Translate this page into:
Required criteria of drug efflux pump inhibitors to disrupt the function of AcrB subunit protein from AcrAB-TolC multidrug efflux pump from Escherichia coli
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
The infections of multidrug-resistant bacteria, particularly the infections caused by Gram-negative bacterial infections, are considered the main global serious health problems. Multidrug-resistant bacteria possess different resistance mechanisms, the most important of which are drug efflux pumps due to their function in recognizing harmful compounds, especially antibiotics, and expelling, them outside the cell, which leads to a decrease in their concentration to sub-toxic levels and losing their effectiveness. One of the most important approaches that can be used to inhibit drug efflux pumps is using small molecule inhibitors targeting specifically pumps to prevent their expelling antibiotics from inside the cell before reaching the required concentration that kills bacteria. The drug efflux pumps inhibition approach is emerging as a new attractive and promising approach in reversing antibiotic resistance in many clinically relevant pathogenic bacteria. This review will discuss the requirements that must be met in inhibitors that target AcrB, which is the main subunit protein responsible for multiple antibiotic resistance in the main pump of AcrAB-TolC which belongs to E. coli, which were specifically chosen due to their various homologs in many Gram-negative bacteria which all of which are implicated in multi-drug resistance (MDR), and the availability of the pump's functions and chemical structures.
Keywords
Required criteria of inhibitors
Inhibitors of drug efflux pump
Multidrug efflux pump
Superfamily efflux pump RND
E. coli
AcrAB-TolC
AcrB subunit protein
1 Introduction
The rapid spread of antibiotic-resistant bacterial infections causes many chronic diseases and currently is considered the most threat to global health (Satta et al., 2022; Paul et al., 2022). This issue was accompanied by a decrease in the effectiveness of various antibiotics that are used as a first defense line to treat antibiotic-resistant bacteria, this problem is concentrated in Gram-negative bacteria because their resistance is much higher than that of Gram-positive bacteria, which makes their treatment more difficult and complex as well (Wang et al., 2021). This also coincided with the reluctance of many pharmaceutical companies to invest in medical research projects related to the search for new effective treatments to confront bacterial-resistant strains, which in turn led to the production of a few new antibiotics for the market. This is due to the lack of economic feasibility from these projects due to the high costs of research and the lack of desired profits compared to treatments for other diseases (Venter, 2019).
1.1 Antimicrobial-resistant bacterial infections
The projections indicate that antimicrobial resistance infections will be responsible for killing one person every 3 s in 2050 (O’Neill, 2016). Bacterial infections annually are responsible for 700,000 deaths around the world. Among these deaths, 230,000 only belong to tuberculosis infections. It is necessary for concerted efforts to confront the infection of bacterial infections, otherwise, the deaths due to these infections will reach 10 million deaths worldwide by 2050, much more than the deaths from diabetes and cancer combined (Wang et al., 2021).
1.2 Gram-negative bacteria are more difficult to treat
Naturally, Gram-negative bacteria have mechanisms that confer resistance against one or more classes of antibiotics that are clinically important (Wang et al., 2021), due to they have an envelope containing three principal layers: (i) an outer membrane, (ii) periplasmic space and (iii) the inner or cytoplasmic membrane (Exner et al., 2017). Together, the inner and outer membranes act as a formidable permeability barrier to prevent potentially toxic compounds, such as antibiotics, from entering the bacteria (Du et al., 2018; Blair et al., 2014; Venter et al., 2015; Nikaido, 2011). The two membranes are one of the reasons that confer intrinsic resistance to Gram-negative bacteria against many different types of antibiotics (Exner et al., 2017). Hence, Gram-negative bacteria infections are highly resistant and more difficult to treat than Gram-positive species (Du et al., 2018; Blair et al., 2014; Venter et al., 2015; Nikaido, 2011; Venter, 2019). World Health Organization (WHO) in 2017 identified the most pathogens (Table 1) that cause various chronic diseases and need urgent solutions to eliminate their risks, most of them were Gram-negative bacteria like E. coli (Candel et al., 2022). Unfortunately, only eight new kinds of antibiotics have been approved for clinical use since 2017 (Terreni et al., 2021).

1.3 AcrAB-TolC multidrug efflux pump and MDR
These pathogens have acquired their importance in the medical field due to their possession of drug efflux pumps such as AcrAB-TolC multidrug efflux pump, which confers MDR because of their capacity to expel a broad range of antibiotics (Nikaido & Pagès, 2012; Venter et al., 2015). For this reason, the pumps have been named multi-drug efflux pumps (Piddock, 2006). Moreover, there are different classes of these pumps, but Resistance-Nodulation-Division (RND) efflux pump is one of the important pumps playing a significant role in resistance, due to its ability to confer a variety of Gram-negative bacteria MDR. The drug efflux pump AcrAB-TolC from RND that belongs to E. coli has various homologs in multiple species of Gram-negative bacteria all of which are implicated in MDR. To understand MDR in these pathogens, the multidrug efflux pump AcrAB-TolC has been chosen to be studied for how to be inhibited by small molecule inhibitors due to the availability of the pump's functions and chemical structures (Darzynkiewicz et al., 2019; Abdali et al., 2017; Davis et al., 2014). Utilizing small molecule inhibitors to inhibit the functions of AcrAB-TolC is considered a unique pathway to reverse MDR in bacterial pathogens. The pharmacological inhibition and the binding mechanisms of the substrate of multidrug efflux pump AcrAB-TolC became more obvious and understandable due to the availability of the data of its structures in complex with inhibitors and substrates by using some techniques such as cryo-electron microscopy and X-ray crystallography. Most efflux pump inhibitors have synergistic activity with antibiotics that are used to treat Gram-positive bacterial strains, while Gram-negative bacterial strains there are no inhibitors currently used against their efflux pumps due to their severe toxicity to bacterial cells and their instability (Alenazy, 2022; Darzynkiewicz et al., 2019; Abdali et al., 2017; Davis et al., 2014). Consequently, in this review, the main topic will be the literature that describes the tools for studying the inhibitors of the AcrB subunit protein from efflux pump AcrAB-TolC from E. coli, because this pump and its homologs play the main role in MDR in Gram-negative bacteria, seeking to find what are the best ways to choose the inhibitors that work specifically on the pump and can fully inhibit its functions without having any off-target activities which indicate indirect activity such as permeabilisation of the inner or outer membranes or possesses off-target effects, aiming to reverse MDR by re-sensitizing the bacterial pathogens to presently utilized antibiotics.
