Translate this page into:
Preparation of polyclonal antibodies against chemically synthesized ApxIA and ApxIVA toxins and their diagnostic efficacy in the experimentally injected mice
⁎Corresponding authors. huanghuajun2004@126.com (Huajun Huang), ahrar1122@yahoo.com (Ahrar Khan), pingliujx@163.com (Ping Liu) Pingliujx@163.com (Ping Liu) Pingliujx@163.com (Ping Liu)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Objectives
ApxIA and ApxIVA are the main virulence factors of Actinobacillus pleuropneumoniae (A. pleuropneumoniae). The preparation of their antibodies is of great significance for diagnosing and treating porcine pleuropneumonia.
Methods
This study used chemically synthesized ApxIA and ApxIVA peptides as immunogens to prepare polyclonal antibodies against ApxIA and ApxIVA. Their titers were determined by ELISA and Dot blotting. Histopathology of apparently infected organs was carried out. Finally, the immunofluorescence method detected the distribution of antigens in the visceral tissues of mice infected with ApxIA and ApxIVA toxins.
Results
The results showed that ApxIA and ApxIVA polyclonal antibodies with titer as high as 1: 200000 and 1: 50000, respectively, were successfully obtained. The immunofluorescence results showed that these polyclonal antibodies could accurately detect ApxIA and ApxIVA antigens in mouse lungs with strong specificity. Histopathologically, there were perialveolar hemorrhages, pulmonary emphysema, and sloughing of bronchiolar epithelium. There was vacuolation and infiltration of chronic inflammatory cells in the liver parenchymal tissue. Atrophy of intestinal villi and glands and sloughing of villus epithelium, and congestion. There was increased urinary space and congestion in the proximal convoluted tubules. Lymphoid follicles increased in size in the spleen, the germinal center enlarged, and congestion in surrounding lymphoid tissue were observed.
Conclusion
This study laid a foundation for the research and development of a rapid detection kit for A. pleuropneumoniae and provided conditions for diagnosing and treating porcine contagious pleuropneumonia.
Keywords
Actinobacillus pleuropneumoniae
ApxIA and ApxIVA toxins
Polyclonal antibodies
Immunofluorescence
1 Introduction
Porcine contagious pleuropneumonia is a highly contagious and lethal respiratory disease in pigs caused by Actinobacillus pleuropneumoniae (A. pleuropneumoniae) (Frey 2019; Hu et al., 2020). Swine of all ages are susceptible to the disease and are difficult to cure the infected animals and they die, with deaths as high as 100 % (Bossé et al., 2002; Shin et al., 2011). The disease was first discovered and reported by Pattison in the United Kingdom in 1957. It is reported that the disease is prevalent and broke out in most areas, such as the United States, Canada, Mexico, South America, South Korea, Japan, Taiwan Province of China, Australia, European countries, and so on. It has caused huge economic losses to the pig industry in various countries (Liu et al., 2017; Dos Santos et al., 2018).
Infectious pleural pneumonia caused by A. pleuropneumoniae is mainly related to virulence factors such as lipopolysaccharide (LPS), outer membrane protein (OMP), and hemolytic toxin (Apx) (Shin et al., 2011; Li et al., 2018). Apx is a toxic substance that causes serious damage to alveolar macrophages, monocytes and lymphocytes (Kim et al., 2019). ApxI has the strongest virulence and hemolytic activity of the four different types of Apx hemolytic toxins (ApxⅠ, ApxⅡ, ApxIII and ApxⅣ), but ApxⅠ cause cross-reaction with other actinomycetes such as porcine actinomycetes and rose actinomycetes, resulting in low accuracy in diagnosis and treatment of the disease (Kamp et al., 1997; Haque et al., 2021). All serotypes of A. pleuropneumoniae can secrete ApxⅣ. Still, the specificity that other actinomycetes or other strains cannot secrete, provides a basis for the differential diagnosis of porcine infectious pleuropneumonia and can assist the combined diagnosis of ApxⅠ, ApxII, and ApxIII toxins (Schaller et al., 1999). In many existing studies, ApxI and ApxIV are used as the starting point for the diagnosis of A. pleuropneumoniae, for example, a quantitative real-time PCR (qPCR) method for detecting ApxIVA gene of A. pleuropneumoniae has been established (Lin et al., 2018). However, the high titer antibodies of ApxI and ApxIV have not been applied to the related research of clinical diagnostic reagents and methods.
