Translate this page into:
RNA editing in chloroplast NADH dehydrogenase (ndhA) of salt stressed wild barley revealed novel type G to A
⁎Corresponding author at: Department of Biological Sciences, Faculty of Science, King Abdulaziz University (KAU), P.O. Box 80141, Jeddah 21589, Saudi Arabia. ahmedramadan782@yahoo.com (Ahmed M. Ramadan), aamara@kau.edu.sa (Ahmed M. Ramadan),
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University. Production and hosting by Elsevier.
Abstract
The ndhA gene of NAD(P)H dehydrogenase in chloroplast DNA is vital for the electron transport chain and needs to be studied, especially in the context of RNA editing and its effects. Wild barley (Hordeum vulgare subsp. spontaneum) plants were studied in 4 groups, with exposure to 500 mM NaCl for 0 h – control (accession no. OM262848), and three treatment groups: 2 h (accession no. OM262849), 12 h (accession no. OM262850) and 24 h (accession no. OM262851) and their RNA was sent for RNA-seq sequencing. The sequences were submitted to NCBI SRA archives and analyzed together after filtering for high quality. This was done to show the RNA editing position and percentages at 10 sites across the ndhA gene. The RNA edits were validated using quantitative-PCR and homology models for the ndhA protein after exposure to salt stress were also generated. The four treatments showed five C to U changes (C50, C303, C563, C1042, C1047), three U to C changes (U550, U592, U1066), and two G to A changes (G111, G1046). We investigated the extent and effect of RNA editing in ndhA transcripts from chloroplasts of barley. We observed high RNA editing percentages after the second hour of salt stress.We have also observed for the first time about ‘U to C’ RNA edits in ndhA gene of wild barley.
Keywords
Chloroplast ndhA gene
RNA editing
Wild barley
1 Introduction
Cellular metabolism and host phenotype in eukaryotes and prokaryotes, especially high organisms such as plants, is controlled at many steps. Epitranscriptomic modifications to RNA transcribed from exons are one of them. One of the recently discovered modifications is RNA editing, which is widespread and has been found to occur predominantly in chloroplasts and mitochondria. RNA editing is present in most organisms, and it involves insertions, deletions and/or base substitutions, leading to the modification of primary RNAs including mRNAs, tRNAs, microRNAs and even long coding RNAs (Picardi et al., 2014). In plants, editing of RNA often involves the deamination process from cytidines to uridines (C-to-U), which is catalyzed by pentatricopeptide repeat proteins (PPR) which have 30–40 amino acid motifs specific for binding to target RNA molecules (Takenaka et al., 2013). Some researchers claim that deciphering the mode of action of PPR proteins might provide novel approaches to find RNA editing sites (Cheng et al., 2016). While RNA editing is primarily used to repair DNA mutations, editing of coding regions in plants has an impact on the protein expression profile (Edera et al., 2018). Recently, numerous studies reported the linkage of RNA editing with cellular stress response in plants and corresponding downstream cellular metabolism including electron transport in plant organelles such as chloroplast and mitochondria (Yuan and Liu, 2012; Yuan et al., 2021) especially in wild plants like wild barley (Ramadan et al., 2023). The chloroplast NDH has Fd-dependent PQ reductase activity which is mainly involved in photosystem I-mediated cyclic electron transport (Shikanai, 2016). NDH complex is coded by at least 11 genes and ndhA is one of the key components. It is possible that this component could become critical to plant synthesis if it is subjected to any RNA editing. Changes in the ndhA mRNA through RNA editing and corresponding ndhA subunit protein, might have a significant effect on electron transport because NDH complex prevents drastic ion changes in stroma during stress (Peng et al., 2011) and thus it might lead to drought sensitivity as shown in Arabidopsis and Soyabean (Rodrigues et al., 2017). Recent advances in genomics such as next-generation sequencing (NGS) and RNA-seq technology have proved to be fast, reliable and high-throughput tools for identifying RNA edits in plants. By using RNA-seq data, as well as improved bioinformatics tools, it has been possible to find accurate edits by avoiding artefacts (Bentolila et al., 2013). This information can be further validated by quantitative PCR and accessory bioinformatics tools for discovering novel edit sites and their effects on the corresponding protein. We studied the RNA editing changes due to salt stress and after salt stress for 2, 12 and 24 h in wild barley plants using RNA-seq data, and further validated by qRT-PCR.
