Translate this page into:
Screening and validation of reference genes using in RT-qPCR for gene expression studies in Paederus fuscipes, a medically and agriculturally important insect
⁎Corresponding authors at: Key Laboratory of Bio-Pesticide Innovation and Application, Guangdong Province, South China Agricultural University, Guangzhou 510642, China. drmusa@scau.edu.cn (Muhammad Musa Khan), baileyqiu@scau.edu.cn (Bao-Li Qiu)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Paederus fuscipes is a medically and agriculturally important insect all over the world. P. fuscipes cause not only a skin condition but is also an aggressive predator in different agro-ecosystems. Prior to performing RT-qPCR under a variety of conditions to investigate the function of target genes in P. fuscipes, it is essential to screen reference genes. However, no P. fuscipes reference gene(s) has yet been discovered. Using the RefFinder software package, which includes ΔCt, geNorm, NormFinder, and BestKeeper, we evaluated the stability of seven housekeeping genes of P. fuscipes under five conditions (developmental stage, diet, temperature, tissue and sex). During the RT-qPCR analysis, geNorm software was used to evaluate pairwise variation in order to identify the most appropriate number of reference genes. The results revealed that for developmental stages RPL-13, EF1A and β-tubulin; for sex-related experiments β-tubulin and Actin, for tissue-related experiments PRS-18, 18S and RPL-13; for temperature-related experiments β-tubulin and PRL-13 and diet-related experiments, β-tubulin and PRS-18 are suitable reference genes. Furthermore, the associated HSP-70 (as the reporter gene) expression patterns differed significantly when the three most stable and three least stable reference genes were selected in different temperature treatments. This is the first time that a complete list of standardized reference genes has been given for P. fuscipes to use in an RT-qPCR study, which could be helpful for potential functional studies of target genes in P. fuscipes.
Keywords
Reference gene
Paederus fuscipes
RefFinder
qRT-PCR analysis
Gene expression
1 Introduction
The clarity, reproducibility, high sensitivity, precision, and cost-effectiveness reverse-transcription quantitative PCR (RT-qPCR) is a well-known technique for gene expression analysis (Schmittgen and Livak, 2008; Wong and Medrano, 2005). Gene expression profiles and the precision of high-throughput transcriptome sequencing (RNA-Seq) data are determined using RT-qPCR (Huggett et al., 2005). Despite the fact that RT-qPCR is a highly accurate tool, a number of experimental factors may cause results that aren't valid gene expression measurements. Integrity and the quantity of RNA and cDNA, pipetting errors and reverse transcription and PCR performance are the factors that must be considered (Bustin et al., 2009). As a result, in RT-qPCR research, reference genes must be utilized to normalize data to remove or at least minimize technical variation across the tested samples and accurately estimate the expression of a specific gene (Kozera and Rapacz, 2013).
Housekeeping genes are supposed to have constant and steady expression under various physiological circumstances and experimental treatments. The genes related to cellular functions are frequently utilized as reference genes in gene expression normalization (Pinheiro and Siegfried, 2020). Numerous investigations have shown that expression of housekeeping gene may not always be consistent and can be profoundly affected by variation in experimental conditions and stress exerted on the organism (Basu et al., 2019; Garciá-Reina et al., 2018; Sagri et al., 2017; Yang et al., 2018; Yu et al., 2018). As a result, selecting appropriate reference genes based on the experimental conditions is critical to obtaining accurate results.
Rice Ragged Stunt Virus (RRSV) is a virus that causes rice ragged stunt disease and belongs to the Oryzavirus genus in the Reoviridae family. In 1976–1977, the virus was first discovered in Indonesia and the Philippines (Hibino et al., 1986; Wu et al., 2010a,b). Nilaparvata lugens is an important sucking pest of rice (Khan et al., 2020b), which acquires the virus by sucking the sap of infected rice plants and transmit it to healthy plants (Huang et al., 2015). Biological control agents play an important role in pest management, an effective and environmentally friendly method. Paederus fuscipes is an important insect predator, belongs to the Staphylinidae family of order Coleoptera (Li and Zhou, 2009). The rove beetle is an outstanding predator because of its high rate of predation on soft-bodied insects different crops. Rove beetles are effective predators of a variety of insect pests, especially brown planthoppers (N. lugens), cotton bollworm (Heliothis armigera), rice moth (Corcyra cephalonica), Spiny bollworm (Earias vittella), Aphid (Aphis gossypii, Aphis glycines), rice leaf folders (Marasmia patnalis), Armyworm (Spodoptera litura), fruitfly and many other dipterous pests (Berglind, 1997; Devi et al., 2003; Khan et al., 2018).
When rove beetles (Paederus fuscipes) are accidentally brushed or crushed on the skin, they release pederin, which causes paederus dermatitis (PD). It tends to affect the exposed body parts, leading to “linear” dermatitis or more serious symptoms. (Rahmah and Norjaiza, 2008; Zagari et al., 2003). With a temporary post-inflammatory hyperchromic patch, the acute vesicular afflictions may take 10–12 days during the healing process. It may cause neuralgia, arthralgia, fever, and vomiting in extreme cases. Erythema will last for several months (Borroni et al., 1991). P. fuscipes invasion is inconvenient since serious cases will result in a loss of person-hours in productivity. Many outbreaks were recorded in various countries, demonstrating the global diversity of P. fuscipes (Khan, 2019).
To determine the best RT-qPCR reference genes, experiments should be carried out systematically to assess the compatibility of various housekeeping genes from various organisms under various laboratory conditions. The level of use of the key reference gene has been reported and aggregated in recent studies (Lü et al., 2018b). In different experiments, 18S ribosomal RNA, a component of ribosomal RNA, was stable in most conditions, and its expression level was used as a template. Actin is a large structural protein that is expressed at different amounts in a variety of cell types and has been extensively used for RT-qPCR research. In previous studies, the primary phosphagen kinase in invertebrates, arginine kinase (AgrK), was also used as a reference gene (Gao et al., 2017; Guo et al., 2020). In insects, elongation factor 1α (EF1A) is used as a normalizer and is active in protein synthesis (Li et al., 2013; Lü et al., 2018b). Many studies used the RPL and RPS families as reference genes since ribosomal protein (RP) is a main component of ribosomes and one of the most conserved proteins in all life forms. Tubulin encodes cytoskeletal function proteins, and several experiments have found that its consistency varies depending on the procedure used on the same animal (Yang et al., 2015).