In recent years, the use of synthetic peptide immunogens as a means of generating a variety of target-specific antibodies has increased markedly (Trier et al., 2019). In this way, not only antibodies can be raised against novel gene products specific to the target site, but synthetic peptides can also be coupled to solid supports to generate affinity matrices for antibody purification (Avendaño et al., 2018; Rodrigues-da-Silva et al., 2019). Before designing peptides, the bioinformatics analysis of the target sequence can better determine the peptides that meet the requirements (Ripoll et al., 2017; Avendaño et al., 2018; Trier et al., 2019). The synthetic peptide antigens overcome the shortcomings of complex preparation of recombinant antigens, multiple non-specific structures, and difficulty in purification (Plewczynski et al., 2007; Lee et al., 2016). Research on Apx has a long history, and there are many reports about using the characteristics of ApxI and ApxIV to diagnose porcine pleuropneumonia. The technology for preparing antibodies from peptide immunogens has also been maturely used (Ripoll et al., 2017; Rodrigues-da-Silva et al., 2019). However, there are still gaps in the research on the use of synthetic peptide immunogens to make ApxIA and ApxIVA polyclonal antibodies.
Therefore, in this study, the polyclonal antibodies of ApxIA and ApxIVA were prepared by using chemically synthesized ApxIA and ApxIVA polypeptide immunogens. The titer of the polyclonal antibody was detected by ELISA and Dot blotting. The expression of antigen in the visceral tissues of mice infected with ApxIA and ApxIVA toxins was detected by Immunofluorescence method, and the sensitivity of the prepared polyclonal antibody was analyzed. It provides a basis for the application of ApxIA and ApxIVA polyclonal antibodies in the diagnosis of A. pleuropneumoniae and also provides a reference for the treatment of porcine pleuropneumonia.
2 Materials and methods
2.1 Experimental animals
Experimental animals (rabbits and mice) were kept in a pathogen-free environment and fed ad libitum. The procedures for the care and use of animals were approved by the Ethics Committee of the Institutional Animal Care and Use Committee of JXAU, China. All applicable institutional and governmental regulations concerning the ethical use of animals were followed. Each experiment was performed in accordance with the recommendations in the ARRIVE guidelines.
2.2 Bioinformatics analysis of ApxIA and ApxIVA
The amino acid sequence of ApxIA and ApxIVA (GenBank accession no. AF240779.1 for ApxIA and no. FJ848574.1 for ApxIVA, respectively) were obtained from National Center for Biotechnology Information (NCBI: https://www.ncbi.nlm.nih.gov/). and homology prediction was carried out by BLAST, a tool in NCBI database. The domains, signal peptides, transmembrane regions, and amino acid composition predictions of ApxIA and ApxIVA proteins were analyzed by using bioinformatics online software (Table 1). The Protean module in DNAStar software was used to predict the secondary structure, hydrophilicity, flexibility, accessibility and antigenicity of proteins.
Online software
Website
Application
NCBI
https://www.ncbi.nlm.nih.gov/
Obtain sequence domain
NCBI (CDD)
https://www.ncbi.nlm.nih.gov Structure/cdd/docs/cdd_scarch.html
Domain prediction
Signall-5.0
https://www.cbs.dtu.dk/services/SignalP/index.php
Signal peptide prediction
TMHMM 2.0
https://www.cbs.dtu.dk/services/TMHMM/
transmembrane region Prediction
ProtParam
https://web.expasy.org/protparam/
amino acid composition Prediction
SOPMA
https://npsa-prabi.ibcp.fr/NPSA/npsa_sopma.html
secondary structure Prediction
SWISS-MODEL
https://swissmodel.expasy.org/ interactive
tertiary structure prediction
2.3 Synthesis of ApxIA and ApxIVA characteristic antigen peptides
The artificially synthesized peptide particles of ApxIA and ApxIVA were synthesized by Shanghai doi:10.1016/j.polymer.2007.07.039. Automatic Synthesizer and coupled with keyhole limpet hemocyanin.
2.4 Preparation of ApxIA and ApxIVA polyclonal antibodies
Two male experimental Japanese big-eared white rabbits (about 2.5 kg), 8 weeks of age, were normally raised for 1 week and were labeled as rabbit 1 and rabbit 2.
For the first immunization, 0.7 mg of ApxIA polypeptide antigen and 1.4 mg of ApxIVA polypeptide antigen were mixed with an equal volume of Freund's complete adjuvant (Sigma, St. Louis, Mo, USA), and respectively injected into rabbits 1 and rabbits 2 via subcutaneous multipoint injections. After an interval of 12 days, a second immunization was performed by emulsifying half of the antigen polypeptide with an equal volume of Freund's incomplete adjuvant (Sigma, St. Louis, Mo, USA). The next several immunizations were injected every two weeks with a mixture of the same amount of antigen and adjuvant as the second immunization.
