Translate this page into:
Role of Acinetobacter baumannii in decolorization of reactive blue 224 dye and functional analysis of azoreductase gene
⁎Corresponding authors. Syed.Zaghum@uniten.edu.my (Syed Zaghum Abbas), mrafatullah@usm.my (Mohd Rafatullah)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Bioremediation is a sustainable and worthy approach for remediating textile industrial wastewater, which poses severe extortions to human and environmental health due to its toxic, mutagenic, and carcinogenic nature. The present study employed Acinetobacter baumannii 1005 isolated from industrial wastewater to emphasize its degradation potential and to characterize the azoreductase (AZA) gene to assess and optimize the biodegradation of reactive blue 224 (RB-224) textile dye. A. baumannii showed significant decolorization against RB-224, resulting in a change in color. Under optimized conditions (pH 6, 37 °C, and 100 mg/L), A. baumannii exhibited 91 % decolorization of RB-224 being analyzed by UV–vis after 24 h of incubation. UV–vis spectroscopy and FTIR analysis have shown the efficiency of A. baumannii for degrading RB-224. In the UV spectrum, the shifting of the peak from560 nm to 400 nm confirmed the decolorization. In the FTIR spectrum, the shift of several significant peaks related to the functional groups S = O C-N, C–H, C = C, and N–H, corresponding to the structure of RB-224 dye along with the emergence of new peaks indicating the formation of metabolites after degradation. The Azoreductase (AZA) gene of ∼600 bp was amplified and cloned in a pTz57R/T cloning vector. Subsequently, the AZA gene was sequenced and submitted to NCBI for accession number. This study explored the potential of A. baumannii isolate suitable for the bio-remediation of textile dyes.
Keywords
Bioremediation
Reactive Blue 224
Acinetobacter baumannii
Decolorization
Biodegradation
Azoreductase
1 Introduction
Industrialization is the leading cause of environmental pollution and imbalanced ecosystems by discharging contaminated wastewater with various toxic organic and inorganic chemicals (Fazal et al., 2021; Mishra & Maiti, 2019). Aquatic pollution is a leading distress and an imminent threat to ecosystems (Rathi et al., 2021). Domestic sewage and industrial wastewater are the roots of water pollution (Deng et al., 2018). Due to large volumes of poisonous, mutagenic, and carcinogenic effluents, the textile industry was ranked as the number one cause of water pollution (Uddin, 2021). The textile industry employs a vast bulk of synthetic dyes as colorants due to the heterogeneity of color shades, high inherent stability, and economic advantages compared to natural dyes (Mustafa et al., 2021b). Annually, a staggering 7 × 105 of commercial synthetic dyes are synthesized and employed in the dyeing practice worldwide (Hanafi & Sapawe, 2020). Synthetic dyes are mainly categorized as azo, reactive, indigo, aromatic methane, anthraquinone, and triphenylmethane dyes (Varjani et al., 2021). These dyes are characterized by their resistance to acid, alkali, and light, exhibiting mutagenic effects and posing carcinogenic risks to life, including humans (Deng et al., 2018).
The global economy is significantly supported by the textile industry, but this is a substantial consumer of freshwater (∼200 L of water per 1 kg of textile dyeing), consequently generating a substantial volume of dyeing wastewater (Teo et al., 2022). About 10–15 % (10–100 mg/L unfixed dye) of total dyes are released into the environment as a result of inefficiency in the dyeing and printing processes of fabric, becoming a threat to public health and natural ecosystems (Jamee & Siddique, 2019; Khaire et al., 2022; Thangaraj et al., 2022). The discharge of crude textile waste into aquifer resources results in several environmental consequences, including reduced sunlight penetration, gas solubility, pH alteration, and eutrophication, as well as the increase in COD, BOD, and TOC levels in water resources. These effects have a detrimental impact on both flora and fauna (Eslami et al., 2019; Thanavel et al., 2019). The use of textile dye-contaminated water in agriculture also affects terrestrial ecosystems by reducing soil fertility and inhibiting seed germination, plant growth, and plant productivity (Pandey et al., 2020). Furthermore, the employ of dye-polluted water for drinking has been analogous with severe effects on human health, including allergy, cutaneous irritation, dermatitis, and an elevated risk of cancer (Mehra et al., 2021).
The widely used remediation approaches include both physicochemical and biological methods. Physicochemical methods, such as photocatalysis, membrane separations, sonication, coagulation/flocculation, ion exchange, electrochemical oxidation, ozonation, activated carbon adsorption, Fenton processes, and irradiation are employed for eliminating dyes from wastewater (Khan et al., 2023). Nevertheless, these techniques have significant limitations, including their high cost, limited potential, inapplicability to textile dyes, and release of subordinate pollutants (Singh et al., 2017). Bioremediation is a sustainable and eco-friendly technology that harnesses various microbial species, including Acinetobacter, Pseudomonas, Bacillus, Aeromonas, Escherichia, Acinetobacter, Proteus, Schewanella, Citrobacter, Alcaligenes, Desulphovibrio, Sphingomonas, Streptococcus and Klebsiella (Mustafa et al., 2023b) to remediate the textile contaminants by consuming them as a source of carbon and nitrogen (Bekhit et al., 2020; Kapoor et al., 2021). Bacterial biodegradation of dyes involves various oxidoreductases, including azoreductases, laccases, peroxidases, and oxygenases (Harish et al., 2023; Mustafa et al., 2022). Azoreductases are a group of flavoenzymes in the genomes of bacteria (aerobic and anaerobic), yeast, and mammals and can be expressed in vitro (Mishra & Maiti, 2019). Cofactors such as NADH, NADPH, and FADH are required for their enzymatic activity. Azoreductases are classified into FMN-dependent reductases, FMN-independent reductases, NADH-catalyzed reductases, NADPH-dependent reductases, and NADH-DCIP reductases based on the cofactor used to reduce azo linkage of azo dyes (Rathod et al., 2022).
Dye decolorization involves the itemization of azo bonds into harmful colorless amines and the bioconversion of the aromatic amines into the least toxic metabolites (Franca et al., 2020; Shah & Rodriguez-Couto, 2022). Azoreductases reported in Klebsiella pneumoniae, Enterococcus faecalis, Bacillus badius, etc., are capable of remediation of textile dyes (Ikram et al., 2022). Acinetobacter baumannii is an aerobic coccobacillus distinguished by its gram-negative, non-motile, urease-negative, gelatin and negative starch hydrolysis, catalase-positive, coagulase-negative nature (Ning et al., 2014). Due to its clinical and ecological importance, Acinetobacter has been used as a model organism for environmental and industrial studies (Zhao et al., 2023). A. baumannii is capable of degrading r reactive 19, reactive black 5, and reactive red 120 (Ameenudeen et al., 2021).