2 Materials and methods
2.1 Insect rearing
Paederus fuscipes were collected from the field area of South China Agricultural University and were reared under laboratory conditions at the temperature of 27 ± 2 °C with 68 ± 5% relative humidity and 16:8h (L:D) photoperiod. The P. fuscipes adults and larvae were reared on an artificial diet described by Khan et al. (2020). The artificial diet was refreshed every day.
2.2 Sample collection of P. Fuscipes
P. fuscipes reference genes were evaluated regarding growth stage, sexes, adult female tissues, adults fed and starved, and temperature. The first and second instar larvae, pupa, and adult stages of P. fuscipes were sampled. The below are the numbers of samples collected: ten eggs, ten larvae in their first instars, ten larvae in their second instars, five pupae, and five adults (five female and five male P. fuscipes adults). P. fuscipes adults of both sexes were sampled, with five females and five males. Ten female adults were dissected to collect their abdomen, thorax, and head to gather various tissues (Thirty females in total). A total of 60P. fuscipes adults were exposed to 5, 15, 25, or 35 °C for 3 h for the temperature treatments. Adults were taken from the reared population and provided with an artificial diet, while another was starved for 24 h. Each sample contained 15 adult insects. Each sample was placed in a centrifuge tube containing TRIzol reagent (50 µL) for RNA isolation (Invitrogen, Carlsbad, CA, USA). Three replicates were used in each experiment.
2.3 Extraction of total RNA and synthesis of cDNA
TRIzol reagent was used to extract RNA from each sample as per the methodology described by Guo et al. (2020). The NanoDrop One spectrophotometer was used to calculate the concentration of RNAs (Thermo Fisher Scientific, Waltham, MA, USA). In 15–30 L of ddH2O, complete RNA was dissolved. According to the manufacturer's instructions, the PrimeScript RT Kit (Takara, Kyoto, Japan) was used to make first-strand cDNAs from 1 g of RNA from different samples. The cDNAs were then diluted tenfold before being used in the RT-qPCR reactions.
2.4 Primer design and gene cloning
Actin, β-tubulin, RPL13, 18S, RPS18, ArgK, and EF1A were tested as insect reference genes widely used in RT-qPCR studies. Pairs of primers based on recently sequenced transcriptomes for P. fuscipes were designed via Primer Premier 5 software. A 25 µL reaction system contained LA Taq DNA polymerase (Takara, Japan). The conditions used for PCR were previously mentioned by Guo et al. (2020); Pinheiro and Siegfried (2020). To sequence, the PCR product, cloned in the pClone007 Blunt vector (TSINGKE, Beijing, China). Finally, the NCBI repository was used to analyze the sequences of the all-reference genes for better understanding and validation of the function of each gene.
2.5 SYBR Green-based RT-qPCR
To make a total reaction volume of 50 µL containing 5 µL of primer (2.5 µL forward and 2.5 µL reverse), SYBR Green Premix 25 µL (Takara, Japan), diluted cDNA template 2.5 µL, and RNase-free water 17.5 µL. The reaction mixture was divided into three technical replications, each containing about 15 µL stock solution. The CFX96 real-time PCR method was used for all reactions (Bio-Rad, Hercules, CA, USA). An initial denaturation phase of 3 min at 95 °C followed by 40 cycles of 95 °C for 10 s and 55 °C for 30 s in the qPCR program. To perform a dissociation curve study, a dissociation phase period (55 °C for 10 s, then 0.5 °C for 10 s until 95 °C) was introduced. RT-qPCR and linear regression models were used to estimate the slope. Serial dilutions of cDNA were used to create standard curves for each primer pair (5-1, 5-2, 5-3, 5-4, 5-5, and 5-6). After that, the RT-qPCR efficiencies (E) were determined by using E=(10[−1/slope] − 1) × 100.
2.6 Stability of reference gene expression
For each of the six experimental conditions, the data were analyzed separately. RefFinder (https://www.heartcure.com.au/reffinder/) was used to assess the stability of each of the seven reference genes. RefFinder combines four statistical programs: geNorm (Vandesompele et al., 2002), NormFinder (Andersen et al., 2004), BestKeeper (Pfaffl et al., 2004), and the ΔCt method (Silver et al., 2006). In geNorm software, pairwise variance (Vn/Vn + 1 ≤ 0.15) was investigated between the standardization factors NFn and NFn + 1. The n reference genes may be called the optimum number during the RT-qPCR study if Vn/Vn + 1 ≤ 0.15.
2.7 Gene expression levels analysis using various reference genes
Heat shock proteins have also been discovered in insects. HSP70 is one of the most strong predictors of insect heat stress tolerance, according to studies, potentially protecting cells from damage (Morammazi and Shokrollahi, 2020). HSP70 analysis could provide insights on P. fuscipes stress responses and contribute to new ideas for natural enemies (Pan et al., 2018). The HSP70 gene of P. fuscipes is used as a target gene to assess the stability of specified reference genes due to their significance. The primers forward (5′-3′) TTCCTTCCAACTCCTTCTGGT and reverse (5′-3′) GAGGCAGCTGACAAGGAAAC were used for gene expression determination. HSP70 expression was quantified at different temperatures using normalization to the three most stable and three least stable genes.
2.8 Statistical analysis
The expression of the HSP70 gene was determined by using the 2-ΔΔCt methodology (Livak and Schmittgen, 2001). One-way ANOVA analyzed gene expression data with Tukey HSD test at P < 0.05 in SPSS software (SPSS Inc., Chicago, IL, USA).
3 Results
3.1 Amplification and performance of candidate reference genes via PCR in Paederus fuscipes
All amplified and expressed reference genes in P. fuscipes are given in Fig. 1. Melting curve analysis was used to check the PCR specificity of reference genes, which revealed a single crest (Fig. 2). Table 1 shows the Efficiency of PCR primer (E), correlation coefficient (R2), and linear regression equation. In addition, Fig. 3 displays the normal curve for each of the reference genes that were analyzed.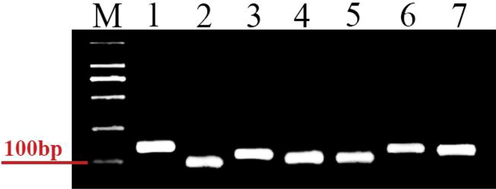
Agarose gel electrophoresis of these seven candidate reference genes. M, Molecular marker. Templates in the polymerase chain reactions (PCRs) were as follows: (1) Actin; (2) ArgK; (3) RPL-13; (4) β-tubulin; (5) 18S; (6) EF1A; and (7) RPS-18.