2 Materials and methods
2.1 RNA-seq data retrieval and analysis
The RNA-seq data was downloaded from NCBI for 0-hour exposure or control (SRR1028012 and SRR1028011), for 2-hour exposure (SRR1055527, SRR1049592, and SRR1049587), for 12-hour exposure (SRR1049626, SRR1049609, and SRR1049570) and 24-hour exposure (SRR1049655, SRR1049648 and SRR1049643).
2.2 RNA editing analysis
Significant RNA editing events were further investigated for linkage to other gene expression characteristics. We used CLC Genomic Workbench 3.6.5. (Qiagen, Denmark) with some modifications for reference assembly with plastome transcripts. Low quality reads were eliminated by filtering reads with parameters for similarity = 0.98 and length fraction = 0.98. The filtered good quality reads were then mapped to chloroplast ndhA gene sequence from Hordeum vulgare subsp. spontaneum (Accession no. NC_042692). The minimum percentage similarity was kept at 80% and the minimum length of the mapping reads was kept at 50%. The parameters for edited nucleotides was chosen as 5% low frequency variance, minimum coverage was set as 20, minimum count was set as 4, and the minimum frequency was set as 5%. Once all the parameters were defined, we could identify the total reads, depth of coverage and RNA editing sites. The frequency of nucleotide conversion between each site due to exposure to salinity as compared to the control was evaluated using the following formula: the number of reads for converted nucleotides divided by the total number of reads.
2.3 RNA extraction
Wild barley (Hordeum vulgare subsp. spontaneum) plants were divided into 4 groups in triplicate, and exposed to 500 mM NaCl salt for 0 (control), 2, 12 and 24 h, under controlled light and humidity conditions. Total RNA was isolated using TRIzol (Thermo, USA) as per the manufacturer’s guidelines. The concentration of RNA was estimated by Nanodrop (Tecan, Finland).
2.4 Quantitative PCR for validation of identified editing sites
The RNA edits identified were selected to be validated by quantitative PCR. The total RNA from each group was isolated using Qiazol (Qiagen, Cat No. 79306) and treated with DNAse kit (Thermo, USA) to remove any contaminating genomic DNA. The amount of total RNA was equalized to 1 μg for each sample and corresponding cDNA was synthesized with 1 μM poly dT oligonucleotides using cDNA synthesis kit (Biolegio, Netherlands), as per manufacturer’s instructions. To run the RT-PCR reaction, 50 l of reaction were setup in a Mx3005P qPCR system (Stratagene, USA) and the CT value was determined. The α‐tubulin gene was used as a housekeeping gene (control) for normalization purposes.
The percentage of RNA editing was determined by the following formula: % RNA edits = 2(CT mean of T variant − CT mean of C variant) ∕ {2(CT mean of T variant − CT mean of C variant) + 1} × 100, where C = control and T = test sample data (Livak and Schmittgen 2001).
2.5 Validation of T and G sites at DNA level
To verify edit sites in identified DNA sequences from the ndhA gene, especially at U edit sites, we extracted DNA from leaves of wild barley using DNeasy Plant Pro Kit (Qiagen, Germany) for use as template. Traditional PCR was run using REDTaq® ReadyMix™ PCR master mix using forward and reverse primers (having original and edited nucleotides) for five sites with two pairs of primers as mentioned (Table 1). PCR conditions used were: initial denaturation at 94 °C for 4 min, followed by 35 cycles of (94 °C for 1 min, 60 °C/30 sec and 72 °C for 1 min), final extension at 72 °C for 5 min and then storage at 4 °C. The PCR products were then run on 1.5% agarose gel and after confirmation of their size, were purified by PCR cleanup kit and sent for Sanger. The sequencing results were further compared by alignment to validate the editing at U/T or G/A sites as mentioned in Table 1.