The current study centers on an eco-friendly bioremediation technology that employs A. baumannii 1005 for the effective biodegradation of RB-224 textile dye. Molecular approaches, including PCR, amplicon sequencing, ligation, and cloning, are employed to identify and amplify the azoreductase gene of A. baumannii, which was cloned in the pTz57R/T cloning vector. This study aims to demonstrate that azoreductase assists in biodegradation by interacting with RB-224. UV–vis analyzed the decolorization and degradation, and Fourier-transformed infrared spectrometry (FTIR). The decolorization process was optimized for dye concentration temperature, pH, and incubation time. The optimization was executed to identify the most effective temperature, incubation period, dye concentration, and pH for potential degradation by A. baumannii.
2 Materials and methods
2.1 Maintenance of bacterial strain
The bacterial strain utilized in this study, Acinetobacter baumannii 1005, was obtained from CMB Lab Dr. Nazir Ahmed School of Biological Science, Government College University Lahore, Pakistan and culture refresh with LB media.
2.2 Textile dye
The RB-224 was obtained from a resident textile industry for the evaluation of degradation. The molecular structure of RB-224 is shown in Fig. 1. A stock solution of RB-224 was prepared for various concentrations 100 mg/L, 200 mg/L, 300 mg/L, 400 mg/L, and 500 mg/L and sterilized by using a membrane filter of 0.22 µm (Pall Life Sciences, USA).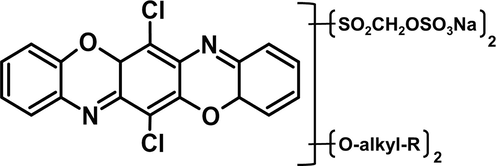
Molecular structure of reactive blue 224, the dye belongs to the Triphenodioxazine Class. Its molecular formula is C26H24Cl2K1N4Na1O20S6.
2.3 Bioremediation potential of A. baumannii 1005
A 1 % of an overnight culture of A. baumannii 1005 to 50 mL of LB broth was inoculated having 100 mg/L of RB-224 and incubated for 24 h at 37 °C in a shaking incubator (120 rpm). The experiment was conducted in triplicate. After 24 h of incubation, the sample (3 mL) was drawn for centrifugation at 10,000 rpm for 5 min and analyzed decolorization % by using a UV–vis spectrophotometer (λmax 600 nm). Here, A1 A2 is the initial and final absorbance, respectively, after the decolorization of the dye (Mustafa et al., 2021b).
2.4 Optimization of the decolorizing potential of a. Baumannii 1005
The dye concentration, pH, and temperature optimized for efficient decolorization of RB-224 by A. baumannii. The temperature (20, 25, 37, 45, 50 °C), pH levels (5–9), yeast extract (0.1 %-0.5 %) and dye concentration (100–500 mg/L). The decolorization percentage was determined on the basis of the absorbance value of RB-224 after 24 h of treatment (Bekhit et al., 2020).
2.5 Removal of textile dyes RB-224
The dye removal kinetics was determined by first-order model (Kurade et al., 2016; Mustafa et al., 2023a; Xiong et al., 2017). Where ‘Co’ and ‘Ct’ is the initial and final dye concentration at time ‘t’ (h), whereas the ‘k’ degradation rate constant (h−1).
2.6 Analysis of metabolites
The cell-free supernatant was used for UV–visible spectra (200–900 nm) of both the control and treated samples (Kilany, 2017; Mustafa et al., 2024). Furthermore, Fourier-transform infrared spectrometry (FTIR, Shimadzu 8400S) analysis was performed in the mid-infrared region (500–4000 cm−1) and facilitated the identification of functional groups of RB-224 and after treatment with A. baumannii (Mustafa et al., 2021b).
2.7 Molecular identification of AZA gene
The gDNA was extracted by phenol–chloroform method, and the AZA gene amplified using the primers AZA-F (5′-GGAGATGCATATGGCTAAAATATTGG-3′) and AZA-R (5′-CAATCAGAGGTACAAGAGTTTCAACT-3′). For amplification the initial denaturation was at 94 °C for 5 min. Subsequently, 35 cycles were carried out, followed by denaturation (94 °C, 50 sec), annealing (56 °C, 1.30 min), elongation (72 °C, 2 min) and ends with final elongation (72 °C,10 min) (Mustafa et al., 2021a). After confirmation of AZA gene amplification, the gene was purified using GeneJet Gel Extraction Kit (Thermo Scientific). The amplicon of the AZA gene was sequenced by First BASE Laboratories Pvt. Ltd., Malaysia. Quality assessment of acquired sequences was conducted using Chromas Lite 2.0 software (https://technelysium.com.au/wp/chromas). BLAST analyses of this sequence (https://blast.ncbi.nlm.nih.gov/Blast.cgi) identified GenBank sequences with maximum identity. The Clustal W program aligned strains with the highest identity to the bacterial isolate and evolutionary relationships were determined using the MEGA 11 program, by neighbor-joining method. A bootstrap analysis with 1,000 pseudo-replicates provided additional robustness to the results (Ali et al., 2022; Singh et al., 2022). A phylogenetic tree of azoreductase of A. baumannii, based on the AZA gene sequence, was constructed using Clustal Omega and the Neighbor-Joining method. Additionally, peptide sequence of azo the molecular structure of azoreductase was determined using SWISS-MODEL (Amera et al., 2020).