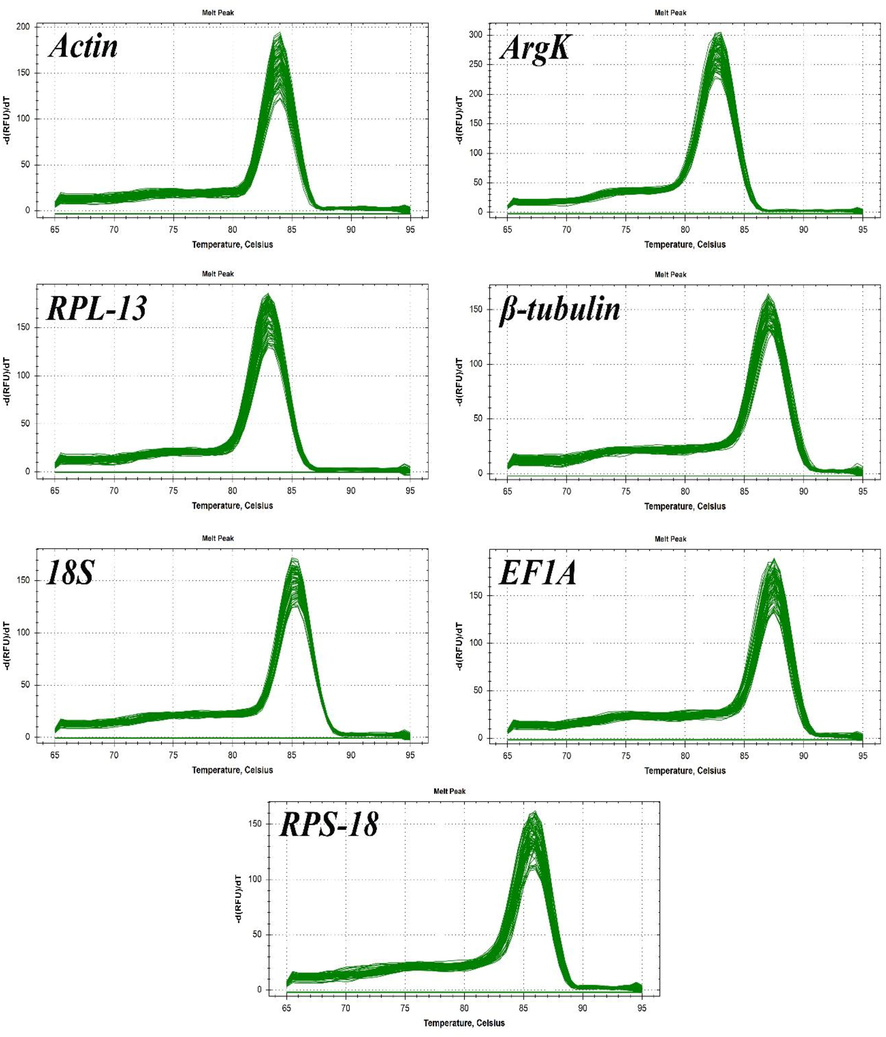
Melting curves of the seven-candidate reference genes in Paederus fuscipes.
Gene
Primer sequence (5′-3′)
Length (bp)
Efficiency (%)
R2
Liner regression
PF-Actin-F
CAGCTCGTTGTAGAAGGTGTGGTG
149
105%
0.9982
Y = -3.2034x + 19.076
PF-Actin-R
TCGGTATGGGACAGAAGGACTCG
PF-ArgK-F
GTGGATGGCTGTCGGTCTTCTTG
90
105%
0.9996
Y = -3.2061x + 20.81
PF-ArgK-R
ATGCTGAGGCTTACACAGTCTTCG
PF-RPL-13-F
GGCTCCAATACAGGCGACTTTCC
118
98%
0.9973
Y = -3.3678x + 20.709
PF-RPL-13-R
TGACTGCATTGGTGTTACGAAGGG
PF-β-tubulin-F
CGTGGAGTTGCCGATGAAGGTG
104
96%
0.9984
Y = -3.4098x + 20.009
PF-β-tubulin-R
GCAGCTACTTCGTCGAGTGGATAC
PF-18S-F
TTGGTCATCTTCCCAGCAACATCG
102
94%
0.9990
Y = -3.4494x + 12.315
PF-18S-R
ACACCGCCCGTCGCTACTAC
PF-EF1A-F
GTAGCCACGACGCAACTCCTTG
128
83%
0.9922
Y = -3.7762x + 18.843
PF-EF1A-R
CCGCCAACCTGACCACTGAAG
PF-RPS-18-F
GTCGGTCGTCGTTACGCCAAC
128
92%
0.9962
Y = -3.521x + 22.461
PF-RPS-18-R
TTGTACTGCCTCGGATTCGTCATG
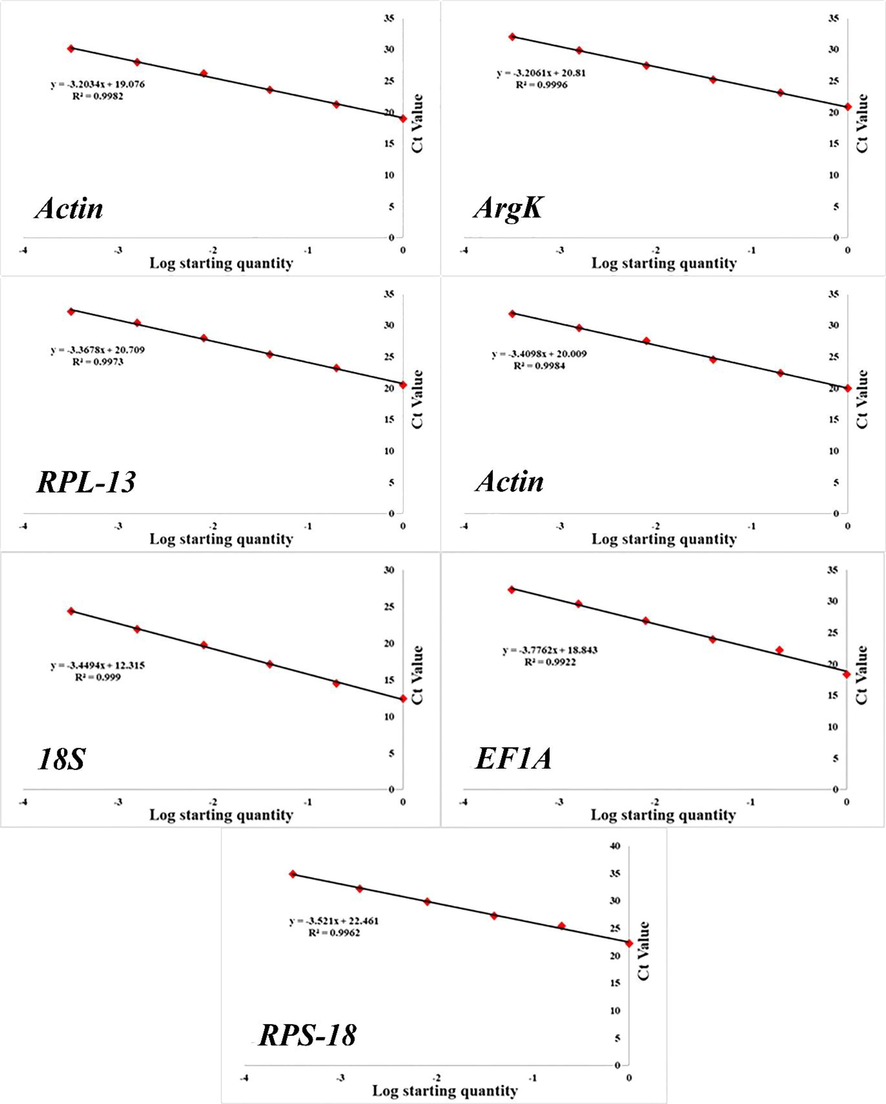
Standard curves of the seven candidate reference genes.
