mPdCIR010 | F: ACCCCGGACGTGAGGTG R: CGTCGATCTCCTCCTTTGTCTC |
(GA)22 | 118–161 | 55.9 |
| PDCAT12 | F: CATCGTTGATTCCTAACCCCTC R:GTTTAGATCTTGCATGGCAACGC |
(CT)19 | 145–167 | 54.3 |
| mPdCIR025 | F: GCACGAGAAGGCTTATAGT R: CCCCTCATTAGGATTCTAC |
(GA)22 | 210–230 | 49.3 |
| mPdCIR050 | F: CTGCCATTTCTTCTGAC R: CACCATGCACAAAAATG |
(GA)21 | 170–210 | 48.5 |
The microsatellite genetic makeup was used to identify the genetic variation and genotypic relationship among 7 different cultivars grown widely in the Sultanate of Oman. Our findings evidences that the Omani date palm associated genomic variation, principal coordinate analysis for seven cultivars of date palm from three different regions results are represented in Fig. 1. As we observe in the Fig. 1 that, the Khasab (KHB) date palm which collected from (Senaw 1a, Bahla 2a and Wadibani Khaled 3a) are close to each other and its morphological and DNA analysis are comparable with each other and seems to be single cultivar. The similar results are seen for Fard (FARD), Khenezi (KHZ) and Madloki (MD) where morphological identification and SSR DNA sequence agree with each other. In addition, Ibra-1e and Tawa-2e of Madloki species are identical to each other. Whereas the analysis for Naghal from Samael-2b is closer to Khasab than Naghal which are from Senaw-1b and Bahla-3b. Also, Nasho (NASH) from Alkamel-3f is closer to the Khenezi. Barni from Senaw-1c is closer to the Khasab than Barni. Based on that five primers which were used in this study, we indicate that Naghal from Samael is maybe not Naghal. Barni from Senaw-1c is seemed to be Khasab and Nasho from Alkamel- 3f is Barni. Sedra et al. (1998) suggested that there might be a general genetic information in the tested varieties but there are chances of variation in their fruit nature and tree structure. There are similar findings in Tunisian date palm cultivars (Zehdi et al., 2004; Wrigley, 1995) have been reported. Studies suggest that the date palm domestication is a long history with the unknown origin (Wrigley, 1995), while the type of cultivation might have played a key role in the genetic composition of cultivars (Elshibi and Korpelainen, 2008). Novel date palm varieties are the results of the selective screening with selective breeding performed by farmers through seed propagation. Interchange of cultivars takes place between farmers which are the mixture of seed and vegetatively propagated date palm varieties. All of these reasons may results mixed genome in date palm in the studied region (Elshibi and Korpelainen, 2008). The present study using SSR analysis revealed the evidence that high level of polymorphism among the tested Omani date palm cultivars. However, classification and nomenclature of date palm in Oman still based on fruit nature like, structural, physical appearance, and chemical composition. In conclusion, Omani cultivated germplasm needs further analysis and application of specific and latest techniques that may help in identifying and characterizing the varieties which are economically and agronomically important (Table 4).
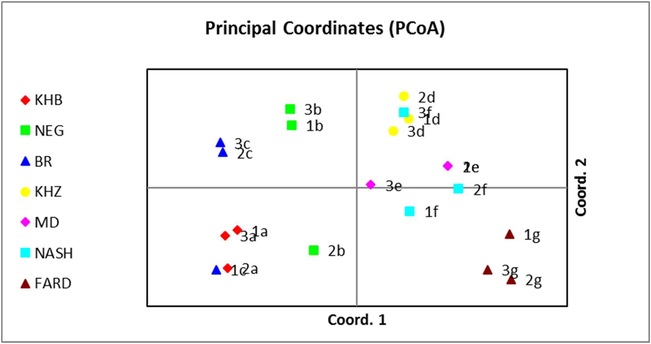
- Principle coordinate analysis among the seven different cultivars between the three different selective locations.