3 Results and discussion
3.1 Evaluation of biodegradation potential of Acinetobacter baumannii 1005
A. baumannii 1005 demonstrated a 91 % biodegradation efficiency of RB-224 (100 mg/L) within 24 h. The decolorization of RB-224 was evident by the change of color from blue to light green at the corresponding time intervals (Fig. 2a, b). A proposed mechanism for the biodegradation of RB-224 post-treatment by A. baumannii 1005 indicates the cleavage of C-N and C-O bonds, subsequently leading to the formation of diverse aromatic compounds (Fig. 2c). This was supported by the reduction and disappearance of peaks corresponding to the C-N and C-O bond in the FTIR spectra graph (Fig. 7a, b).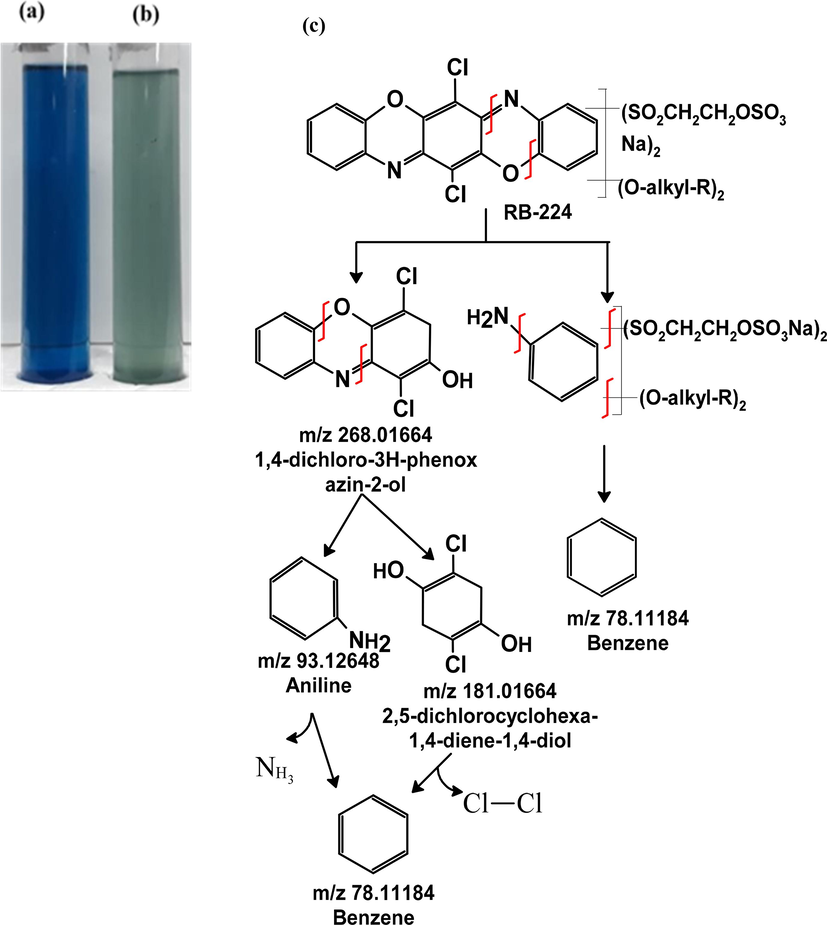
Decolorization of RB-224 by 1 % inoculum of A. baumannii 1005 at 24 h, (a) control RB-224, (b) RB-224 after treatment by A. baumannii 1005, (c) Proposed biodegradation mechanism of RB-224 by A. baumannii 1005.
This bacterial strain achieved sequential decolorization of 52 ± 2.3 % followed by 72 ± 4.9 %, 76 ± 4.2 %, and 91 ± 1.8 % decolorization after 6, 12, 18 and 24 h, respectively (Fig, 3a). Various Acinetobacter species have been documented to participate in the decolorization of colored compounds. A. baumannii isolated from sea sediment, degraded 500 mg/L of reactive red with an efficiency of 96.02 % after 72 h of treatment at 37 °C (Unnikrishnan et al., 2018). A. baumannii YNWH 226, demonstrated a remarkable 99 % decolorization of Congo red under aerobic conditions (Ning et al., 2014). In a parallel investigation, significant decolorization rates were documented, with 98.8 %, 96 %, and 96.2 % decolorization of B-GDN, RP, and RNB respectively, observed after a 48 h treatment with a marine isolate of A. baumannii (Ameenudeen et al., 2021). A. baumannii MN3, demonstrated the efficiency in degrading Congo red and gentian violet at rates of 89 % and 90 %, respectively (Kuppusamy et al., 2017). The A. baumannii VITVB strain exhibited efficient aerobic degradation, achieving decolorization efficiency 90 % and 87 % for RB-221 and RB-5, respectively, at a concentration of 500 mg/L after 48 h (Sreedharan et al., 2021).
3.2 Biodegradation optimization of a. Baumannii 1005
RB-224 showed decolorization rates of 52 %, 72 %, 76 %, and 91 % after 6, 12 16, and 24 h of incubation, respectively (Fig. 3a). The result clearly indicates that the decolorization of RB-224 by A. baumannii 1005 increases with prolonged incubation time. The decolorization of RB-224 increases to 72 % within 12 h, 76 % in 18 h, and ultimately reaches 91 % at 24 h. The extended incubation period led to an increase in bacterial biomass, resulting in enhanced decolorization (Mustafa et al., 2021b). These results are supported by the study that shows that an incubation period of three days is an ideal period for maximum biodegradation of dyes due to rapid bacterial growth and increased bacterial biomass. After this time, bacterial degradation efficacy decreased due to enzyme saturation and decreased growth due to limited nutrients (Ikram et al., 2022).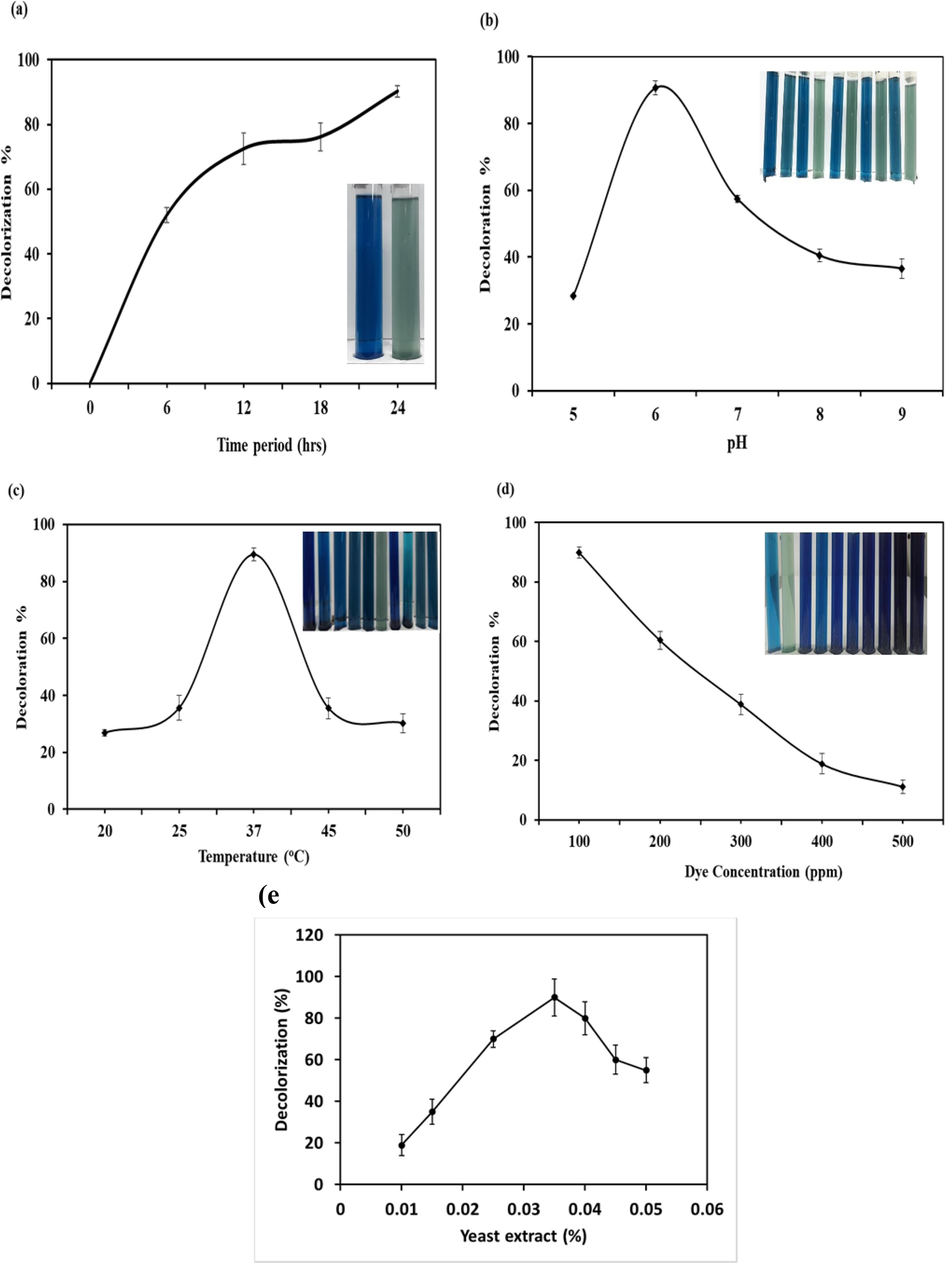
Graph representing optimization of physiochemical parameter for biodegradation: (a) Incubation period (b) pH (c) Temperature (d) dye concentration, and (e) yeast extract (0.01%-0.05%) effects on biodegradation of RB-224 by A. baumannii 1005.