1.4 Multi-drug resistance in pathogenic bacteria
During the antimicrobial discovery, there was great optimism that antimicrobials would eliminate infectious diseases soon. Unfortunately, many species of bacteria began to develop resistance to different classes of antimicrobials (Murugaiyan et al., 2022). This is known as multi-drug resistance (MDR). MDR is classified as the non-susceptibility of pathogens to one or more antimicrobials from three or more antimicrobial classes (Rafailidis & Kofteridis, 2022). In 2021, the World Health Organization (WHO) issued a statement warning that antibiotics and antimicrobials have increasingly become ineffective due to the resistance of bacteria to antibiotics and antimicrobials, which conferred treatment difficulties to many diseases that were previously easily treatable to diseases that are difficult or impossible to treat (WHO, 2021). MDR infections, in particular by Gram-negative pathogens, play an important role in increasing mortality and morbidity rates and the length of stay in hospitals, which causes an increase in financial burdens as well (Russo et al., 2022). While antibiotics have different classes with diverse chemical structures and different modes of action pathogenic bacteria have acquired resistance mechanisms to face the effect of these antibiotics and other toxic substances. Pathogenic bacteria possess different resistance mechanisms including i) overexpressing multidrug efflux pumps, which is considered the main mechanism, ii) modification/alteration of drug targets by producing specific enzymes that can protect the drug targets inside the bacterial cell, iii) preventing antibiotic entry by reducing the size of the porins that reside on the surface of the outer membrane of a bacterial cell, iv) inactivation of antibiotics (Zając et al., 2022). One of the most important mechanisms that bacteria use in resistance is multi-drug efflux pumps which confer MDR to pathogens via recognizing and expelling a variety of nonrelated antimicrobial agents to the outside bacterial cell, thus leading bacteria to have resistance to more than one class of antimicrobial. Hence, the concentration of these agents will be reduced to subtoxic levels. Overexpression of efflux pumps leads to the recognition and expelling of a large number of structurally dissimilar compounds (Casalone et al., 2022). Currently, inhibitors of drug efflux pumps are emerging as a promising and alternative treatment to counteract antimicrobial resistance in many pathogens, in light of the severe shortage of new therapies and the narrow antibiotic pipeline. Efflux pump inhibitors (EPIs) can restore the activities of many antimicrobials by inhibiting the functions of pumps which causes an increase in the concentration and accumulation of the antimicrobials inside microbial cells leading to reaching the required concentrations for killing bacteria (Casalone et al., 2022). Although no efflux pump inhibitors have been officially approved yet for clinical use, there are increased investigations to discover appropriate inhibitors that may be approved for medical use. The reason for this is the difficulties facing the development of inhibitors, whether natural or synthetic, the structural and functional complexity of the inhibitors and the binding way with the pumps, and most importantly, their toxicity and their expected side effects on humans (Casalone et al., 2022). In this review, we will discuss the mechanisms, and tools of EPIs testing, whether phytochemical or synthetic with mention of some examples of experiments that have been done before of some inhibitors on Gram-negative bacteria to give clear imaging of how could be choosing the novel inhibitors to reach fully inhibition of drug efflux pumps of pathogenic bacteria. This one approach can simultaneously disrupt virulence, pathogenesis, and biofilm formation in addition to reversing antibacterial resistance.
2 Drug efflux pump resistance-nodulation-cell division (RND)
Generally, bacteria possess five superfamilies of efflux pumps, and which RND efflux pump superfamily is one of them. RND efflux pump superfamily, to which AcrAB-TolC belongs, plays an important role in intrinsic, acquired resistance and MDR in many species of Gram-negative bacteria by recognizing and expelling a variety of unrelated antimicrobials with different chemical structures (Fig. 1) (Plé et al.,2022; Hillman et al., 2022). RND has a feature not found in other pumps, which is recognizing many different classes of antibiotics like tetracycline, novobiocin, fusidic acid, chloramphenicol, fluoroquinolones, and some β-lactams then expel them outside the cell (Munita & Arias, 2016), not only that, these pumps can expel the determinants of virulence including toxins and adhesins moreover some important proteins in the colonization processes of host cells (Huang et al., 2022). RND substrates are diverse and not limited to those mentioned above; but also include detergents, biocides, metals, and bile salts synthesized, compounds that synthesize by the bacteria including iron-chelating siderophores and virulence factors (Munita & Arias, 2016). RND possesses a tripartite protein complex that extends vertically, starting from the outer membrane of the envelope of bacteria to the inner membrane, passing through the periplasmic space (Hillman, 2022).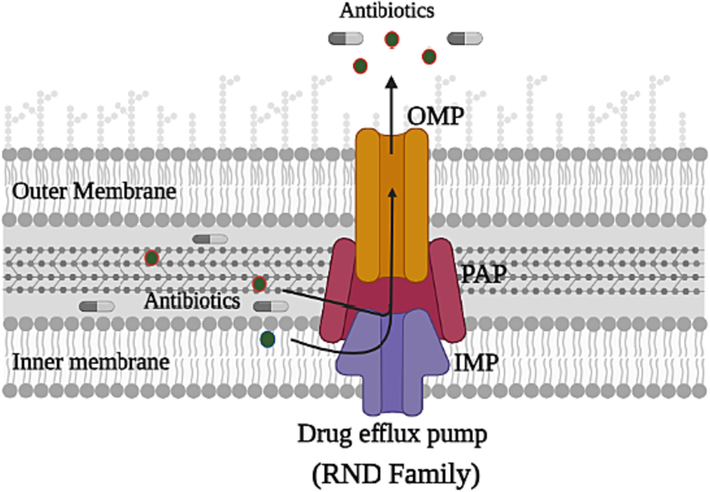
Schematic diagram illustrating representative structures and the system of multidrug efflux, the recognizing and expelling of the antibiotics outside the bacterial cell of the RND superfamily. RND possesses three main subunit proteins; the periplasmic adapter protein (PAP), which connects the two other proteins; the inner-membrane protein (IMP) with the outer-membrane protein (OMP).
The tripartite complex drug efflux pump AcrAB-TolC comprising three separate protein subunits: (i) the inner membrane protein (IMP), AcrB, which acts as a recognizer of substrates and then pushes them out of the pump utilizing the proton motive force (pmf); (ii) the periplasmic adapter protein (PAP), AcrA, which its function is linking AcrB and TolC also works as a protection for the transporting of the substrate that is captured through the periplasm and (iii) the outer membrane protein channel (OMP), TolC, which acts as a channel facilitating the expulsion of substrates outside the pump (Vargiu et al., 2022). AcrB is the most important and largest protein in the complex due to its responsibility for multi-drug poly-specificity also the translocation of the proton responsible for the movement of the substrate (Eicher et al., 2012). Therefore, this review will focus on studying the required criteria of inhibitors for the inhibition of AcrB.