position
F 5′ 3′ →
R 5′ 3′ →
C50
CTATCAATTCTTTTTCGAAATT
CCAGCATATTCAGGACCAATA
CTATCAATTCTTTTTCGAAATC
G111
CCATTTTGACCCTCCTTTTA
AAATAAAGAAATATCTCCTC
CCATTTTGACCCTCCTTTTG
C303
CTTTATTTAGCATTGGACCT
TAGACCGCCTGAAAAAGAAT
CTTTATTTAGCATTGGACCC
U550
TGTGCTAGCAATATCTCTAC
GGTAACCTGCTACTAATTCT
TGTGCTAGCAATATCTCTAT
C563
TCTCTATTATCTAACAGTTT
GAATATTCAGTTTGGTAACC
TCTCTATTATCTAACAGTTC
U592
TGATATAGTTGAAGCACAGC
CAAGATAAAATAAACCATAT
TGATATAGTTGAAGCACAGT
C1042
TTTTGATTTTTTCCAAATGA
GTTAATAATAGATTGCCCAA
GTTAATAATAGATTGCCCAG
C1046
TTTTCCAAATGAATAAAGC
GGTTGTTAATAATAGATTAT
GGTTGTTAATAATAGATTGC
C1047
TTTTCCAAATGAATAAAGC
GGTTGTTAATAATAGATTAT
GGTTGTTAATAATAGATTGC
U1066
GCAGTTGGAATTTTGGAAAT
AGTGAAACAAGTTGAGAAGG
AGTGAAACAAGTTGAGAAGA
α‐tubulin
TCCATGATGGCCAAGTGTGA
GACATCCCCACGGTACATGAG
2.6 Analysis for amino acids deduced for ndhA gene
The correspondence amino acids for RNA editing sits are identified by CLC genomic workbench 3.6.5. The predictions for any change in the tertiary (3D) and secondary structure of each protein are generated by the same program.
2.7 Statistical validation and analysis
In order to analyze the data, SPSS used the analysis of variance (ANOVA) test. Using Tukey's HSD, several comparisons were made to test our hypothesis and statistically significant levels .
3 Results
3.1 ndhA characterization
cDNA and genomic DNA were recovered for the ndhA gene from H. vulgare subsp. spontaneum under control (acc. no. OM262848), after two hours (acc. no OM262849), after twelve hours (acc. no. OM262850) and after twenty-four hours (acc. no OM262851) of salt stress. 105,526,154 pair-end-RNA reads were used to identify the ndhA gene transcripts at zero hour, which were mapped to the ndhA gene (acc. no. NC_042692) as a reference, 159,631,812 at two hours, 161,793,598 at twelve hours, and 159,631,812 at 24 h (Table 2).
Transcriptome data file
Time
Mapped Reads
Total Reads
SRR1028012
SRR1028011Control
915,629
105,526,154
SRR1055527
SRR1049592
SRR10495872H
1,179,688
159,631,812
SRR1049626
SRR1049609
SRR104957012H
1,751,987
161,793,598
SRR1049655
SRR1049648
SRR104964324H
2,814,196
162,762,776
3.2 Analysis of RNA edits and modifications in amino acids
Comparison of ndhA genome sequence with its transcripts from each of the four treatments showed five C to U changes (C50, C303, C563, C1042, C1047), three U to C changes (U550, U592, U1066), and two G to A changes (G111, G1046). RNA editing did not differ significantly between all treatments at U550 and C1042. At other sites, salt stress appeared to have significantly increased RNA editing at positions G111, C303, and C563. Positions G1046, C1047, and U1066 showed significant decreases in RNA editing after salt exposure for 24 h. In positions C50 and U592, however, RNA editing in control group was similarly high to other salt exposure times (Fig. 1).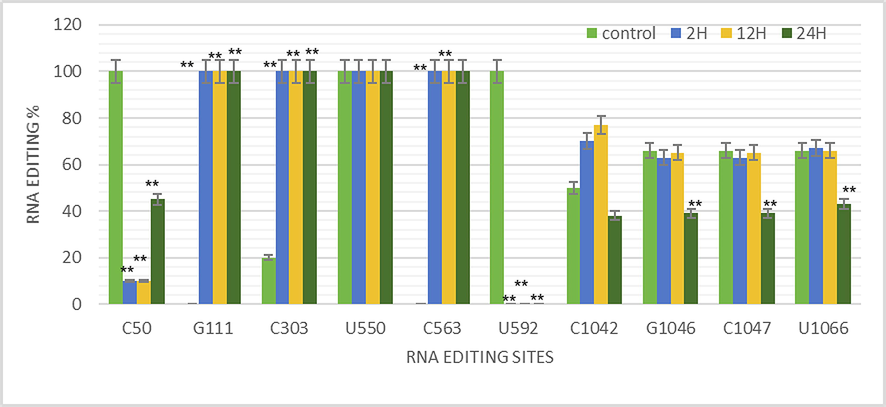
The efficiency of RNA editing for ndhA at indicated positions, as compared with the control, through the sequences identified from the RNA-seq reads. (C or U) nucleotide site. Data has been expressed as Mean ± SD (solid bars) for biological replicates (n = 3) of each sample group. ** indicates significant difference between the compared groups at significance level of p < 0.01.