The decolorizing potential of A. baumannii for RB-224 was observed at 28 %, 91 %, 57 %, 40 %, and 37 % at different pH levels 5, 6, 8, 9, and 10, respectively. The A. baumannii exhibited maximum decolorization of RB-224 at pH 6 (Fig. 3b). At pH 5, the decolorization of RB-224 was reduced due to low bacterial growth rate. This underscores the impact of pH on the decolorization process, as it affects the binding of azo dyes with fibers through a substitution mechanism. Effective decolorization of reactive azo dyes was observed at slightly acidic and alkaline pH levels (Lalnunhlimi & Krishnaswamy, 2016). It is worth noting that our findings differ from another study that explained A. baumannii JC359 showed maximum decolorization at pH 7 of reactive azo dyes (Ameenudeen et al., 2021; Amera et al., 2020).
A range of temperatures, 20 °C, 25 °C, 37 °C, 45 °C, and 50 °C, the A. baumannii 1005 showed 27 %, 36 %, 89 %, 35 %, and 30 % decolorization, respectively. The maximum decolorization of 89 % was observed at 37 °C (Fig. 3c). Below 30 °C and above 45 °C, the decolorization rate decreases due to less bacterial growth rate (Mustafa et al., 2021b). The findings align with a prior study on the decolorization of B-GDN, RP, and RNB by A. baumannii JC359 at 37 °C (Ameenudeen et al., 2021). In another study, the maximum degradation of Congo red (100 mg/L) was observed at 37 °C by bacterial consortium. At low temperatures, precisely 30 °C and 20 °C, lower degradation rates of 27 % and 36 % were observed, respectively. In comparison, high temperatures of 45 °C and 50 °C were not suitable for degradation because of enzyme denaturation (Etezad & Sadeghi-Kiakhani, 2021).
At different concentrations 100, 200, 300, 400, and 500 mg/L of RB-224, the decolorization rates were 91 %, 60 %, 39 %, 19 % and 11 %, respectively. The maximum degradation of RB-224 was observed at 100 mg/L, and the decolorization rates decreased as the concentration of RB-224 increased (Fig. 3d). The decolorization efficiency of A. baumannii dropped drastically to 60, 39 %, 19 %, and 11 % with increases in concentration 200, 300, 400, and 500 mg/L RB-224, respectively. These findings are in accordance with Ameenudeen et al. (2021), who described that the decolorization potential of reactive azo dyes reduced by A. baumannii JC359 at higher concentration, the decolorization rate was decreased (Ameenudeen et al., 2021). Another study also suggested that the decolorization potential of bacteria decreases at high dye concentration (Roy et al., 2018). The reduction in the degradation potential of A. baumannii 1005 with increasing dye concentration is attributed to the toxic effects of dye, low bacterial concentration, or reduced enzymatic activity due to the unavailability of active sites. (Singh et al., 2017). The toxicity of dyes spiked as the concentration of dyes increases, that results in low growth rate and decolorization efficiency of bacterial strain. The reduction in the degradation potential of A. baumannii 1005 with increasing dye concentration is attributed to the toxic effects of dye, low bacterial concentration, or reduced enzymatic activity due to the unavailability of active sites (Singh et al., 2017).
The samples were inoculated with different percentages of yeast extract (0.01–0.05 %). The best decolorization rate was obtained with 0.035 % of yeast extract with 100 mg/L concentration of the dye and 24 h of incubation (Fig. 3e). Acinetobacter junii FA10, effectively decolorized the direct and reactive textile azo dyes (150 mg/L) at a 0.03 % solution of yeast extract, was identified and described (Anwar et al., 2014; Imran et al., 2016). The high concentrations of yeast extract decrease the rate of decolorization (Bhatt et al., 2005).