3 Inhibition of AcrB by small molecules; efflux pump inhibitors (EPI)
The functional rotation mechanism is used to translocate the substrates through AcrAB-TolC which needs to work all protein subunits. Therefore, inhibiting any protein from the proteins that belong to the AcrB homotrimer by small molecule inhibitors leads to the entire inactivation of the AcrAB-TolC pump (Eicher et al., 2012). The small molecules could be used as adjuvants or chemical sensitizers with antibiotics to improve the effectiveness of the antibiotics in case these molecules show effectiveness in inhibiting the pump (Spengler et al., 2017; Venter, 2019). AcrB has a distal binding pocket (DBP) which has a main contribution to the design of small molecule inhibitors. DBP has a large cavity that is lined by various residues such as I277 which contributes significantly to binding and stabilizing various substrates inside the AcrB complex (Vargiu & Nikaido, 2012). This pocket has two features: (i) has multi-drug binding sites also has a channel used to translocate hydrophilic substrates and (ii) a hydrophobic trap located inside the distal binding pocket and branches out from it (Fig. 2). The hydrophobic trap has a significant role in the inhibition of AcrB which was first determined when Nakashima and his research team in 2013 published the first co-crystal structure of the inhibitor, which was pyridopyrimidine EPI D13-9001, that was bound to AcrB from E. coli and MexB from P. aeruginosa (Nakashima et al., 2013). The co-crystal structure of D13-9001 showed that there was a strong binding interaction between the phenylalanine-rich hydrophobic trap and D13-9001. Moreover, this binding interaction led to the inactivation of the functional rotation mechanism which inhibited the activity of AcrB and prevented the movement of the substrates to AcrA (Nakashima et al., 2013; Sjuts et al., 2016). Hydrophobic trap discovery is considered a new promising approach that could lead to improving the development of inhibitors by utilizing the structure-guided design (Aron & Opperman, 2018).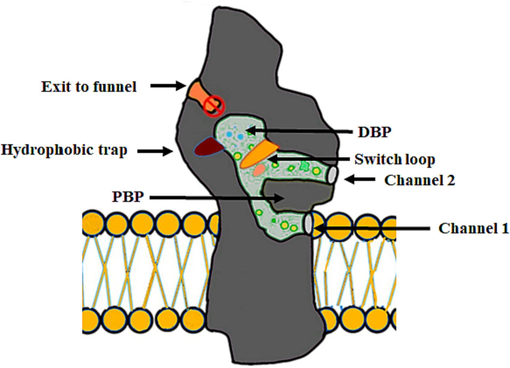
Schematic diagram showing the drug pathways and their pockets, and the two substrates transport channels in AcrB. The diagram also shows the periplasmic entry sites; i) channel 1, which directs the movement of substrates from the leaflet of the inner membrane, and ii) channel 2 which has the proximal binding pocket (PBP). Channel 2 has the proximal binding pocket which includes the hydrophobic trap which branches out from the channel of the main substrate-translocation, and a switch loop that separates the proximal and distal binding pockets and helps the substrates in their unidirectional movement. Also, the diagram shows the connected two substrates' entry channels which lead to the distal binding pocket with the channel of exit which connects the central cavity with TolC.
4 Criteria for studying efflux pump inhibitors
To determine the inhibitors that could act as appropriate to inhibit multidrug efflux pumps in pathogenic bacteria specifically Gram-negative bacteria, they must have some specific criteria (Fig. 3) (Lomovskaya and Bostian, 2006).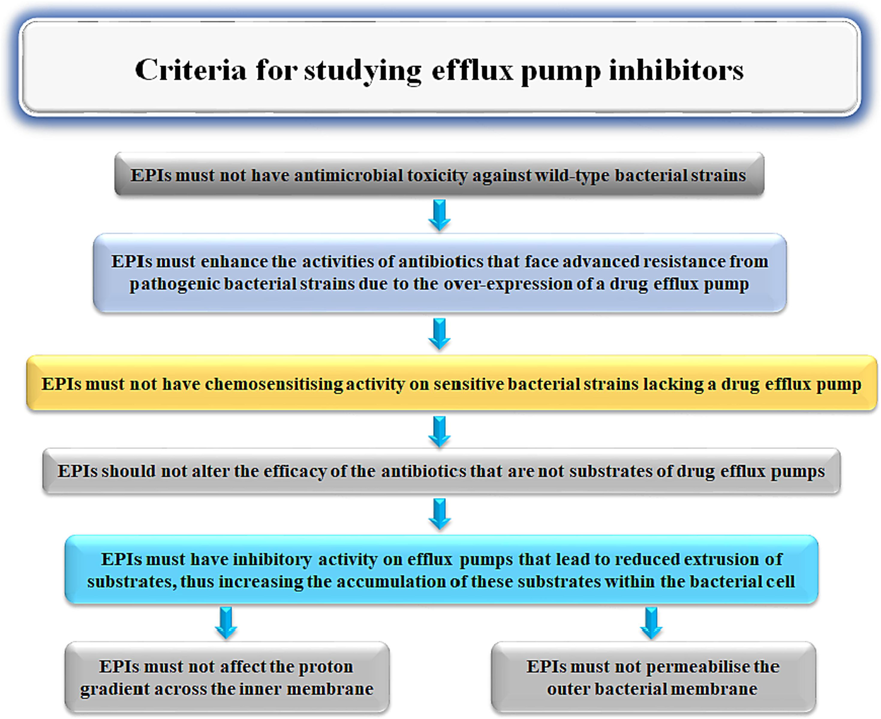
The required criteria for potential compounds to be effective EPIs.
5 Compounds progression plan
To investigate the required criteria for potential compounds that can act as effective EPI, the following well-developed techniques must be employed, which is called A Compounds Progression Plan, and the potential compounds must have the ability to pass all these Biological assays (Fig. 4).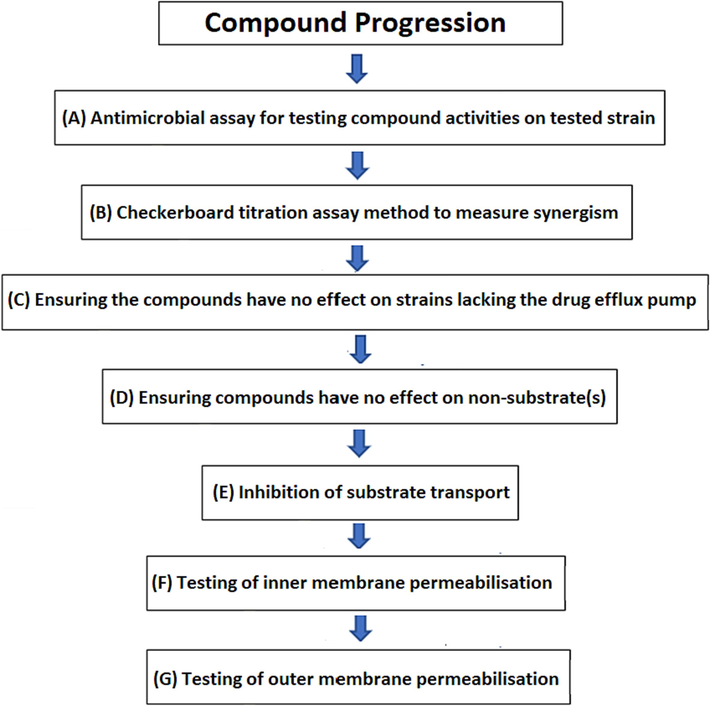
Compound progression plan for potential compounds which can act as effective EPIs. Shown several biological assays to identify the ability of the biological activity of tested compounds to satisfy the required criteria for potential compounds EPIs.