3.3 Validation of ndhA editing
As a way to verify editing sites and confirm the value of bioinformatics tool analysis, four salt exposure intervals were measured to quantify and measure editing positions (C50, G111, C303, U550, C563, U592, C1042, G1046, C1047, and U1066) by qRT-PCR (Fig. 2). Likewise, uncommon RNA edits G111, U550, U592, G1046, and U1066 were confirmed on the DNA level (Ramadan et al., 2021) to exclude any errors of published sequence (NC_042692) using the original and edited nucleotides in 3 prime ends (Fig. 3).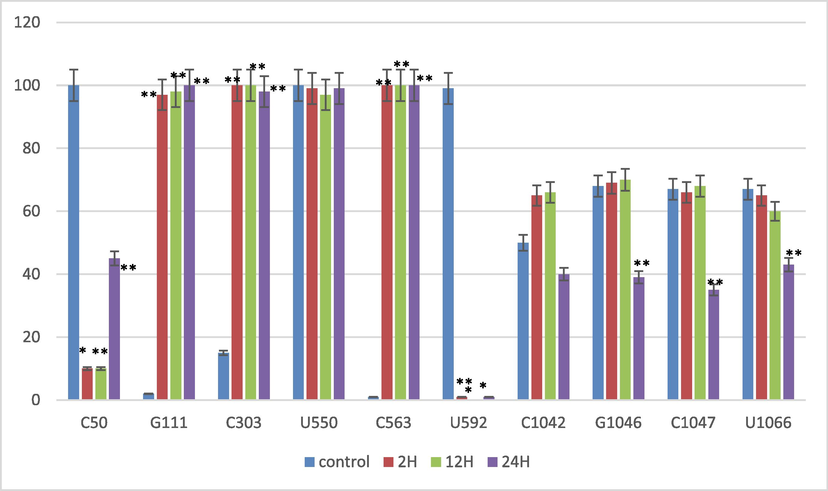
The qRT-PCR analysis confirmed the wild barley edited sites that were predicted by the CLC genomic workbench in different salinity stress situations (control, 2 h after salinity exposure, 12 h after salinity exposure, 24 h after salinity exposure). Biological replicates are expressed as means with standard deviations (black bars). Differences between treatments that are significant indicate ** P < 0.01.
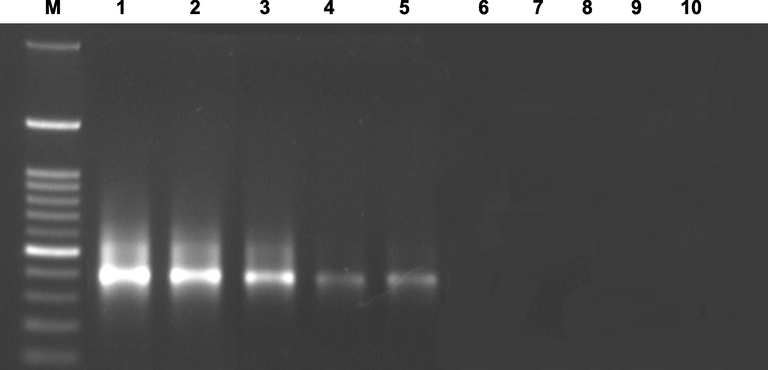
PCR analysis for genomic ndhA DNA using (M) 100 bp ladder bioronTM. (1, 6) ndhA- G111, (2, 7) ndhA – U550, (3, 8) ndhA – U592, (4, 9) ndhA – G1046, (5, 10) ndhA- U1066. Primers are designed using original and alternate nucleotides, respectively.