3.3 Molecular characterization of azoreductase (AZA)
The AZA gene in A. baumannii 1005 was successfully amplified (Fig. 4a), and the size of the AZA gene ∼ 600 bp was subsequently confirmed via agarose gel electrophoresis (Fig. 4b). The AZA gene was cloned in E. coli strain DH5α (Fig. 4c). The cloning was confirmed by Miniprep and double digestion with restriction enzymes (Nde1 and EcoR1) (Fig. 4d). The azoreductase gene of similar size was characterized from Bacillus sp., K. pneumonia, and Halomonas sp., for dyes decolorization (Abbas et al., 2020; Tian et al., 2019). Azoreductase gene of A. baumannii 1005 illustrating its genetic relationship with azoreductase gene from other Acinetobacter sp. belonging to the Moraxellacea family (Fig. 4e). AZA gene-based phylogenetic tree showed that A. baumannii 1005 has 100 % homology with azoreductase gene already reported in A. baumannii (CP043180) available at NCBI GenBank, respectively. The three-dimensional structure of AZA protein, resolved via the Swiss Model, reveals that AZA protein consists of ten α helices connected by eight β-pleated sheets and seven random coils in the complete globular structure (Fig. 4f).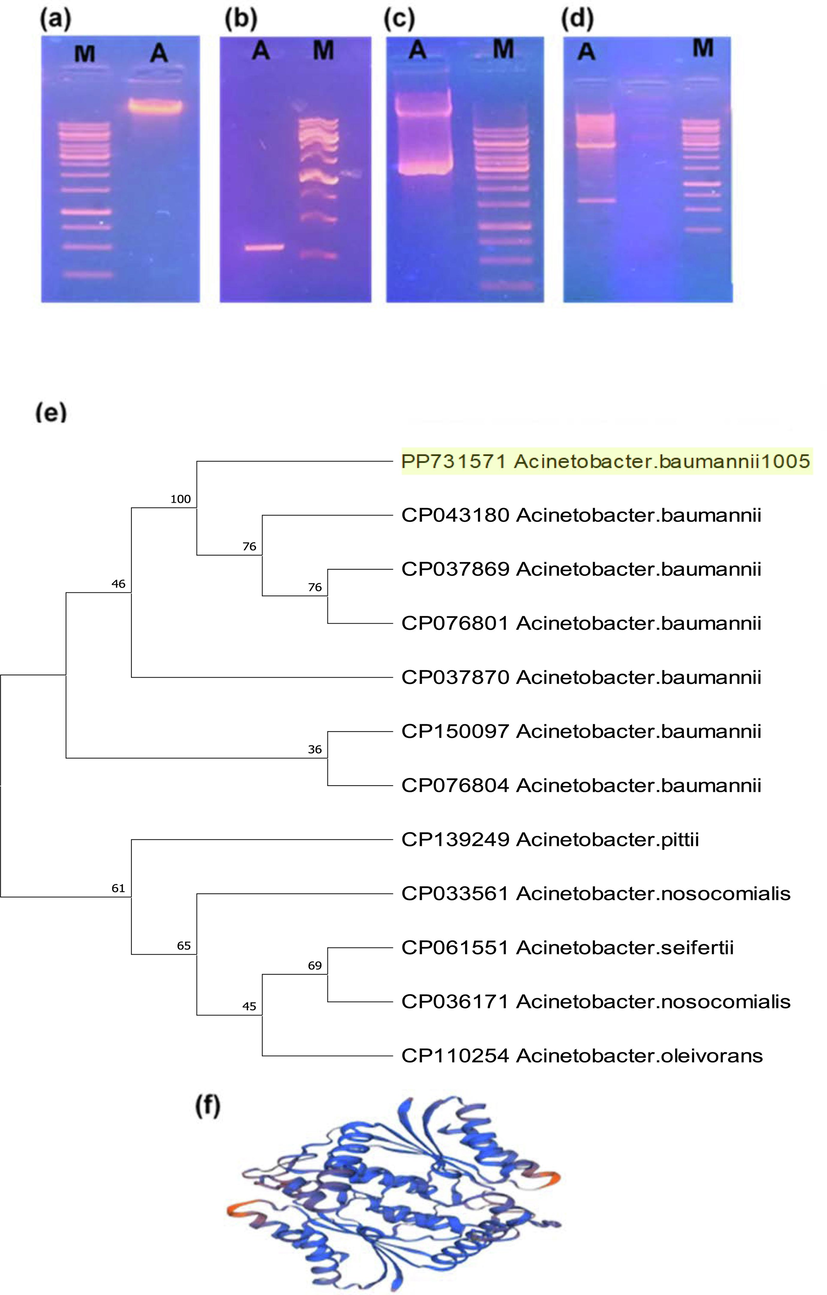
Molecular characterization of Azoreductase AZA gene: (a) Agarose gel electrophoresis of genomic DNA; “M” is the DNA marker, and lane A shows genomic DNA of A. baumannii 1005, (b) Lane A showing amplification of AZA Gene of A. baumannii 1005 and M DNA Marker, (c) Recombinant pTZ57R cloning vector (pTZ57R-AZA). Lane A shows a recombinant pTZ57R cloning vector (pTZ57R-AZA), (d) Double digestion of recombinant pTZ57R cloning vector (pTZ57R- AZA). Lana A showing restricted bands of recombinant pTZ57R cloning vector at 3000 bp and AZA gene band appeared at ∼ 600 bp, (d) AZA gene-based phylogenetic nucleotide tree of azoreductase gene present in A. baumannii 1005, (f) Three-dimensional globular structure of AZA protein of A. baumannii 1005.