5.1 Antimicrobial assay for testing compound activities on tested strain
Antimicrobial assays are performed to confirm that these compounds do not have antimicrobial activities against wild-type strains of bacteria. The minimum inhibitory concentration (MIC) of the test compound is used and measured by utilizing assays of the standard broth dilution to ensure that the tested compounds do have not any negative biological activity against tested strains (Lomovskaya et al., 2001; Belanger & Hancock, 2021; Ohene-Agyei et al., 2012).
5.2 Checkerboard titration assay method to measure synergism
Test compounds should be assayed at concentrations usually 4 × lower than their MICs (if they have antibacterial effects) to determine their synergistic activity against a panel of antibiotics that are known substrates for the efflux system. Initially, checkerboard titration assays should be used as the preliminary screening examination to measure synergism (Lomovskaya and Bostian, 2006; Orhan et al., 2005; Venter et al., 2015). These examinations are conducted utilizing 96-well plates with serially diluted concentrations of antibiotics used vertically down the plate (eight 2-fold serial dilutions) whereas the serially diluted concentrations of test compounds are used horizontally across the plate (six 2-fold serial dilutions) (Lomovskaya and Bostian, 2006; Orhan et al., 2005; Ohene-Agyei et al., 2014). The index of the fractional inhibitory concentration (FIC) is then will be used to quantitate antibiotic-EPI interactions. The index of FIC (ΣFIC) is defined as the total of the FIC of each of the antibiotics, which is determined by dividing the MIC of each antibiotic in combination with the MIC of the antibiotic alone. The antibiotic-EPI interaction could be considered synergistic interaction if the ΣFIC is < 0.5, and could be indifferent interaction if the ΣFIC is ≥ 0.5 to < 2, whereas it could be antagonistic interaction if the ΣFIC is ≥ 2 (Fig. 5).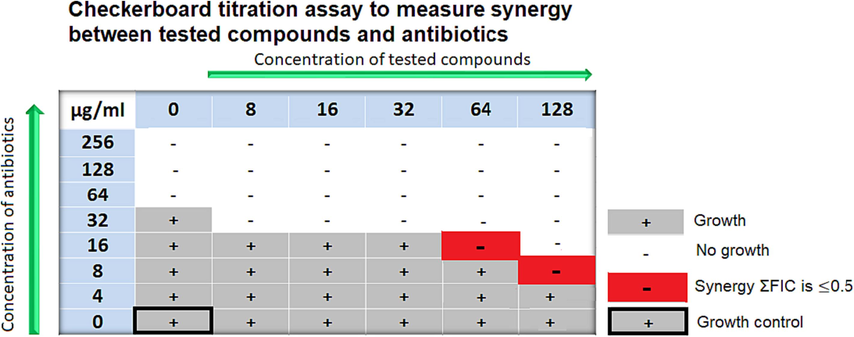
The figure shows an example of the synergistic interaction of tested compounds with different antibiotics with different concentrations against wild-type strain E. coli.
5.3 Ensuring the compounds have no impact on bacterial strains that do not have the drug efflux pump
One effective method for defining whether EPIs function specifically on the target efflux system is to test for antibacterial activity on a bacterial strain containing a deletion for the gene encoding that pump. Those compounds that reduce the MIC in a pump-deleted strain should be excluded from further development, as they likely have off-target activities.
5.4 Ensuring compounds do not affect non-substrate(s)
To rule out false positives, checkerboard titration assays were used on wild-type strain E. coli using an antibiotic (Rifampicin), which is considered a non-substrate of the AcrAB TolC pump and will not be transported outside the pump, to measure their effect on rifampicin and confirm that the compounds act specifically on the target efflux system. The change in the MIC of rifampicin could be indicated as an indirect activity such as permeabilisation of the inner or outer membranes or possessing off-target effects (Ohene-Agyei et al., 2014).
5.5 Inhibition of substrate transport
Substrate efflux assays provide an effective approach to directly measuring the inhibitory activity of EPI. These assays are performed using different amounts of tested compounds (50 & 100 µM) on pump-deleted strains (-AcrB), and intact cells (+AcrB), which often use bacteria recombinantly over-expressing an efflux pump (Venter et al., 2015; Ohene-Agyei et al., 2014). A fluorescent substrate, such as Nile Red, is employed for the measurement of the activity of the efflux of AcrAB-TolC. Nile Red accumulates inside cells and is fluorescent in hydrophobic environments, but its fluorescence is quenched as it is exported into an aqueous environment (Bohnert et al., 2010). The difference in fluorescence is used to monitor substrate efflux in real time and, hence, presents an ideal way to directly observe the inhibition of substrate efflux (Fig. 6).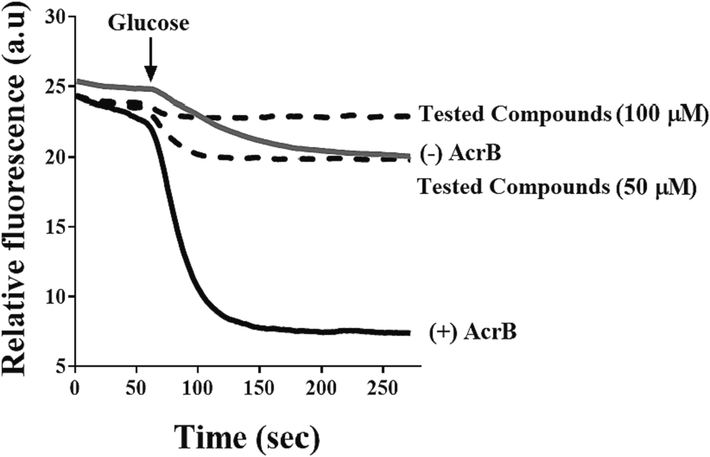
The picture shows an example of the inhibition of Nile Red efflux through AcrB protein. Wild-type-resistant cells expressing AcrB (black line), devoid of AcrB (grey line), while the dotted lines indicate AcrB-expressing cells with Nile Red that were added to them the tested compounds. At 60 secs, the measurement of the efflux was started by adding 0.2% glucose (black arrow). This assay is repeated three times with different batches of cells to obtain precise results. Adapted from Wang et al. (2021). *(This experiment and some following experiments have been done by our group (Wang et al., (2021), where I was the first co-author in this work).