3.4 Analysis for conserved domains and predicted structure of ndhA protein
RNA editing has been assessed at the protein level by measuring the current influence of ndhA (CDD accession number CHL00032; Figure S2) on the NADH subunit 1 structure. The homology modelling of the proteins showed no difference in the tertiary structure of protein under stress at four times (Fig. 4), but minor changes in alpha helices and beta sheets were observed in the secondary structure (Fig.S2).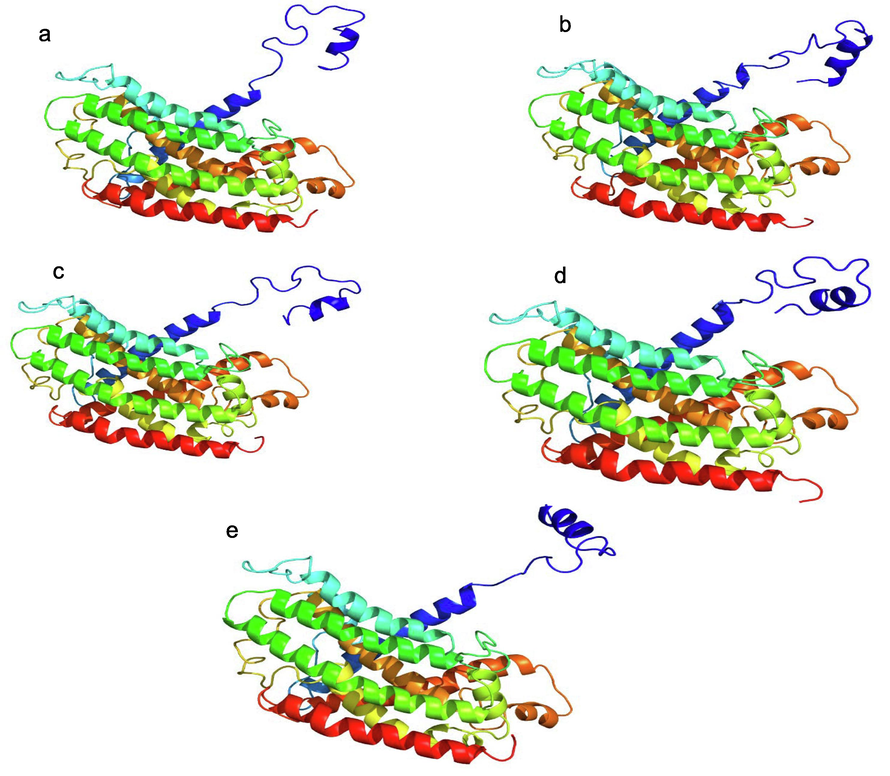
Model of the 3D structure of the ndhA protein exposed to various levels of salt at different times. Proteins with no editing (a), controls (b), 2 h after salt exposure (c), 12 h after salt exposure (d), and 24 h after salt exposure (e).
4 Discussion
In higher plants, RNA editing is a vital process, which occurs increasingly through deamination reactions that are catalyzed by pentatricopeptide repeat proteins (PPRs) (Takenaka et al., 2013). This is more relevant in the context of genes involved in vital functions for plant survival such as those encoding proteins involved in cellular transpiration and photosynthesis. The NDH complex of chloroplasts has many subunits, is coded by at least 11 chloroplastic genes (ndhA to ndhK), and it plays a vital role in assisting cyclic transport of electrons through photosystem I, and maintenance of homeostasis during diverse environmental conditions (Shen et al., 2021). Changes in the ndhA transcripts through RNA editing might affect electron transport through disruption of NDH complex which prevents drastic ion changes in stroma during stressful environmental conditions (Peng et al., 2011) and it showed induction of drought sensitivity in Soyabean and Arabidopsis (Rodrigues et al., 2017). Thus, we focused on studying RNA editing events in ndhA transcripts of wild barley, and to avoid false positives during editing analysis, we excluded any effect of heteroplasmy during nucleotide exchange (Makki et al., 2019). In our study, we found RNA editing at 10 sites: C50, G111, C303, C563, U550, U592, C1042, G1046, C1047 and U1066, in ndhA gene of H. vulgare subsp. spontaneum (Fig. 1). The editing ratios of these sites were significantly different following four salt stress cycles (except U550 and U1042) and qPCR validation was conducted on each of the edited sites (Fig. 2). Several trends were observed in our study, including 'C to U' edits at five positions (C50, C303, C563, C1042 and C1047) and 'U to C' edits at three positions (U550, U592, U1066) and first report edits on plants “G to A” at two positions (G111, G1046). The editing of some of these proteins appears to be related to salt stress tolerance in the barley plant, such as G111, C303, and C563. After salt exposure, editing increases significantly after 2, 12, and 24 h. However, RNA editing percentage was significantly decreased during salt stress periods in positions C50, U592 which indicate that RNA editing in these sites may inhibit salinity tolerance. RNA editing percentages increase in 2 and 12 h after salt exposure in C1042, which agrees with our previous report (Ramadan et al., 2021). The significantly decrease in RNA editing after 24 h like, G1046, C1047 and U1066 might be indicative of adaptation of edited proteins at higher salt stress in the barley plant or lost tolerance due to the extreme salinity and may initiate programmed cell death (Katsuhara, 1997; Katsuhara and Shibasaka, 2000; Lin et al., 2005; Li et al., 2007a; He et al., 2018) in yeasts (Li et al., 2007b) and algae (Huh et al., 2002). The fixed editing percentage of U550 across all sample groups indicates essential function of this site in the ndhA gene.