3.4 Removal of textile dyes
The dye removal capabilities of A. baumannii 1005 was maximum at 100 mg/L showed 3.7 mg h−1 removal of RB-224 by 24 h of incubation. However, this removal efficacy reduced to 2.375 mg h−1, 1.625 mg h−1, 0.875 mg h−1 and 0.58 mg h−1 of RB-224 at a concentration of 200, 300, 400, and 500 mg/L, respectively, when treated with A. baumannii 1005 (Fig. 5). The decreased removal of dye at high dye concentrations because of low bacterial growth rate (Kurade et al., 2023). The removal constant (k) (0.0516–0.0005 h−1) determined by first-order kinetic model (Table 1). This removal rate decreased because of high dye concentrations, even though, A. baumannii 1005 was less effective at higher concentrations. These results are consistent with a study suggesting that an increase in dye concentration exerts an inhibitory effect on the enzyme azoreductase, a key player in the biodegradation of dyes (Azam et al., 2022). High concentrations of dye inhibit bacterial activity and biodegradation potential, primarily due to intrinsic toxicity blocking the active site of the enzyme (Zhuang et al., 2020). Moreover, the presence of a sulfonic group in reactive blue-224 serves as a potent inhibitor of bacterial growth, as the number of these groups increases with higher concentrations (Velusamy et al., 2022). k — kinetic removal rate constant (d-1). T1/2 — degradation half-life (d). R2 — correlation coefficient.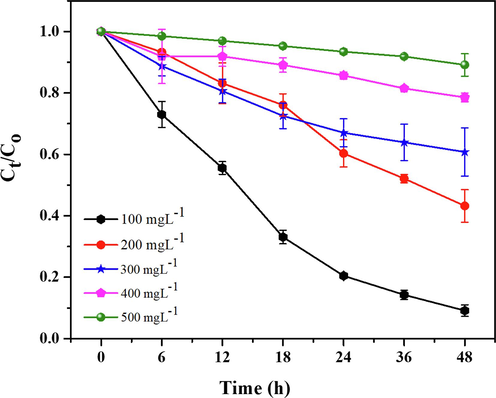
Total removal of textile dyes RB-224 by Acinetobacter baumannii. Error bars represent the standard deviation (SD) of the mean (n = 3).
Dye Concentration (mg L-1)
k
R2
T1/2
Removal (%)
100
0.0516
0.9719
13.4331
91
200
0.0073
0.9735
94.9517
57
300
0.0028
0.868
247.553
39
400
0.0011
0.9385
630.134
21
500
0.0005
0.9879
1386.29
11
3.5 Uv–visible spectrophotometry
The decreased absorbance in UV visible light spectra (200 nm- 800 nm) of RB-224 after treatment with bacterial strain suggested the formation of new metabolites (Fig. 6). The parent dye peaks disappeared, and new peaks appeared, indicating the formation of new metabolites. Similarly, the maximum peaks of B-GDN, RP, and RNB dye in the visible range shifted to the UV range. Following decolorization, there was a shift in the absorbance wavelength in the visible spectra from 560 nm to 400 nm in the experimental culture (Ameenudeen et al., 2021).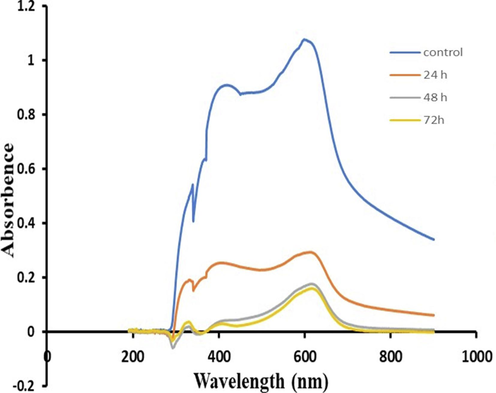
Graph showing UV–vis spectroscopy of RB-224 before and after treatment with 1005.
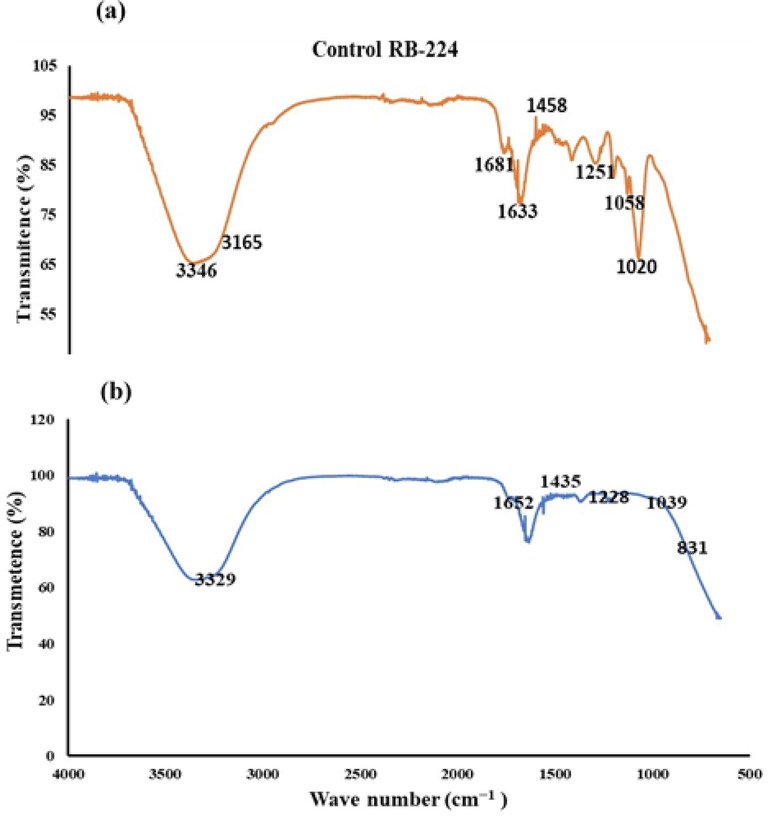
FTIR analysis of RB-224 a) Control RB-224b) after treatment with A. baumannii 1005.
3.6 Fourier transforms infrared spectrometry analysis
The characterizations of the functional groups of RB-224 dye and its metabolites after treatment, using FTIR, provide further evidence of biodegradation. The FTIR spectrum of RB-224 showed peaks at 1020, 1251,1458, 1681, and 3346 cm− 1 representing S = O, C-N, C–H, C = C, and N–H bond, respectively. A peak at 1020 cm− 1 for S = O and 1251 cm− 1 for C-N confirmed the aromatic structure and sulfonic nature of RB- 244 (Fig. 7a). After treatment with A. baumannii, the FTIR spectrum showed disappearance of peak at 1020 cm− 1 and a reduction in intensity at 1251, 1458, 1681, and 3346 cm- 1, respectively, confirming the biodegradation of RB-224 (Fig. 7b). In the FTIR spectrum, the disappearance of the peak at 1020 cm−1 represents S = O, and reduced peaks at 1251, 1458, 1681, and 3346 cm- 1, indicating C-N, C–H, C = C, and N–H, respectively, provide strong evidence of biodegradation of RB-224. Peaks below 900 cm−1 showed non-aromaticity of metabolites formed. The disappearance of peaks at 1060 cm−1 and the reduction in peaks at 631 cm−1 indicate the removal of O-SO3 that is present in RB-224 (Kalpana et al., 2012).