3.2 Reference gene Ct values
The expression of the seven reference genes was estimated using cycle threshold (Ct) values under five different experimental settings. These reference genes had Ct values ranging from 8.87 to 30.90 in different studies (Fig. 4). Furthermore, 18S had the lowest Ct value, making it the most expressed gene in all experimental conditions. Ct values of about 20 were found in most of the remaining six genes, with Actin and ArgK being the least expressed genes among them.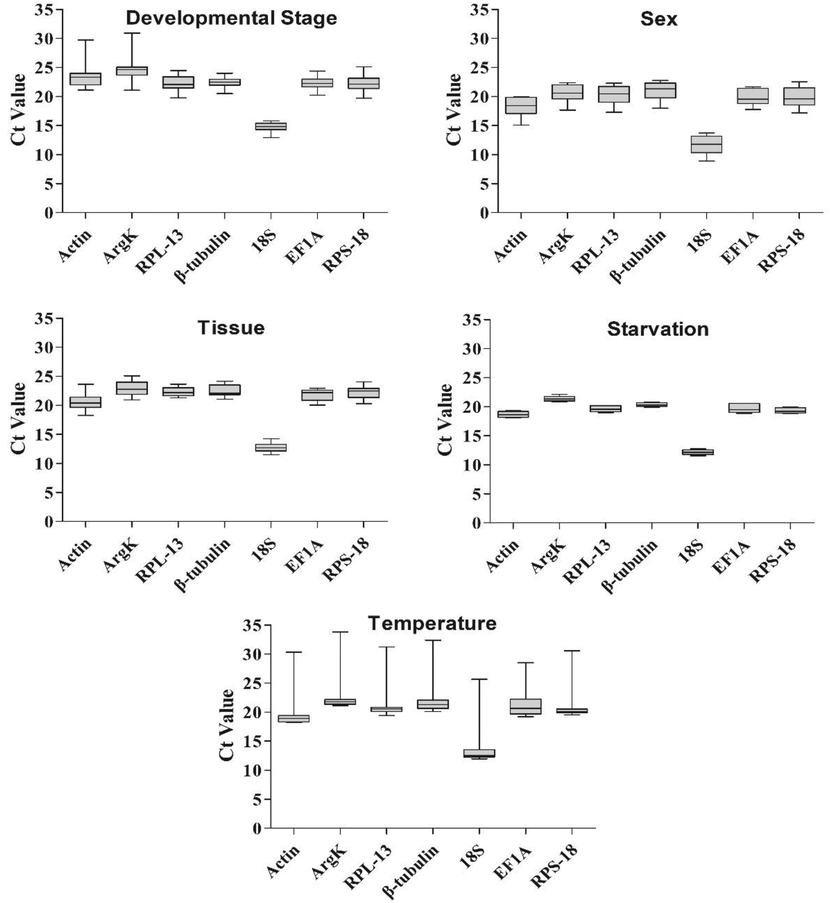
Expression profiles of the seven candidate reference genes in all six experiments for P. fuscipes. The expression levels of the reference genes are shown in terms of the Ct-value for each experimental condition: Developmental stage, sex, tissue, dietary and temperature.
3.3 Stability of the reference genes under specific experimental conditions
The ΔCt methods, BestKeeper, geNorm, and NormFinder, were used to measure the stability of reference genes in all experimental settings. (Table 2).