Next, we observed the first 'U to C' RNA edits at positions U550, U592 and U1066 of the ndhA gene. This is considered rare in higher plants and reported only in hornworts, lycophytes and ferns (Gerke et al., 2020). However, a few recent reports have shown such edits in some mitochondrial genes in Arabidopsis thaliana and Hordeum vulgare (Ramadan et al., 2021, Ruchika et al., 2021). In our case, we argue that these edits, which are considered reverse changes might have occurred to restore the activity of the evolutionarily conserved domains of ndhA protein as a corrective response against a possibly undesired mutation. Other unexpected editing G to A is found in this chloroplast gene in positions G111 and G1046, this type of editing not reported before in plant but reported in animal and human nuclear APOBEC3A and ribonucleoprotein K genes (hnRNP K) (Niavarani et al., 2015; Christofi and Zaravinos, 2019; Tao et al., 2021).
Our third observation was the presence of higher percentage RNA editing in the control group at two positions (C50 and U592) when compared to the groups receiving salt stress. This might be due to a requirement for these edits in natural state which might not have been necessary for the plant when it is exposed to higher salt stress.
With regard to effects due to the RNA edits on structure of the resulting protein, minor changes are observed in alpha helix and beta sheet in the secondary structure (Fig. 3c, d) for salt stress exposure time of 2, 12 and 24 h. However, this effect did not affect the final 3D structure of the protein.
Unlike our previous work, there was high editing frequency without any salt stress conditions which accept other investigators in Arabidopsis, Allium cepa, Helianthus annus and Cucumis sativus (Edera et al., 2018). Our observations indicate towards the complex landscape of RNA editing and its role especially in photosynthesis when compared among higher plants which are adapted to different environmental conditions. This also directly suggests that some wild cereals might have different photosynthesis needs when compared to cultured plants, to cope with harsh environmental conditions including salt stress. Many investigators have linked the increasing in salt stress with increased RNA editing, especially in mitochondria (Ramadan, 2014; Ramadan et al., 2021), but in this gene we did not find a specific pattern at all sites. Also, the lack of effect of RNA editing on 3D protein makes us conclude that the release at some sites has not related to salt stress, but to restore the conserved protein (Fig. 1). This has been previously reported to increase plant responsiveness to environmental stress, and substantiates previous claims that improved photosynthesis performance of plants leads to mitigate salt stress (Ramadan et al., 2021). However, the decrease in editing percentages after 24 h in some sites might also mean that the plant has lost salt tolerance and might have initiated cell senescence, which has been seen in many plants (Katsuhara and Shibasaka, 2000; Lin et al., 2005) including algae (Li et al., 2007b) and yeast (Huh et al., 2002).
5 Conclusions
RNA editing in photosynthesis I ndhA gene of wild barley is explored under salt stress and clearly observed RNA editing in 10 sites. the major finding in this study is un-expecting G to A RNA editing which not reported before in flowering plant. This foundation opens the door to many questions around the mechanism of this type of editing. Furthermore, we think this is the first time to report ‘U to C’ RNA edits in ndhA gene of barley chloroplast. All sites have no same pattern under salt stress.
Acknowledgments
This project was funded by the Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, under grant no. G: 318-130-1442. The authors, therefore, acknowledge and thank DSR's technical and financial support.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genet.. 2013;9:e1003584.
- [Google Scholar]
- Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J.. 2016;85(4):532-547.
- [Google Scholar]
- RNA editing in the forefront of epitranscriptomics and human health. J. Transl. Med.. 2019;17:319.
- [CrossRef] [Google Scholar]
- Towards a comprehensive picture of C-to-U RNA editing sites in angiosperm mitochondria. Plant Mol. Biol.. 2018;97(3):215-231.
- [Google Scholar]
- Towards a Plant Model for Enigmatic U-to-C RNA Editing: The Organelle Genomes, Transcriptomes, Editomes and Candidate RNA Editing Factors in the Hornwort Anthoceros Agrestis. New Phytol.. 2020;225:1974-1992.