5.6 Testing of inner membrane permeabilisation
Another important property of an ideal inhibitor is to not disrupt the inner membrane of a bacterial cell, as this could also be an indication of cytotoxicity. Many fluorescent dyes can be employed by measuring the ΔΨ component of the proton motive force (pmf) to confirm the integrity of the inner membrane. The reagent 3,3′ diethyloxacarbocyanine iodide (DiOC2(3)), is considered a commonly used fluorescent membrane potential probe used to confirm the integrity of the inner membrane as well. (Venter et al., 2003; Venter et al., 2015; Ohene-Agyei et al., 2014). When DiOC2(3) is added to bacteria, the bacteria show an increase in fluorescence when the pmf is established by adding glucose. If the inner membrane is disrupted by a test compound, it will lead to a decrease in fluorescence, as the cells are unable to establish a chemical gradient. The untreated cells are compared to treated cells to know if the tested compounds have disrupted the inner membrane (Fig. 7).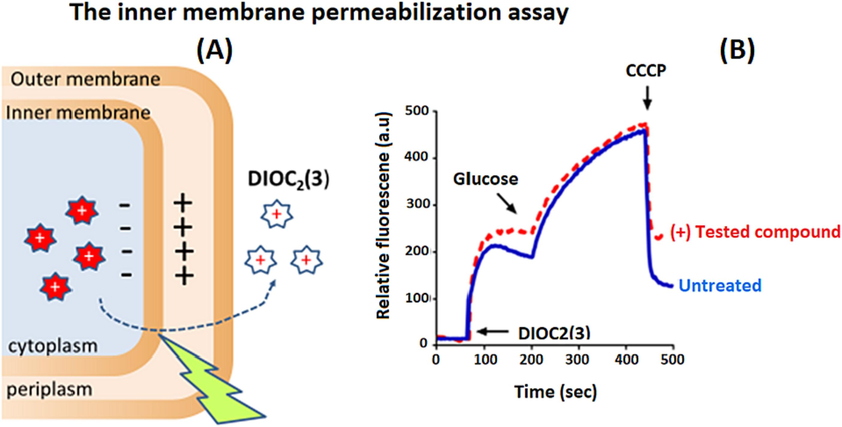
The figure shows an example of how the tested compounds affect pmf over the inner membrane, while the solid blue line indicates untreated bacterial suspensions, whereas the broken red line indicates the amounts of tested compounds (8–128 µg/mL) that were added to suspensions of bacteria for 10 min after that the potentiometric probe, DiOC2(3) was added, this is followed by monitoring the fluorescence until it has completely plateaued. To re-energize the cells, 0.5 % glucose is added followed by measuring the membrane potential establishment (inside negative), which refers to the increase in fluorescence until it has completely plateaued. The proton ionophore CCCP then is added to disrupt the membrane potential, which can be observed by a significant drop in the intensity of the fluorescence. To obtain precise results, these results should be done in three independent experiments with different batches of cells on different days. .
Adapted from Wang et al. (2021)
5.7 Testing of outer membrane permeabilisation
Similarly, an ideal EPI should not act off-target to disrupt the outer membrane. Nitrocefin hydrolysis assay could be used to measure outer membrane permeabilisation (Wang et al., 2018; Wang et al., 2017). Nitrocefin is a chromogenic beta-lactam that could be changed when it is hydrolyzed by adding periplasmic β-lactamase from yellow (∼380 nm) to red (∼490 nm). The nitrocefin diffuses fast through the outer membrane and comes into contact with periplasmic β-lactamase if the tested compound causes permeabilization of the outer membrane. In this matter, polymyxin B which uses as a known membrane disruptor and positive permeabilization control. The rate of nitrocefin hydrolysis could be measured spectrophotometrically after incubation of the cells with EPIs and compared to untreated cells and cells treated with polymyxin B (Lomovskaya and Bostian, 2006; Ohene-Agyei et al., 2014) (Fig. 8).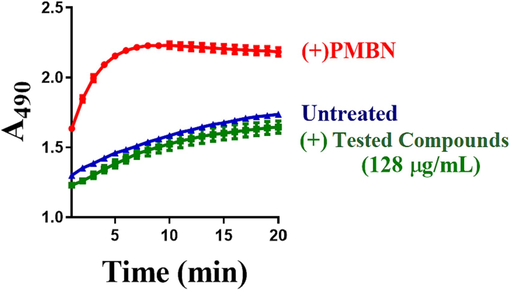
The figure shows whether the tested compounds have an effect on the permeability of the outer membrane. Bacterial cells are treated with 10 µM CCCP to inhibit the efflux of nitrocefin. Nitrocefin is added to either wild-type cells that did not receive the tested compounds (blue triangles), or treated cells with the test compounds (green squares), while red circles indicate cells that were treated with polymyxin B (PMBN) which uses as a positive permeabilization control and membrane disruptor. At 490 nm, the periplasmic β-lactamase causes an increase in the absorbance leading to an observed increase in the hydrolysis of Nitrocefin. To obtain precise results, these results should be done on three independent experiments with different batches of cells. .
Adapted from Wang et al. (2021)
6 Testing the cytotoxicity of the EPIs candidate compounds on mammalian cells
Having identified compounds with bioactivity in microbiological assays, the next step will be to test their efficacy in animal models. Before commencing such experiments, however, it is important to first establish the compounds are likely to be well tolerated in vivo and are devoid of acute toxicity. This will first be tested in vitro using cytotoxicity assays on mammalian cell lines. It is proposed that those compounds that do damage the bacterial inner membrane are likely to also damage mammalian structures such as the cellular or mitochondrial membranes, but this does need to be verified. Cytotoxicity assays are considered essential for any compound being considered for in vivo studies to confirm safety (Abraham et al., 2008). This assay can be performed using commercial kits such as the Real-time Glo MT Cell Viability assay kit from Promega. If the tested compounds have no cytotoxic activity, the cells remain metabolically active and produce a chromogenic product that can be measured spectrometrically. If the compounds have toxic activity, the cells do not produce a measurable product (Wang et al., 2017). An expanded panel of mammalian cell lines should be tested derived from organs that are important for the metabolism and excretion of the test compounds or derivatives thereof, such as the liver (HepG2) and kidney (HEK293) (Wang et al., 2017). These assays provide a rapid way to readily identify toxic compounds and eliminate them from a project before animal testing.