| KHASAB | NEGHAL | BARNI | KHENEZI | MADLOKI | NASHO | FARD | |
|---|---|---|---|---|---|---|---|
| 0.000 | KHASAB | ||||||
| 0.238 | 0.000 | NEGHAL | |||||
| 0.142 | 0.184 | 0.000 | BARNI | ||||
| 0.322 | 0.207 | 0.267 | 0.000 | KHENEZ | |||
| 0.300 | 0.287 | 0.315 | 0.174 | 0.000 | MADLOKI | ||
| 0.251 | 0.175 | 0.266 | 0.098 | 0.141 | 0.000 | NASHO | |
| 0.378 | 0.344 | 0.423 | 0.267 | 0.261 | 0.144 | 0.000 | FARD |
5 Conclusion
The maxi kit method is best of suitable method of DNA extraction from date palm leaf, because this method is safe, provides high yield and purity and less time consuming as compered as Edward method. In this study, SSR markers have been used to determine the genetic diversity among the selected cultivars from different locations in Oman and to analyze the significance of variants among the tested cultivars. Present results provide evidence of a genetic diversity among the studied Omani date palm cultivars and ability of SSR marker to detect the genetic diversity in date palm and the possibility of using these powerful markers as descriptors in the certification and the control of origin labels of date-palm material. The similarity coefficient value of Omani date palm is 0.74. Genetic variance could regroup the Omani date palm in a way that Khenezi and Nasho cultivars were much closed and could be considered as same cultivar. Our present study clearly documents the genomic sequence associated genetic variability in date palm, an important cash crop in Oman. The findings of this research may be useful in future taxonomical characterization to find out the genetic variations based on morphological characters and environmental variations for better planning of identification strategies. However, more exhaustive sampling covering wider geographical areas and more cultivars with additional molecular markers may foster a better understanding on genetic diversity of Phoenix dactylifera L.
Funding
This work was supported by University of Technology and Applied Sciences-Higher College of Technology, Muscat, Sultanate of Oman. This study was also financially supported by the Researchers Supporting Project number (RSP-2021/371), King Saud University, Riyadh, Saudi Arabia.
Acknowledgements
The authors are highly thankful to the management of University of Technology and applied Science-Higher College of Technology, Al Khuwair for providing facilities and also to Directorate General of Agriculture & Livestock Research in AL-Rumaisse, Sultanate of Oman. The authors would also like to acknowledge the funding support by the Researchers Supporting Project number (RSP-2021/371), King Saud University, Riyadh, Saudi Arabia. Sincere thanks and appreciation are also due to all the staff members of the Applied Sciences Department. In addition, we express our sincere gratitude to Dr Syed Najmul Hejaz Azmi for his constructive feedback and suggestions on improving the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Genomic diversity in date palm (Phoenix dactylifera L.) as revealed by AFLPs in comparison to RAPDs and ISSRs. Arab. J. Biotechnol.. 2005;8(1):99-114.
- [Google Scholar]
- De novo genome sequencing and comparative genomics of date palm (Phoenix dactylifera) Nat. Biotechnol.. 2011;29(6):521-527.
- [Google Scholar]
- Biochemical classification of date palm male cultivars. J. Hortic. Sci.. 1990;65(6):725-729.
- [Google Scholar]
- Molecular phylogeny of date palm (Phoenix dactylifera L.) cultivars from Saudi Arabia by DNA fingerprinting. Theor. Appl. Genet.. 2003;107(7):1266-1270.
- [Google Scholar]
- SSRs for marker -assisted selection for blast resistance in rice (Oryza sativa L.) Plant Mol. Biol. Rep.. 2012;30(1):79-86.
- [Google Scholar]
- Systematic studies in Phoenix L. (Palmae: Coryphoideae) In: Henderson A., Borchsenius F., eds. Evolution, variation and classification of palms. New York: The New York Botanical Garden Press; 1999.
- [Google Scholar]
- Nuclear microsatellite markers for the date palm (Phoenix dactylifera L.): characterization, utility across the genus Phoenix and in other palm genera. Mol. Ecol. Notes. 2004;4:256-328.