4 Conclusion
Wastewater from textile industries poses a significant risk to the ecosystem due to its toxic and mutagenic nature. Acinetobacter baumannii 1005 has demonstrated effective decolorization activity against RB-224. Under optimal conditions, this bacterium achieved the highest decolorization of 91 % for RB-224 at pH 6, yeast extract 0.035 %, a temperature of 37 °C, and a dye concentration of 100 mg/L. The confirmation of biodegradation was supported by UV–visible spectroscopy and FTIR analysis. Furthermore, the azoreductase gene (AZA) of ∼ 600 bp was successfully isolated from A. baumannii 1005 and subsequently cloned with cloning vector pTZ57R in E. coli DH5α. The phylogenetic nucleotide sequence of the AZA gene of A. baumannii 1005 showed that it has 100 % homology with azoreductase gene in A. baumannii (CP043180). A. baumannii can be used for biodegradation of textile effluents at pilot scale with cheap, eco-friendly and efficient output.
CRediT authorship contribution statement
Faheem Ullah: Writing – original draft. Ghulam Mustafa: Conceptualization, Validation. Muhammad Tariq Zahid: Resources, Supervision. Ihtisham Jamil: Data curation, Formal analysis. Syed Zaghum Abbas: Investigation, Writing – review & editing. Byong-Hun Jeon: Validation, Visualization. Abdulrahman H Alessa: Investigation, Software. Mohd Rafatullah: Formal analysis, Methodology, Supervision.
Acknowledgments
The authors are thankful to Universiti Tenaga Nasional, Malaysia for providing financial support J510050002 - IC-6 BOLDREFRESH2025 - CENTRE OF EXCELLENCE for this study.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Heterologous expression of azoreductase-encoding gene azrS of Bacillus sp. MR-1/2 for enhanced azo dye decolorization and wastewater treatment. Arch. Microbiol.. 2020;202:2135-2145.
- [Google Scholar]
- Identification and characterization of a new Serratia proteamaculans strain that naturally produces significant amount of extracellular laccase. Front. Microbiol.. 2022;13:878360
- [Google Scholar]
- Statistical optimization for the efficacious degradation of reactive azo dyes using Acinetobacter baumannii JC359. J. Environ. Manage.. 2021;279:111512
- [Google Scholar]
- Prioritization of Mur family drug targets against A. baumannii and identification of their homologous proteins through molecular phylogeny, primary sequence, and structural analysis. J. Genet. Eng. Biotechnol.. 2020;18(1):1-22.
- [Google Scholar]
- Characterization of reactive red-120 decolorizing bacterial strain Acinetobacter junii FA10 capable of simultaneous removal of azo dyes and hexavalent chromium. Water Air Soil Pollut.. 2014;225:1-16.
- [Google Scholar]
- Aerobic Biological Units in Dye Removal. In: Biological Approaches in Dye-Containing Wastewater. Vol Volume 1. Springer; 2022. p. :57-94.
- [Google Scholar]
- Decolorization and degradation of the Azo dye by bacterial cells coated with magnetic iron oxide nanoparticles. Environ. Nanotechnol. Monit. Manage.. 2020;14:100376
- [Google Scholar]
- Decolorization of diazo-dye reactive blue 172 by Pseudomonas aeruginosa NBAR12. Journal of Basic Microbiology: an International Journal on Biochemistry, Physiology, Genetics, Morphology, and Ecology of Microorganisms. 2005;45(6):407-418.
- [Google Scholar]
- Decolorization and biodegradation of reactive Red 198 Azo dye by a new Enterococcus faecalis–Klebsiella variicola bacterial consortium isolated from textile wastewater sludge. World J. Microbiol. Biotechnol.. 2019;35:1-10.
- [Google Scholar]
- Decolorization of malachite green dye solution by bacterial biodegradation. Progress in Color, Colorants and Coatings. 2021;14(2):79-87.
- [Google Scholar]
- Integrating bioremediation of textile wastewater with biodiesel production using microalgae (Chlorella vulgaris) Chemosphere. 2021;281:130758
- [Google Scholar]
- Oerskovia paurometabola can efficiently decolorize azo dye Acid Red 14 and remove its recalcitrant metabolite. Ecotoxicol. Environ. Saf.. 2020;191:110007
- [Google Scholar]
- A review on the water problem associate with organic pollutants derived from phenol, methyl orange, and remazol brilliant blue dyes. Mater. Today:. Proc.. 2020;31:A141-A150.
- [Google Scholar]
- Heterogeneous biocatalytic system for effective decolorization of textile dye effluent. 3 Biotech. 2023;13(6):165.
- [Google Scholar]
- Bacillus subtilis: As an efficient bacterial strain for the reclamation of water loaded with textile azo dye, orange II. Int. J. Mol. Sci.. 2022;23(18):10637.
- [Google Scholar]
- Yeast extract promotes decolorization of azo dyes by stimulating azoreductase activity in Shewanella sp. strain IFN4. Ecotoxicol. Environ. Saf.. 2016;124:42-49.
- [Google Scholar]
- Biodegradation of synthetic dyes of textile effluent by microorganisms: an environmentally and economically sustainable approach. European Journal of Microbiology and Immunology. 2019;9(4):114-118.
- [Google Scholar]
- Biodecolorization and biodegradation of reactive Levafix Blue E-RA granulate dye by the white rot fungus Irpex lacteus. J. Environ. Manage.. 2012;111:142-149.
- [Google Scholar]
- Exploration of microbial factories for synthesis of nanoparticles–a sustainable approach for bioremediation of environmental contaminants. Front. Microbiol.. 2021;12:658294
- [Google Scholar]
- The Role of Microbes in Environmental Contaminants’ Management. Environmental Management Technologies: Challenges and Opportunities; 2022.