Conditions
Rank
geNorm
NormFinder
BestKeeper
ΔCt
Gene
Stability
Gene
Stability
Gene
Stability
Gene
Stability
Developmental period
1
RPL-13/EF1A
0.416
β-tubulin
0.551
18S
0.58
RPL-13
1.26
2
RPL-13
0.628
β-tubulin
0.71
EF1A
1.27
3
β-tubulin
0.472
EF1A
0.695
EF1A
0.80
β-tubulin
1.27
4
RPS-18
0.558
RPS-18
0.837
RPL-13
0.92
RPS-18
1.38
5
18S
0.868
18S
1.496
RPS-18
1.12
18S
1.82
6
Actin
1.376
Actin
1.859
Actin
2.01
Actin
2.09
7
ArgK
1.616
ArgK
2.054
ArgK
2.13
ArgK
2.22
Sex
1
Actin/β-tubulin
0.242
β-tubulin
0.275
EF1A
1.11
β-tubulin
0.62
2
RPL-13
0.321
ArgK
1.15
Actin
0.63
3
ArgK
0.277
ArgK
0.322
β-tubulin
1.20
RPL-13
0.65
4
RPL-13
0.366
Actin
0.347
Actin
1.21
ArgK
0.66
5
RPS-18
0.472
RPS-18
0.468
18S
1.29
RPS-18
0.77
6
18S
0.537
18S
0.576
RPL-13
1.34
18S
0.81
7
EF1A
0.796
EF1A
1.390
RPS-18
1.44
EF1A
1.44
Tissue
1
RPL-13/RPS-18
0.503
RPL-13
0.407
18S
0.59
RPL-13
0.83
2
RPS-18
0.444
RPL-13
0.63
RPS-18
0.86
3
EF1A
0.518
EF1A
0.644
RPS-18
0.83
EF1A
0.94
4
18S
0.708
β-tubulin
0.653
β-tubulin
0.84
β-tubulin
1.00
5
β-tubulin
0.806
18S
0.894
EF1A
0.84
18S
1.11
6
Actin
0.943
Actin
0.944
ArgK
0.99
Actin
1.17
7
ArgK
1.016
ArgK
0.988
Actin
1.06
ArgK
1.20
Dietary
1
RPL-13/RPS-18
0.109
β-tubulin
0.157
β-tubulin
0.26
β-tubulin
0.54
2
RPS-18
0.339
ArgK
0.33
PRS-18
0.56
3
β-tubulin
0.232
RPL-13
0.381
18S
0.35
RPL-13
0.58
4
18S
0.412
18S
0.483
RPS-18
0.41
18S
0.69
5
Actin
0.519
Actin
0.506
RPL-13
0.47
Actin
0.70
6
ArgK
0.584
ArgK
0.612
Actin
0.49
ArgK
0.77
7
EF1A
0.68
EF1A
0.817
EF1A
0.60
EF1A
0.92
Temperature
1
RPL-13/RPS-18
0.305
β-tubulin
0.271
RPS-18
0.29
β-tubulin
0.63
2
ArgK
0.345
RPL-13
0.40
ArgK
0.65
3
β-tubulin
0.409
RPL-13
0.405
ArgK
0.47
RPL-13
0.66
4
ArgK
0.425
18S
0.427
18S
0.48
RPS-18
0.69
5
18S
0.512
RPS-18
0.474
β-tubulin
0.61
18S
0.72
6
EF1A
0.662
EF1A
0.850
Actin
0.65
EF1A
0.99
7
Actin
0.767
Actin
0.908
EF1A
0.94
Actin
1.03
ΔCt method. When considering sex, dietary, and temperature conditions, RPL-13 was the most stable reference gene, while for developmental and tissue-related experiments, β-tubulin was the most stable gene.
BestKeeper. When comparing the opposite sexes, the SD values of EF1A, β-tubulin, ArgK, RPL-13, 18S, and RPS-18 were all greater than 1, excluding them from being used as reference genes. Similarly, in development stage comparisons, ArgK, Actin, and RPS-18 could not be used as reference genes, and Actin should not be used to research tissue variations. The most stable genes for dietary and temperature comparison were β-tubulin and RPS-18, while EFIA was the least stable gene for dietary and temperature comparison. However, in comparisons of tissue, sex, developmental stage, and temperature, 18S was the most optimal gene.
NormFinder. Regarding development stage comparisons, sexes, dietary and temperature, β-tubulin was the optimal gene. The expression of RPL-13 was the most consistent across tissues.
geNorm. RPL-13 and RPL-18 were determined to be the best genes (Table 2). When comparing RPL-13 and EF1A, they were the suitable genes for the developmental period experiments, while Actin and β-tubulin had the best stability in the sexes. In addition, geNorm was used to calculate pairwise variance to find the smallest number of reference genes for optimal normalization. Our findings suggest that in sexes, dietary, and temperature comparisons, the V-value of V2/3 was<0.15, indicating that 2 housekeeping genes are sufficient for expression normalization for temperature, dietary and sex-related experiments investigated here (Fig. 5). However, the V-value (V3/4) was smaller than 0.15 in the developmental stage, which implied that three reference genes are enough for normalization, while the V-value (V6/7) was smaller than 0.15 in tissue which implied that six reference genes must be used to optimize the normalization of gene expression (Fig. 5).Fig. 6.Fig. 7.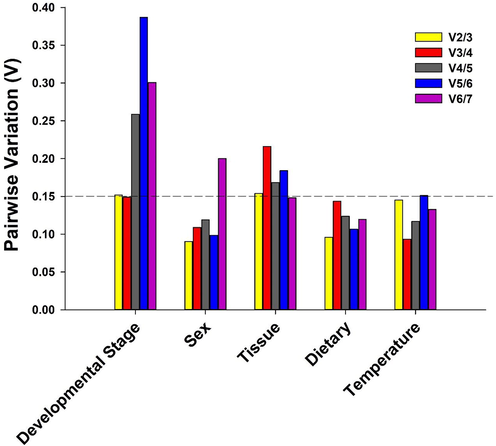
Pairwise variation (V) values using geNorm based on different comparisons: developmental stage, sex, tissue, temperature and dietary.