7 Conclusion
In conclusion, using inhibitors targeting the actions of drug efflux pumps is one of the most important solutions proposed to eliminate the multiple antibiotic resistance in many pathogenic bacteria, especially Gram-negative ones. This review discussed the standards required for inhibitors of efflux pumps, RND efflux pumps AcrAB-TolC that belongs to E. coli, and the problems facing the use of inhibitors such as their effect on the internal or external membrane of bacteria. One of the suggestions discussed in the review is that when searching for new inhibitors, we should take into account the size and chemical elongation of the possible compounds. This will help to identify new derivatives with extended bonding and make higher levels of inhibition. Crucially, the inhibitors need to have a clearly defined mode of action, target the drug efflux pumps exclusively, be readily soluble and cellular permeable so they can penetrate bacterial cells, not cause any unintended side effects, and not damage the outer or inner membranes of bacteria, and not be harmful to mammalian cells. Moreover, this review suggests that the inhibitors should be active against a broad spectrum of Gram-negative bacteria. To find the mutations that led to the establishment of multi-drug resistance, this review also suggests conducting a thorough search of the genome sequence. One of the recommendations of this review is that ease some of the barriers that prevent potential inhibitors from being used as therapeutic aids after confirming their effectiveness and releasing them from potential toxicity.
8 Discussion
The literature reviewed above explores the difficulty of treating the infections related to MDR in Gram-negative infections. Unfortunately, only two novel families of antibiotics have been brought into clinical usage in the last 30 years, which highlights the difficulty in finding new solutions to combat infectious diseases (Lamut et al., 2019; Tyers and Wright, 2019; Pouch et al., 2019). This is a serious concern source, due to the high rates of mortality and morbidity worldwide caused by these infections (Pouch et al., 2019). Gram-negative bacteria possess a higher tolerance to antimicrobial risks than Gram-positive bacteria due to containing a thick outer membrane that acts as a barrier that prevents harmful substances from entering the bacterial cell, which makes negative bacteria difficult to treat (Slipski et al., 2018; Willyard, 2017). Likewise, Gram-negative have over-expression efflux pumps which can be used in order to face harmful chemicals such as antibiotics (Arzanlou et al., 2017; Du et al., 2018). Drug efflux pumps were recognized for their role in MDR, due to their responsibility to export a wide spectrum of antibiotics and harmful compounds, thereby contributing to bacterial virulence and biofilm formation. Drug efflux pumps are one of the main essential mechanisms of protecting bacteria from external dangers because of possessing the systems of efflux and the poly-specificity of drugs leading to extruding a wide spectrum of harmful compounds to the outside before reaching their targets (Slipski et al., 2018). Moreover, Gram-negative bacteria possess all types of efflux pumps; the most important of which is the efflux pumps of the Resistance-Nodulation-Division (RND) superfamily which has significance in a clinical field, to which the multi-drug efflux pump AcrAB-TolC belongs to it. AcrAB-TolC is considered an important example that can be representative of many efflux pumps because it has highly similar characteristics between it and many escape pumps, as well as the availability of its structural and functional data (Wang et al., 2017; Opperman et al., 2014; Törnroth-Horsefield et al., 2007). Importantly, these studies have revealed the important role of the hydrophobic trap's site located in DBP in inhibitor binding and pharmacological targeting having a main part in improving the designing and synthesizing of the inhibitors by using structure-guided design. Inhibition of efflux pumps by appropriate EPIs provides the most promising strategy to combat global MDR, as EPIs can (a) make good synergizing with antibiotics that are currently used and lost their efficacy, (b) revive the effectiveness of antibiotics that lost their affecting on pathogenic bacteria which become highly resistant, (c) contribute to reducing the emergence of new infectious pathogens, (d) decrease the infectivity of pathogens, and (e) leading to a decrease in the formation of highly drug-resistant biofilms.
This review suggests that to design suitable inhibitors to target AcrB, firstly it should know the structure of this protein clearly and understand its functions. This knowledge guided the small molecule inhibitors design and directed the biological experiments necessary to fully characterize these EPIs (Ruggerone et al., 2013). The mechanisms of binding and expulsion of substrates also the symmetric and asymmetric structures of trimeric of AcrB were defined by X-ray crystallography and cryo-electron microscopy (Murakami et al., 2002; Murakami et al., 2006; Seeger et al., 2006; Sennhauser et al., 2006; Zgurskaya, 2009). Together, these studies revealed that the protomers in the trimeric structure have three conformations – loose (L), tight (T), and open (O) – that play unique roles in the mechanism of functional rotation of AcrB to translocate substrates through the efflux machinery (Du et al., 2018; Ruggerone et al., 2013; Pos, 2009; Yamaguchi et al., 2015). Moreover, in these studies, it was discovered that AcrB has two well-defined substrate binding pockets, namely the proximal site accommodates high-molecular-mass drugs whereas the distal site accommodates low-molecular-mass drugs (Ababou & Koronakis, 2016; Nakashima et al., 2011). A switch loop separates the distal and proximal pockets and contributes to the movement of substrates between the two pockets, which is a unidirectional movement (Eicher et al., 2012; Nakashima et al., 2011; Vargiu & Nikaido, 2012; Jamshidi et al., 2018). Furthermore, the distal pocket contains a hydrophobic trap located in the main substrate-translocation channel and branches off from this channel. As described in Fig. 2, Structural biology has contributed significantly to a clear understanding of the AcrB's functions. Moreover, the discovery of the hydrophobic trap and the importance of its site in the binding process of the inhibitors contributed significantly to improving the design of small molecules that selectively target AcrB (Nakashima et al., 2013), which this review recommends to design suitable EPIs that can specifically target it due to its significant role in the binding process of the inhibitors.
Furthermore, the numerous aromatic amino acids that line the hydrophobic trap, located next to the DBP, have an essential role in the binding of poly-substrates via the interactions of the hydrophobic and π–π stacking (Nakashima et al., 2013; Sjuts et al., 2016). Of the suggested things substances or inhibitors that attach to this pocket either stop other substances from passing through the tunnel that is responsible for the translocation of substrates or prevent substrate translocation as well via blocking the conformational changes (Aron and Opperman, 2018; Vargiu et al., 2018). As a result, this discovery has significantly aided in the efficient design of inhibitors with high efficacy through the use of structure-guided design (Aron & Opperman, 2018). This finding will eventually help to precisely target the major drug efflux pump. Finding novel, safer, and non-toxic preclinical candidates with known and clear functions may be a significant advantage of this strategy. This is crucial since many EPIs in the literature frequently experience toxicity issues as a result of unwanted off-target effects. Additionally, it will help discover several types of inhibitors, hence enhancing the pool of substances available for chemical optimization programs Fig. 9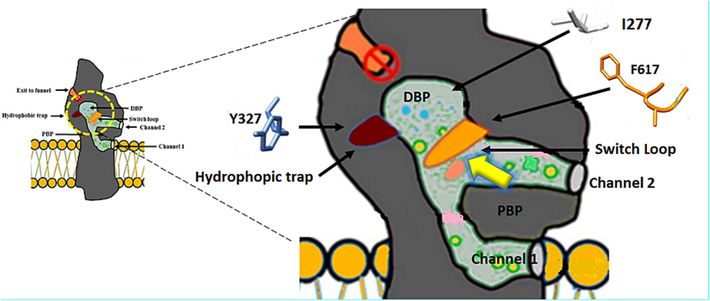
Schematic diagram illustrating the hydrophobic trap, which is represented by (Y327, which lies deep inside the trap), and its position which is located between the distal binding pocket (DBP) and the proximal binding pocket (PBP). Also, the diagram exhibits the switch loop, which is represented by (F617), its location between the PBP and DBP, and its function which is the separation between PBP and DBP. The diagram shows also I277 which is located in the tunnel of translocation. Whilst the yellow arrow represents the direction of the movement of the substrates inside AcrB.