- Current perspectives, recent advancements, and efficiencies of various dye-containing wastewater treatment technologies. J. Water Process Eng.. 2023;53:103579
- [Google Scholar]
- Isolation, screening and molecular identification of novel bacterial strain removing methylene blue from water solutions. Appl Water Sci. 2017;7(7):4091-4098.
- [Google Scholar]
- Biodecolourization of textile dyes by novel, indigenous Pseudomonas stutzeri MN1 and Acinetobacter baumannii MN3. J. Environ. Chem. Eng.. 2017;5(1):716-724.
- [Google Scholar]
- Insights into microalgae mediated biodegradation of diazinon by Chlorella vulgaris: microalgal tolerance to xenobiotic pollutants and metabolism. Algal Res.. 2016;20:126-134.
- [Google Scholar]
- Integrated phycoremediation and ultrasonic-irradiation treatment (iPUT) for the enhanced removal of pharmaceutical contaminants in wastewater. Chem. Eng. J.. 2023;455:140884
- [Google Scholar]
- Decolorization of azo dyes (Direct Blue 151 and Direct Red 31) by moderately alkaliphilic bacterial consortium. Braz. J. Microbiol.. 2016;47:39-46.
- [Google Scholar]
- Adverse impact of textile dyes on the aquatic environment as well as on human beings. Toxicol. Int. 2021;28(2):165.
- [Google Scholar]
- Optimization of process parameters to enhance the bio-decolorization of Reactive Red 21 by Pseudomonas aeruginosa 23N1. Int. J. Environ. Sci. Technol.. 2019;16:6685-6698.
- [Google Scholar]
- Mustafa, G., Tariq Zahid, M., Ali, S., Zaghum Abbas, S., Rafatullah, M. 2021a. Biodegradation and discoloration of disperse blue-284 textile dye by Klebsiella pneumoniae GM-04 bacterial isolate. J King Saud University Sci. 2021; 33 (4): 101442.
- Mustafa, G., Zahid, M.T., Ullah, F., Zia, I., Younas, A., Batool, T., Zahid, I. 2023b. Bacterial tools for the removal and degradation of synthetic dyes from the wastewater. in: Current Developments in Bioengineering and Biotechnology, Elsevier, pp. 339-370.
- Biodegradation and discoloration of disperse blue-284 textile dye by Klebsiella pneumoniae GM-04 bacterial isolate. Journal of King Saud University-Science. 2021;33(4):101442
- [Google Scholar]
- Bacterial Extracellular Polymeric Substances for Degradation of Textile Dyes. In: Polymer Technology in Dye-Containing Wastewater: Volume 2. 2022. p. :175-191.
- [Google Scholar]
- Microalgal and activated sludge processing for biodegradation of textile dyes. Environ. Pollut.. 2024;123902
- [Google Scholar]
- Decolorization and biodegradation of the azo dye Congo red by an isolated Acinetobacter baumannii YNWH 226. Biotechnol. Bioprocess Eng.. 2014;19:687-695.
- [Google Scholar]
- A study on the utility of immobilized cells of indigenous bacteria for biodegradation of reactive azo dyes. Prep. Biochem. Biotech.. 2020;50(4):317-329.
- [Google Scholar]
- A review on effective removal of emerging contaminants from aquatic systems: Current trends and scope for further research. J. Hazard. Mater.. 2021;409:124413
- [Google Scholar]
- Homologous overexpression of azoreductase (azoA) in Enterococcus sp. L2 moderated growth and azo dye decolorization while gaining an oxidative and heavy metal stress resistance: A trade-off. Environ. Technol. Innov.. 2022;27:102531
- [Google Scholar]
- Assessment on the decolourization of textile dye (Reactive Yellow) using Pseudomonas sp. immobilized on fly ash: Response surface methodology optimization and toxicity evaluation. J. Environ. Manage.. 2018;223:185-195.
- [Google Scholar]
- Development in Wastewater Treatment Research and Processes: Microbial Degradation of Xenobiotics Through Bacterial and Fungal Approach. Elsevier; 2022.
- Biodegradation of Reactive Yellow-145 azo dye using bacterial consortium: A deterministic analysis based on degradable Metabolite, phytotoxicity and genotoxicity study. Chemosphere. 2022;300:134504
- [Google Scholar]
- Role of azoreductases in bacterial decolorization of azo dyes. Toxicol Int. 2017;21(2):160-166.
- [Google Scholar]
- Dye degradation potential of Acinetobacter baumannii strain VITVB against commercial azo dyes. Biorem. J.. 2021;25(4):347-368.
- [Google Scholar]
- Sustainable toxic dyes removal with advanced materials for clean water production: A comprehensive review. J. Clean. Prod.. 2022;332:130039
- [Google Scholar]
- Combined biological and advanced oxidation process for decolorization of textile dyes. SN Applied Sciences. 2019;1:1-16.
- [Google Scholar]
- Biodegradation of Reactive Red 198 by textile effluent adapted microbial strains. Arch. Microbiol.. 2022;204:1-13.
- [Google Scholar]
- Isolation, cloning and characterization of an azoreductase and the effect of salinity on its expression in a halophilic bacterium. Int. J. Biol. Macromol.. 2019;123:1062-1069.
- [Google Scholar]
- Environmental hazard in textile dyeing wastewater from local textile industry. Cellul.. 2021;28(17):10715-10739.
- [Google Scholar]
- Dye-tolerant marine Acinetobacter baumannii-mediated biodegradation of reactive red. Water Sci. Eng.. 2018;11(4):265-275.
- [Google Scholar]
- Trends in dye industry effluent treatment and recovery of value added products. J. Water Process Eng.. 2021;39:101734
- [Google Scholar]
- Transformation of aqueous methyl orange to green metabolites using bacterial strains isolated from textile industry effluent. Environ. Technol. Innov.. 2022;25:102126
- [Google Scholar]
- Ciprofloxacin toxicity and its co-metabolic removal by a freshwater microalga Chlamydomonas mexicana. J. Hazard. Mater.. 2017;323:212-219.
- [Google Scholar]
- A comprehensive genomic analysis provides insights on the high environmental adaptability of Acinetobacter strains. Front. Microbiol.. 2023;14:1177951.
- [Google Scholar]
- Azo dye degrading bacteria tolerant to extreme conditions inhabit nearshore ecosystems: optimization and degradation pathways. J. Environ. Manage.. 2020;261:110222
- [Google Scholar]