- [Google Scholar]
- Two pivotal RNA editing sites in the mitochondrial atp1 mRNA are required for ATP synthase to produce sufficient ATP for cotton fiber cell elongation. New Phytol.. 2018;218:167-182.
- [CrossRef] [Google Scholar]
- Salt causes ion disequilibrium-induced programmed cell death in yeast and plants. Plant J.. 2002;29:649-659.
- [Google Scholar]
- Apoptosis-like cell death in barley roots under salt stress. Plant Cell Physiol.. 1997;38:1091-1093.
- [Google Scholar]
- Cell death and growth recovery of barley after transient salt stress. J. Plant Res.. 2000;113:239-243.
- [Google Scholar]
- Lanthanum prevents salt stress-induced programmed cell death in rice root tip cells by controlling early induction events. J Integr Biol.. 2007;49:1024-1031.
- [CrossRef] [Google Scholar]
- Salt stress-induced programmed cell death in rice root tip cells. J. Integr. Plant Biol.. 2007;49:481-486.
- [CrossRef] [Google Scholar]
- Salt stress-induced programmed cell death via Ca2+-mediated mitochondrial permeability transition in tobacco protoplasts. Plant Growth Regul.. 2005;45:243-250.
- [Google Scholar]
- Analysis of relative gene expression datausing real-time quantitative PCR and the 2(-Delta Delta C(T)) Methods. 2001;25(4):402-408.
- [CrossRef] [Google Scholar]
- Single nucleotide polymorphism analysis in plastomes of eight Catharanthus roseus cultivars. Biotechnol. Biotechnol. Equip.. 2019;33:419-428.
- [CrossRef] [Google Scholar]
- APOBEC3A is implicated in a novel class of G-to-A mRNA editing in WT1 transcripts. PLoS One. 2015;10
- [CrossRef] [Google Scholar]
- Structure and biogenesis of the chloroplast NAD(P)H dehydrogenase complex. Biochim Biophys Acta – Bioenerg.. 2011;1807:945-953.
- [Google Scholar]
- Uncovering RNA editing sites in long non-coding RNAs. Front. Bioeng. Biotechnol.. 2014;2:64.
- [Google Scholar]
- RNA editing in Calotropis procera mitochondrial nadh-dehydrogenase subunit 3 gene. Egypt J Genet Cytol.. 2014;43:353-364.
- [Google Scholar]
- The first report of RNA U to C or G editing in the mitochondrial NADH dehydrogenase subunit 5 (Nad5) transcript of wild barley. Mol. Biol. Rep.. 2021;48:6057-6064.
- [CrossRef] [Google Scholar]
- Ramadan, A., Alnufaei, A.A., Fiaz, S., Khan, T.K., Hassan, S.M., 2023. Effect of salinity on ccmfn gene RNA editing of mitochondria in wild barley and uncommon types of RNA editing. Funct Integr Genomics. 23(1):50. doi: 10.1007/s10142-023-00978-5. PMID: 36707470.
- Salt stress affects mRNA editing in soybean chloroplasts. Geneticsand molecular biology. 2017;40:200-208.
- [CrossRef] [Google Scholar]
- Genome-Wide Identification of U-To-C RNA Editing Events for Nuclear Genes in Arabidopsis thaliana. Cells. 2021;10:635.
- [CrossRef] [Google Scholar]
- Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare. Nature. 2021;601:649-654.
- [CrossRef] [Google Scholar]
- Chloroplast NDH: a different enzyme with a structure similar to that of respiratory NADH dehydrogenase. BBA. 2016;1857:1015-1022.
- [Google Scholar]
- Transcriptome-Wide Identification of G-to-A RNA Editing in Chronic Social Defeat Stress Mouse Models. Front. Genet.. 2021;12:680548
- [CrossRef] [Google Scholar]
- ZmPPR26, a DYW-type pentatricopeptide repeat protein, is required for C-to-U RNA editing at atpA-1148 in maize chloroplasts. J. Exp. Bot.. 2021;72(13):4809-4821.
- [CrossRef] [Google Scholar]
- Functional disruption of the pentatricopeptide protein SLG1 affects mitochondrial RNA editing, plant development, and responses to abiotic stresses in Arabidopsis. Plant J.. 2012;70(3):432-444.
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2023.102755.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