In silico docking studies have contributed to understanding the molecular basis of binding and inhibition, however, these studies have not been followed up with the common co-complex structures of compounds that bind to AcrB yet. Moreover, amino acids such as Tyr 327, Met 573, and Val 517 which line the hydrophobic trap contribute significantly to forming deep contacts with compounds having elongated structures (Vargiu & Nikaido, 2012). These then protrude out into the substrate translocation tunnel towards amino acids Gln 151, Gly 179, and Ile 277 (Vargiu & Nikaido, 2012). By partially or completely occupying the tunnel, the inhibitors may sterically block the movement of multiple substrates as they transverse through AcrB, thus preventing the movement of substrates. Thus, one of the main strategies that should be taken in the future designing of inhibitors is 1) the compounds' length and 2) the compounds should fully span along the hydrophobic trap. Furthermore, the ability to bind both ends of the binding site must contribute to potency. Therefore, to find high-efficiency inhibitors to fully inhibit the activity of drug efflux pumps in pathogenic bacteria, these inhibitors must be characterized by their compliance with the required criteria that were discussed above and pass the biological tests required as well, and work specifically and selectively on these pumps without having any off-target activities.
Acknowledgments
The author would like to thank the Deanship of Scientific Research at Shaqra University for supporting this work.
Funding
This research received no external funding.
Institutional review board statement
Not applicable.
Informed consent statement
Not applicable.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Structures of gate loop variants of the AcrB drug efflux pump bound by erythromycin substrate. PLoS One. 2016;11(7):e0159154-e160254.
- [Google Scholar]
- Reviving antibiotics: efflux pump inhibitors that interact with AcrA, a membrane fusion protein of the AcrAB-TolC multidrug efflux pump. ACS Infect. Dis.. 2017;3(1):89-98.
- [Google Scholar]
- Application of a High-Content Multiparameter Cytotoxicity Assay to Prioritize Compounds Based on Toxicity Potential in Humans. J. Biomol. Screen.. 2008;13(6):527-537.
- [Google Scholar]
- Drug Efflux Pump Inhibitors: A Promising Approach to Counter Multidrug Resistance in Gram-Negative Pathogens by Targeting AcrB Protein from AcrAB-TolC Multidrug Efflux Pump from Escherichia coli. Biology. 2022;11(9):1328.
- [Google Scholar]
- The hydrophobic trap—the Achilles heel of RND efflux pumps. Res. Microbiol.. 2018;169(7):393-400.
- [Google Scholar]
- Intrinsic, adaptive and acquired antimicrobial resistance in Gram-negative bacteria. Essays Biochem.. 2017;61(1):49-59.
- [Google Scholar]
- Testing physiologically relevant conditions in minimal inhibitory concentration assays. Nat. Protoc.. 2021;16(8):3761-3774.
- [Google Scholar]
- Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol.. 2014;13:42-51.
- [Google Scholar]
- Optimized Nile Red Efflux Assay of AcrAB-TolC Multidrug Efflux System Shows Competition between Substrates. Antimicrob. Agents Chemother.. 2010;54(9):3770-3775.
- [Google Scholar]
- In vitro activity of the novel siderophore cephalosporin, cefiderocol, in Gram-negative pathogens in Europe by site of infection. Clin. Microbiol. Infect.. 2022;28(3):447-e1.
- [Google Scholar]
- Characterization of substituted piperazines able to reverse MDR in Escherichia coli strains overexpressing resistance-nodulation-cell division (RND) efflux pumps. J. Antimicrob. Chemother.. 2022;77(2):413-424.
- [Google Scholar]
- Identification of binding sites for efflux pump inhibitors of the AcrAB-TolC component AcrA. Biophys. J .. 2019;116(4):648-658.
- [Google Scholar]
- General platform for systematic quantitative evaluation of small-molecule permeability in bacteria. ACS Chem. Biol.. 2014;9(11):2535-2544.
- [Google Scholar]
- Multidrug efflux pumps: structure, function and regulation. Nat. Rev. Microbiol.. 2018;16(9):523-539.
- [Google Scholar]
- Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc. Natl. Acad. Sci.. 2012;109(15):5687-5692.
- [Google Scholar]
- Antibiotic resistance: What is so special about multidrug-resistant Gram-negative bacteria? GMS Hygiene and Infection Control. 2017;12
- [CrossRef] [Google Scholar]
- Reducing bacterial antibiotic resistance by targeting bacterial metabolic pathways and disrupting RND efflux pump activity. Iberoamerican Journal of Medicine. 2022;4(1):60-74.
- [Google Scholar]
- Bacterial Multidrug Efflux Pumps at the Frontline of Antimicrobial Resistance: An Overview. Antibiotics. 2022;11(4):520.
- [Google Scholar]
- Mapping the dynamic functions and structural features of AcrB efflux pump transporter using accelerated molecular dynamics simulations. Sci. Rep.. 2018;8(1):10470.
- [CrossRef] [Google Scholar]
- Efflux pump inhibitors of clinically relevant multidrug resistant bacteria. Med. Res. Rev.. 2019;39(6):2460-2504.
- [Google Scholar]
- Practical applications and feasibility of efflux pump inhibitors in the clinic—A vision for applied use. Biochem. Pharmacol.. 2006;71(7):910-918.
- [Google Scholar]
- Mechanisms of antibiotic resistance. Virulence Mechanisms of Bacterial Pathogens 2016:481-511.
- [Google Scholar]
- Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419(6907):587-593.
- [Google Scholar]
- Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443(7108):173-179.
- [Google Scholar]
- Progress in alternative strategies to combat antimicrobial resistance: Focus on antibiotics. Antibiotics. 2022;11(2):200.