- [Google Scholar]
- The utility of single nucleotide polymorphisms in inferences of population history. Trends Ecol. Evol.. 2003;18(5):249-256.
- [Google Scholar]
- Identification of date cultivars in California using AFLP markers. Hortic. Sci.. 2002;37:966-998.
- [Google Scholar]
- A simple and rapid method for the preparation of genomic plant DNA for PCR analysis. Nucleic Acids Res.. 1991;19:1349.
- [Google Scholar]
- Genetic analyses of date palms (Phoenix dactylifera L.) from Egypt using fluorescent-AFLP markers. Hortic. Sci.. 2003;38:733-774.
- [Google Scholar]
- Genetic analysis of Egyptian date (Phoenix dactylifera L.) accessions using AFLP markers. Genet Res. Crop Evol.. 2005;52(5):601-607.
- [Google Scholar]
- Phenotypic diversity of date-palm cultivars (Phoenix dactylifera L.) from Morocco. Genet. Resour. Crop Evol.. 2002;49:483-490.
- [Google Scholar]
- Phenotypic diversity of datepalm cultivars (Phoenix dactylifera L.) from Morocco. Genet. Resour. Crop Evol.. 2002;49:483-490.
- [Google Scholar]
- Microsatellite markers reveal high genetic diversity in date palm (Phoenix dactylifera L.) Germplasm from Sudan. Genetica. 2008;134:251-260.
- [Google Scholar]
- Identity of date palm (Phoenix dactylifera L.) germplasm in Sudan: from the morphology and chemical characters to molecular markers. Acta Hort.. 2010;859:143-153.
- [Google Scholar]
- Genetic relationships and evolution in Cucurbita pepo (pumpkin, squash, gourd) as revealed by simple sequence repeat polymorphisms. Theor. Appl. Genet.. 2011;124(5):875-891.
- [Google Scholar]
- Microsatellites and other simple sequences: genomic context and mutational mechanisms. In: Goldstein D.B., SchlÖttern C., eds. Microsatellites: Evolution and Applications. Oxford, UK: Oxford University Press; 1997. p. :1-9.
- [Google Scholar]
- Assessment of AFLP - based genetic relationships among date palm (Phoenix dactylifera L.) varieties of Iraq. J. Am. Soc. Hort. Sci.. 2005;130(3):442-447.
- [Google Scholar]
- Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol. Biol. Evol.. 2001;18:1161-2117.
- [Google Scholar]
- Edwards D Simple sequence re peat (SSR) and GC distribution in the Arabidopsis thaliana genome. J. Plant Biotechnol.. 2005;7:17-25.
- [Google Scholar]
- Morphological variability of Mauritanian date- palm (Phoenix dactylifera L.) cultivars as revealed by vegetative traits. Acta Bot. Croat.. 2008;67(1):81-90.
- [Google Scholar]
- Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 1989.
- Identification and genetic diversity analysis of date palm (Phoenix dactylifera L.) varieties from Morocco using RAPD markers. Euphytica. 1998;103:75-82.
- [Google Scholar]
- Identification and genetic diversity analysis of date palm (Phoenix dactylifera L.) varieties from Morocco using RAPD markers. Euphytica. 1998;103:75-82.
- [Google Scholar]
- Genetic comparisons of Egyptian date palm cultivars (Phoenix dactylifera L.) by RAPD-PCR. Afr. J. Biotechnol.. 2003;2:86-87.
- [Google Scholar]
- Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res.. 2000;10(7):967-981.
- [Google Scholar]
- Phylogenetic relationships in Tunisian date palm (Phoenix dactylifera L.) germplasm collection using DNA amplification fingerprinting. Agronomie. 2000;20(6):665-671.
- [Google Scholar]
- The Evolution of Crop Plants, Date palm (Phoenix dactylifera L.). London, UK: Longman; 1995. p. :399-403.
- Genetic diversity of Tunisian date palms (Phoenix dactylifera L.) revealed by nuclear microsatellite polymorphism. Hereditas. 2004;141:278-287.
- [Google Scholar]