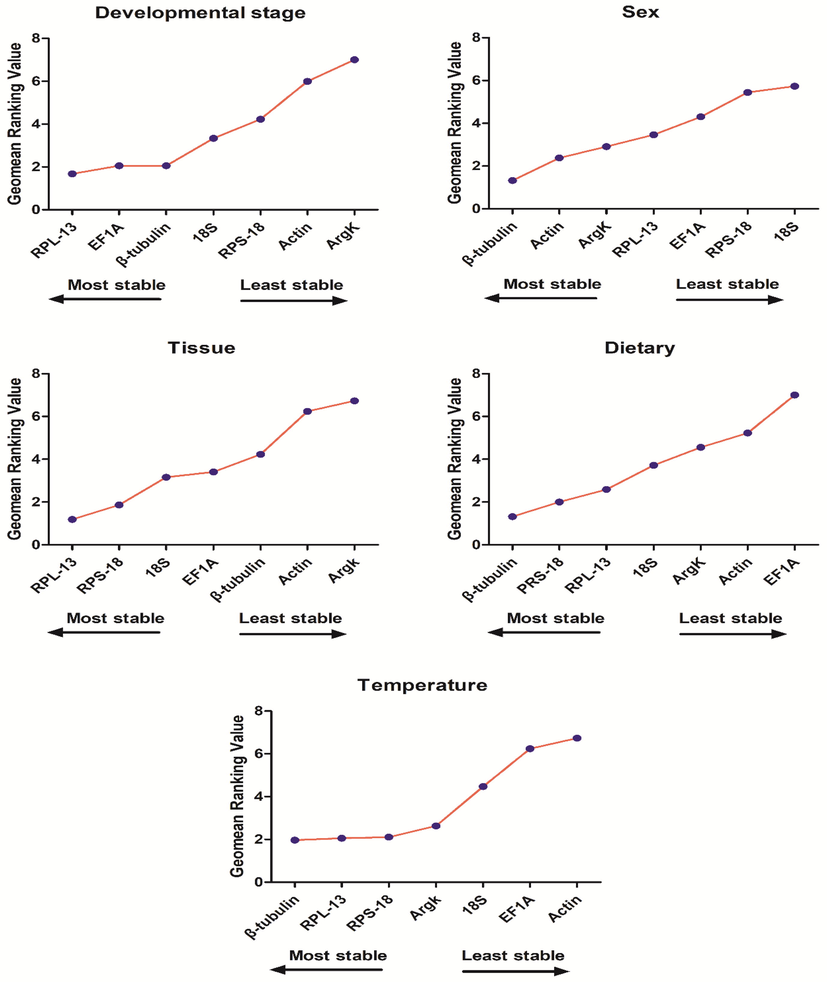
Stability of the seven candidate reference gene expressions in P. fuscipes under different treatment conditions analyzed using RefFinder. A lower Geomean value indicates a more stable expression based on RefFinder.
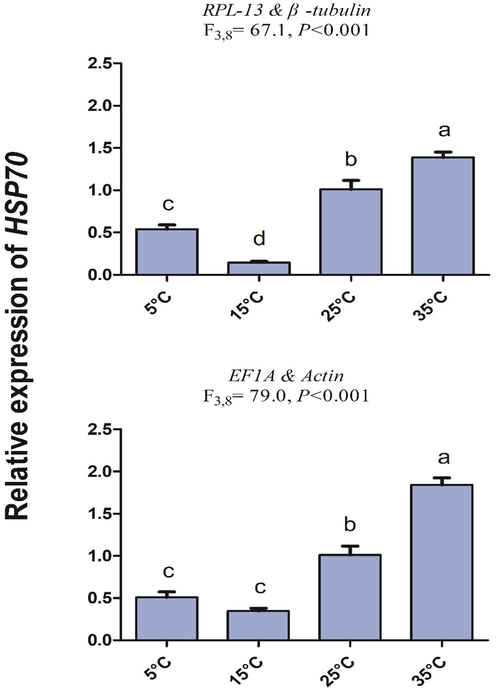
Relative gene expression of HSP70 in different temperatures. The relative abundance of HSP70 in different temperatures were normalized to the most stable (A: RPL-13 and β-tubulin) and least stable (B: EF1A and Actin) reference genes, respectively. The values are means ± SE (Standard error). The significant differences are indicated by different letters, e.g., a, b, c (P < 0.05).
3.4 The overall ranking for reference gene expression stability in RefFinder
The standardized reference genes for the development stage, according to RefFinder, are rated from the most stable to the least stable, were RPL-13, EF1A, β-tubulin, 18S, RPS18, Actin, and ArgK. Similarly, the overall rating for sex was β-tubulin, Actin, ArgK, RPL-13, EF1A, RPS-18 and 18S, while that for different tissues was RPL-13, RPS-18, 18S, EF1A, β-tubulin, Actin and ArgK. For dietary experiments, β-tubulin and RPS-18 were ranked most stable genes, while β-tubulin along with RPL-13 appeared to be most stable under different temperature conditions.
3.5 Validation of reference genes
The HSP70 gene expression was more clearly shown when more stable genes (RPL-13 and β-tubulin) were used than the least stable genes (EF1A and Actin). Results also showed that variation in gene expression due to temperature variation was significant in most stable (F3,8 = 67.1, P < 0.001) and least stable (F3,8 = 79.0, P < 0.001) housekeeping genes were used.
4 Discussion
RT-qPCR is a technique used in molecular science to measure the expression of a target gene under various conditions (Huggett et al., 2005; Lü et al., 2018b). For this purpose, among all the cell types, housekeeping genes are considered the most stable. However, under all conceivable test conditions, there are no stably expressing “universal” reference genes. Furthermore, most gene expression experiments measure gene expression values using just one reference gene; consequently, the data and its interpretation have serious flaws and inaccuracies. Verifying the chosen reference genes under complex experimental treatments is important and indispensable in addressing this problem. The chosen reference genes were selected due to their extensive use in different gene expression studies. Moreover, many studies have already reported these genes as reference genes for their high gene expression stability. Insects from the Hemiptera, Lepidoptera, Coleoptera and Diptera families have been screened for reference genes in recent years (Li et al., 2013; Lü et al., 2018b). However, the reference gene(s) of parasitoid and predator animals have largely been ignored to date (Gao et al., 2017; Lü et al., 2018b).
In previous studies, we explained the biological traits of Paederus fuscipes, including the impact of insecticide on development and biology (Khan et al., 2018) and the behavioral response of P. fuscipes towards different synthetic compounds (Khan et al., 2020a). However, P. fuscipes reference genes are still to be identified, making it impossible to use RT-qPCR to investigate variations in the expression of the genes in this predator. The level of transcription of reference genes differs depending on the experimental conditions, according to our findings, which supports previous findings from a number of studies that no one reference gene will function as a “universal” reference gene (Dai et al., 2017; Lü et al., 2018b, 2018a; Lv et al., 2018; Yang et al., 2014). For instance, in the sex comparison of P. fuscipes, we discovered that ArgK is the second most stable gene; In the sex comparison of Lysiphlebia japonica, it was also discovered to be the most stable reference gene (Gao et al., 2017). Guo et al. (2020) reported the instability of the ArgK gene in sex comparison of Tamarixia radiate.