- [Google Scholar]
- Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature. 2011;480(7378):565-569.
- [Google Scholar]
- Structural basis for the inhibition of bacterial multidrug exporters. Nature. 2013;500:102.
- [CrossRef] [Google Scholar]
- Structure and mechanism of RND-type multidrug efflux pumps. Adv. Enzymol. Relat. Area Mol. Biol.. 2011;77:1-60.
- [Google Scholar]
- Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol. Rev.. 2012;36(2):340-363.
- [Google Scholar]
- Tackling Drug-Resistant Infections Globally: Final Report and Recommendations-The Review on Antimicrobial Resistance Chaired by Jim O’Neill. Wellcome Trust; HM Government: London, UK; 2016.
- Mutations in MexB that affect the efflux of antibiotics with cytoplasmic targets. FEMS Microbiol. Lett.. 2012;333(1):20-27.
- [Google Scholar]
- Phytochemicals increase the antibacterial activity of antibiotics by acting on a drug efflux pump. Microbiology Open. 2014;3(6):885-896.
- [Google Scholar]
- Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob. Agents Chemother.. 2014;58(2):722-733.
- [Google Scholar]
- Synergy tests by E test and checkerboard methods of antimicrobial combinations against Brucella melitensis. J. Clin. Microbiol.. 2005;43(1):140-143.
- [Google Scholar]
- Antimicrobial Resistance Traits and Resistance Mechanisms in Bacterial Pathogens. In: Antimicrobial Resistance. Singapore: Springer; 2022. p. :1-27.
- [Google Scholar]
- Clinically Relevant Chromosomally Encoded Multidrug Resistance Efflux Pumps in Bacteria. Clin. Microbiol. Rev.. 2006;19(2):382-3402.
- [Google Scholar]
- Pyridylpiperazine-based allosteric inhibitors of RND-type multidrug efflux pumps. Nat. Commun.. 2022;13(1):1-11.
- [Google Scholar]
- Drug transport mechanism of the AcrB efflux pump. Biochim. Biophys. Acta (BBA) – Proteins and Proteomics. 2009;1794(5):782-793.
- [Google Scholar]
- Multidrug-resistant Gram-negative bacterial infections in solid organ transplant recipients—Guidelines from the American Society of Transplantation Infectious Diseases Community of Practice. Clin. Transplant. 2019:e13594.
- [Google Scholar]
- Proposed amendments regarding the definitions of multidrug-resistant and extensively drug-resistant bacteria. Expert Rev. Anti Infect. Ther.. 2022;20(2):139-146.
- [Google Scholar]
- RND Efflux Pumps: Structural information translated into function and inhibition mechanisms. Curr. Top. Med. Chem.. 2013;13(24):3079-3100.
- [Google Scholar]
- The role of Gram-negative bacteria in skin and soft tissue infections. Curr. Opin. Infect. Dis.. 2022;35(2):95-102.
- [Google Scholar]
- Advancing bacteriophages as a treatment of antibiotic-resistant bacterial pulmonary infections. Curr. Opin. Pulm. Med.. 2022;28(3):225-231.
- [Google Scholar]
- Seeger, MA, Schiefner, A, Eicher, T, Verrey, F, Diederichs, K., Pos, KM 2006, 'Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism', Science, 313, vol. 313, no. 5791, pp. 1295-1298.
- Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol.. 2006;5(1):e7.
- [Google Scholar]
- Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proc. Natl. Acad. Sci.. 2016;113(13):3509-3514.
- [Google Scholar]
- Biocide selective TolC-independent efflux pumps in Enterobacteriaceae. J. Membr. Biol.. 2018;251(1):15-33.
- [Google Scholar]
- New roads leading to old destinations: efflux pumps as targets to reverse multidrug resistance in bacteria. Molecules. 2017;22(3):468.
- [Google Scholar]
- New antibiotics for multidrug-resistant bacterial strains: latest research developments and future perspectives. Molecules. 2021;26(9):2671.
- [Google Scholar]
- Crystal structure of AcrB in complex with a single transmembrane subunit reveals another twist. Structure. 2007;15(12):1663-1673.
- [Google Scholar]
- Drug combinations: a strategy to extend the life of antibiotics in the 21st century. Nat. Rev. Microbiol.. 2019;17(3):141-155.
- [Google Scholar]
- Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc. Natl. Acad. Sci.. 2012;109(50):20637-20642.
- [Google Scholar]
- Computer simulations of the activity of RND efflux pumps. Res. Microbiol.. 2018;169(7):384-392.
- [Google Scholar]
- The structural and functional study of efflux pumps belonging to the RND transporters family from gram-negative bacteria. Antibiotics. 2022;11(4):429.
- [Google Scholar]
- An ABC transporter with a secondary-active multidrug translocator domain. Nature. 2003;426(6968):866-870.
- [Google Scholar]
- RND-type drug efflux pumps from Gram-negative bacteria: molecular mechanism and inhibition. Front. Microbiol.. 2015;6:377.
- [Google Scholar]
- Venter, H 2019, 'Reversing resistance to counter antimicrobial resistance in the World Health Organisation’s critical priority of most dangerous pathogens', Bioscience Reports, vol. 39, no. 4, p. BSR20180474.
- Evaluation of a series of 2-napthamide derivatives as inhibitors of the drug efflux pump AcrB for the reversal of antimicrobial resistance. Bioorg. Med. Chem. Lett.. 2017;27(4):733-739.
- [Google Scholar]
- Design, synthesis and biological activity evaluation of novel 4-subtituted 2-naphthamide derivatives as AcrB inhibitors. Eur. J. Med. Chem.. 2018;143:699-709.
- [Google Scholar]
- Design and structural optimization of novel 2H-benzo [h] chromene derivatives that target AcrB and reverse bacterial multidrug resistance. Eur. J. Med. Chem.. 2021;213:113049
- [Google Scholar]
- Willyard, C 2017, 'The drug-resistant bacteria that pose the greatest health threats', Nature, vol. 543, no. 7643, pp. 15-15.
- World Health Organization – WHO, 2017 [viewed 27 February 2020]. WHO publishes list of bacteria for which new antibiotics are urgently needed [online]. Geneva: WHO. Available from: https://www.who.int/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed.
- Structural basis of RND-type multidrug exporters. Front. Microbiol.. 2015;6(327)
- [CrossRef] [Google Scholar]
- Phenotypic and molecular characteristics of the MDR efflux pump gene-carrying stenotrophomonas maltophilia strains isolated in Warsaw, Poland. Biology. 2022;11(1):105.
- [Google Scholar]
- Covalently linked AcrB giant offers a new powerful tool for mechanistic analysis of multidrug efflux in bacteria. J. Bacteriol.. 2009;191(6):1727-1728.
- [Google Scholar]