Previous research has proposed that several reference genes (at least two) should be included in the RT-qPCR study to prevent biased normalization (Bustin et al., 2009). The quantity of reference genes needed to normalize RT-qPCR data varies depending on the elective treatment (Li et al., 2013; Lü et al., 2018a; Yang et al., 2014, 2018). For reliable normalization, no less than five reference genes were recommended for various growth stages of Coleomegilla maculata; three reference genes were needed (Yang et al., 2015). Using too many reference genes, on the other hand, would make the study's execution and activity more difficult; therefore, choosing only two or three accurate reference genes for reliable normalization would be optimal. Our research found that three reference genes were sufficient for development-related experiments, and two reference genes were adequate for sex, dietary, and temperature-related experiments, based on pairwise variations determined by geNorm. According to the study, two reference genes should suffice for target gene normalization (Lü et al., 2018a). As a result of the combined RefFinder rating, the most stable genes for P. fuscipes were β-tubulin, EF1A and RPL-13 for developmental stages, β-tubulin and Actin for sexes, PRS-18, 18S and RPL-13 for tissues, β-tubulin and PRS-18 for dietary experiments, and β-tubulin and PRL-13 for temperature conditions. The requirement of more than two genes for gene expression normalization for developmental and tissue was reported by (Pan et al., 2015b, 2015a).
RPL-13 was the gene that remained the most stable through developmental stages in our research for P. fuscipes. For Tetranychus urticae development studies, RPL-13 was reported as one of the best reference genes. (Morales et al., 2016), RPL-13 was chosen as best in development stages studies for Tetranychus cinnabarinus (Sun et al., 2010). RP-encoding genes were known to be the optimum reference genes in previous studies (Morales et al., 2016); RPL-13 and RPS-18, for example, are ideal reference genes for Henosepilachna vigintioctopunctata when it comes to developmental level, tissue, temperature, and host plant (Lü et al., 2018a). Based on our findings, we believe that RPL-13 was the most stable reference gene for developmental and tissue-related studies. ArgK is unsuitable for use as a reference gene in many studies (Yang et al., 2016, 2015; Yifan et al., 2014). According to our results, ArgK was an optimal reference gene for sex and temperature-related experiments. β-tubulin was the most stable sex, Diet and temperature-related experiments in our study. According to Lü et al. (2018a) and Guo et al. (2020), When insects' temperature conditions are regulated, β-tubulin is frequently used as a reference gene, confirming our findings. Guo et al. (2020) reported that Actin is not a stable gene among any of our six experiment conditions for T. radiata. The current study results confirmed that Actin was the second most stable reference gene for sex-related studied for P. fuscipes.
The relative levels of gene expression of HSP-70 were analyzed at various temperatures to confirm the reference genes in P. fuscipes further. These findings also show that selecting the best reference gene for RT-qPCR analysis is important. Due to the irrational use of reference genes, incorrect target gene expression patterns would affect clear target gene comprehension (Guo et al., 2020).
5 Conclusions
In this research, we tested the stability of seven P. fuscipes housekeeping genes under five different conditions. To the best of our knowledge, this is the first research to define stable RT-qPCR reference genes for P. fuscipes. As a result, these results should aid future studies of target gene expression profiles in P. fuscipes, especially in light of its significance as a medical pest and an effective predator of N. lugens.
Funding
Special fund for the Key Laboratory of Bio-Pesticide Innovation and Application of Guangdong Province (2019–2021). This work was funded by Researchers Supporting Project number (RSP-2021/165), King Saud University, Riyadh, Saudi Arabia.
Acknowledgement
The authors also thank Dr. Andrew G. S. Cuthbertson (York, UK) for his critical comments on manuscript.This work was funded by Researchers Supporting Project number (RSP-2021/165), King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res.. 2004;64(15):5245-5250.
- [CrossRef] [Google Scholar]
- Evaluation of reference genes for real-time quantitative PCR analysis in southern corn rootworm, Diabrotica undecimpunctata howardi (Barber) Sci. Rep.. 2019;9:10703.
- [CrossRef] [Google Scholar]
- Riparian beetles, biodiversity, and stream flow regulation - the examples of Svartån and Mjällån streams, Central Sweden. Entomol. Tidskr.. 1997;118:137-154.
- [Google Scholar]
- Paederus fuscipes dermatitis: A histopathological study. Am. J. Dermatopathol.. 1991;13(5):467-474.
- [CrossRef] [Google Scholar]
- The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem.. 2009;55:611-622.
- [CrossRef] [Google Scholar]
- Selection and validation of reference genes for qRT-PCR analysis during biological invasions: The thermal adaptability of Bemisia tabaci MED. PLoS One. 2017;12(3):e0173821.
- [CrossRef] [Google Scholar]
- Biology of Paederus fuscipes Curtis (Coleoptera: Staphylinidae) Pest Manag Econ Zool. 2003;10:137-147.
- [Google Scholar]
- Comprehensive evaluation of candidate reference genes for gene expression studies in Lysiphlebia japonica (Hymenoptera: Aphidiidae) using RT-qPCR. Gene. 2017;637:211-218.
- [CrossRef] [Google Scholar]
- Validation of reference genes for quantitative real-time PCR in tiger beetles across sexes, body parts, sexual maturity and immune challenge. Sci. Rep.. 2018;8:10743.
- [CrossRef] [Google Scholar]
- Comprehensive Assessment of Candidate Reference Genes for Gene Expression Studies Using RT-qPCR in Tamarixia radiata, a Predominant Parasitoid of Diaphorina citri. Genes (Basel).. 2020;11:1178.
- [CrossRef] [Google Scholar]
- Rice ragged stunt virus-induced apoptosis affects virus transmission from its insect vector, the brown planthopper to the rice plant. Sci. Rep.. 2015;5:1-14.
- [CrossRef] [Google Scholar]
- Real-time RT-PCR normalisation; strategies and considerations. Genes Immun.. 2005;6(4):279-284.
- [CrossRef] [Google Scholar]
- Risk assessment of emamectin benzoate and chlorantraniliprole for the rove beetle Paederus fuscipes, a general predator of the brown planthopper Nilaparavata lugens. Huazhong Agricultural University; 2019.
- Behavioral response of Nilaparvata lugens (Stål), Cyrtorhinus lividipennis Reuter and Paederus fuscipes Curtis to three synthetic volatile chemical compounds. J. Asia. Pac. Entomol.. 2020;23(2):269-276.
- [CrossRef] [Google Scholar]
- Insecticide resistance and detoxification enzymes activity in Nilaparvata lugens Stål against neonicotinoids. J. Agric. Sci.. 2020;12:24.
- [CrossRef] [Google Scholar]
- Lethal and sublethal effects of emamectin benzoate on the rove beetle, Paederus fuscipes, a non-target predator of rice brown planthopper, Nilaparvata lugens. Ecotoxicol. Environ. Saf.. 2018;165:19-24.
- [CrossRef] [Google Scholar]
- Reference Gene Selection for qRT-PCR Analysis in the Sweetpotato Whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae) PLoS One. 2013;8(1):e53006.
- [CrossRef] [Google Scholar]
- A review of Chinese species of the subgenus Paederus s. str. (Coleoptera: Staphylinidae: Paederinae) with description of a new species. Zootaxa. 2009;2083(1):46-64.
- [Google Scholar]
- Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25(4):402-408.
- [CrossRef] [Google Scholar]
- Selection and validation of reference genes for RT-qPCR analysis of the ladybird beetle Henosepilachna vigintioctomaculata. Front. Physiol.. 2018;9:1-11.
- [CrossRef] [Google Scholar]
- Selection of reference genes for the normalization of RT-qPCR data in gene expression studies in insects: A systematic review. Front. Physiol.. 2018;9:1560.
- [CrossRef] [Google Scholar]
- Reference gene selection for RT-qPCR analysis in two invasive whiteflies after the acquisition of vectored or non-vectored viruses. J. Asia. Pac. Entomol.. 2018;21(1):19-24.
- [CrossRef] [Google Scholar]
- Selection of reference genes for expression studies of xenobiotic adaptation in Tetranychus urticae. Int. J. Biol. Sci.. 2016;12(9):1129-1139.
- [CrossRef] [Google Scholar]
- The pattern of HSP70 gene expression, flight activity and temperature in Apis mellifera meda colonies. J. Therm. Biol.. 2020;91:102647.
- [CrossRef] [Google Scholar]
- Pan, H., Yang, X., Bidne, K., Hellmich, R.L., Siegfried, B.D., Zhou, X., 2015a. Selection of reference genes for RT-qPCR analysis in the monarch butterfly, Danaus plexippus (L.), a migrating bio-indicator. PLoS One 10. https://doi.org/10.1371/journal.pone.0129482.
- A comprehensive selection of reference genes for RT-qPCR analysis in a predatory lady beetle, Hippodamia convergens (Coleoptera: Coccinellidae) PLoS One. 2015;10(4):e0125868.
- [CrossRef] [Google Scholar]
- Identification and characterization of heat shock proteins in a parasitic wasp Chouioia cuneae (Hymenoptera: Eulophidae) Entomol. Res.. 2018;48(3):145-155.
- [CrossRef] [Google Scholar]
- Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pair-wise correlations. Biotechnol. Lett.. 2004;26(6):509-515.
- [CrossRef] [Google Scholar]
- Selection of reference genes for normalization of RT-qPCR data in gene expression studies in Anthonomus eugenii Cano (Coleoptera: Curculionidae) Sci. Rep.. 2020;10:5070.
- [CrossRef] [Google Scholar]
- An outbreak of Paederus dermatitis in a primary school, Terengganu. Malaysia. Malays. J. Pathol.. 2008;30:53-56.
- [Google Scholar]
- Housekeeping in Tephritid insects: The best gene choice for expression analyses in the medfly and the olive fly. Sci. Rep.. 2017;7:45634.
- [CrossRef] [Google Scholar]
- Analyzing real-time PCR data by the comparative CT method. Nat. Protoc.. 2008;3(6):1101-1108.
- [CrossRef] [Google Scholar]
- Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol.. 2006;7:1-9.
- [CrossRef] [Google Scholar]
- Suitable reference gene selection for different strains and developmental stages of the carmine spider mite, Tetranychus cinnabarinus, using quantitative real-time PCR. J. Insect Sci.. 2010;10(208):1-12.
- [CrossRef] [Google Scholar]
- Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol.. 2002;3:1-12.
- [CrossRef] [Google Scholar]
- Identification of Pns6, a putative movement protein of RRSV, as a silencing suppressor. Virol. J.. 2010;7:2-7.
- [CrossRef] [Google Scholar]
- Rice ragged stunt virus segment S6-encoded nonstructural protein Pns6 complements cell-to-cell movement of Tobacco mosaic virus-based chimeric virus. Virus Res.. 2010;152(1-2):176-179.
- [CrossRef] [Google Scholar]
- Selection of reference genes for expression analysis using quantitative real-time PCR in the pea aphid, Acyrthosiphon pisum (Harris) (hemiptera, aphidiae) PLoS One. 2014;9(11):e110454.
- [CrossRef] [Google Scholar]
- Selection of reference genes for RT-qPCR analysis in a predatory biological control agent, Coleomegilla maculata (Coleoptera: Coccinellidae) Sci. Rep.. 2015;5:18201.
- [CrossRef] [Google Scholar]
- Selection of reference genes for RT-qPCR analysis in Coccinella septempunctata to assess un-intended effects of RNAi transgenic plants. Front. Plant Sci.. 2016;7:1-10.
- [CrossRef] [Google Scholar]
- Reference gene selection for RT-qPCR analysis in Harmonia axyridis, a global invasive lady beetle. Sci. Rep.. 2018;8:2689.
- [CrossRef] [Google Scholar]
- Identification and validation of reference genes for Quantitative Real-Time PCR in Drosophila suzukii. PLoS One. 2014;9
- [CrossRef] [Google Scholar]
- Reference genes selection for quantitative gene expression studies in tea green leafhoppers. Empoasca onukii Matsuda. PLoS One. 2018;13(10):e0205182.
- [CrossRef] [Google Scholar]
- Paederus dermatitis in northern Iran: a report of 156 cases. Int. J. Dermatol.. 2003;42:608-612.
- [Google Scholar]