After each immunization, 1 mL of blood was collected from rabbit ear veins to obtain anti-ApxIA serum and anti-ApxIVA serum for ELISA and Dot blotting detection. When the antibody was detected, the antiserum of anti-ApxIA and anti-ApxIVA were isolated from the blood of immunized rabbit one week after the last immunizations were performed.
2.5 Titer determination of anti-ApxIA and anti-ApxIVA protein antiserum by ELISA and Dot blotting
ApxIA and ApxIVA antiserum were purified by antigen affinity purification using the peptides C-DRVKGLIDSLNQHTKSAAK and C-RNSVIDAGAGNDTVNGGNGDD, respectively. The above synthesized ApxIA and ApxIVA specific antigen peptides were used as antigens. ApxIA and ApxIVA polyclonal antibodies were used as primary antibodies, and the HRP-labeled goat anti-rabbit IgG (Wanliebio, Shenyang, China) was used as secondary antibodies. The titers of ApxIA and ApxIVA polyclonal antibodies were determined by indirect ELISA and Dot blotting.
2.6 Histopathology and immunofluorescence detection
The internal organs such as lung, heart, liver, intestine, kidney, and spleen were collected from the mice infected with ApxIA and ApxIVA toxins of A. pleuropneumoniae, respectively, fixed in 4 % paraformaldehyde (Bakr et al., 2020; Mujahid et al., 2021) and then made into paraffin blocks. The paraffin blocks enclosing visceral tissue were cut into pathological sections (Lin et al., 2021) and observed under the microscope after hematoxylin-eosin staining (Al-Samawy et al., 2020). After dewaxing the pathological sections, ApxIA and ApxIVA polyclonal antibodies were used as primary antibodies, and Cy3-conjugated Goat anti-Rabbit IgG (Servicebio, Wuhan, China) was used as secondary antibodies for immunofluorescence detection (Mousa et al., 2019; Chen et al., 2020).
3 Results
3.1 Peptides synthesis based on bioinformatics analysis of ApxIA and ApxIVA proteins
The ApxIA and ApxIVA amino acid sequences obtained from GeneBank were homology predicted by BLAST tools in NCBI, and the results showed that the amino acid sequences of ApxIA and ApxIVA were relatively conservative (Figs. 1-A1/B1). ApxIA and ApxIVA proteins have a large number of conserved domains (Figs. 1-A2/B2). In the 200–420 positions of the ApxIA amino acid sequence, there may be a transmembrane region (Figs. 1-A3), while ApxIVA protein does not have a transmembrane region (Figs. 1-B3). In the prediction of ApxIA and ApxIVA protein signal peptides by SignalP 5.0 Server online analysis software, neither of these two proteins had signal peptides (Figs. 1-A4/B4). The amino acid composition of ApxIA and ApxIVA proteins were predicted and analyzed online by ProtParam. The amino acid contents of ApxIA and ApxIVA proteins were Glycine (Figs. 1-A5) and Aspartic acid (Figs. 1-B5), respectively. The results of SOPMA's prediction of secondary structure of protein (Figs. 1-A6/B6) show that their secondary structure consists of α-helix (blue), extension chain (red), β-corner (green) and irregular crimp (purple). These structures account for 45.11, 19.37, 7.73 and 27.79% of ApxIA proteins, respectively. In the secondary structure of ApxIVA protein, they accounted for 26.37, 26.15, 8.98 and 38.50%, respectively. SWISS-MODEL was used to predict the tertiary structure of ApxIA and ApxIVA proteins, and the results were shown in Fig. 1A7/B7. In the prediction results of the Protean module in DNAStar software, the ApxIA and ApxIVA protein secondary structure predicted by Gamier-Robson, Chou-Fasman, and Eisenberg methods were almost the same as those predicted by SOPMA software (Figs. 1-A8/B8). The hydrophilic and flexible regions of ApxIA and ApxIVA proteins were evenly distributed in the proteins (Kyte-Doolittle method and Karplus-Schulz method in Figs. 1-A8/B8). The potential antigenic epitope sites of ApxIA and ApxIVA proteins were analyzed by Jameson-Wolf method. It was found that ApxIA and ApxIVA proteins contained many regions with high antigen index. In addition, taking the surface accessibility index > 1 as the selection criterion, multiple regions in ApxIA and ApxIVA proteins were more accessible (Emini method of Figs. 1-A8/B8). Based on the above results, the 7-25aa sequence of ApxIA protein and the 1367-1387aa sequence of ApxIVA protein showed hydrophilicity and good plasticity. The prediction results of antigen index and surface accessibility showed that the two fragments had great antigenicity (Figs. 1-A9/B9).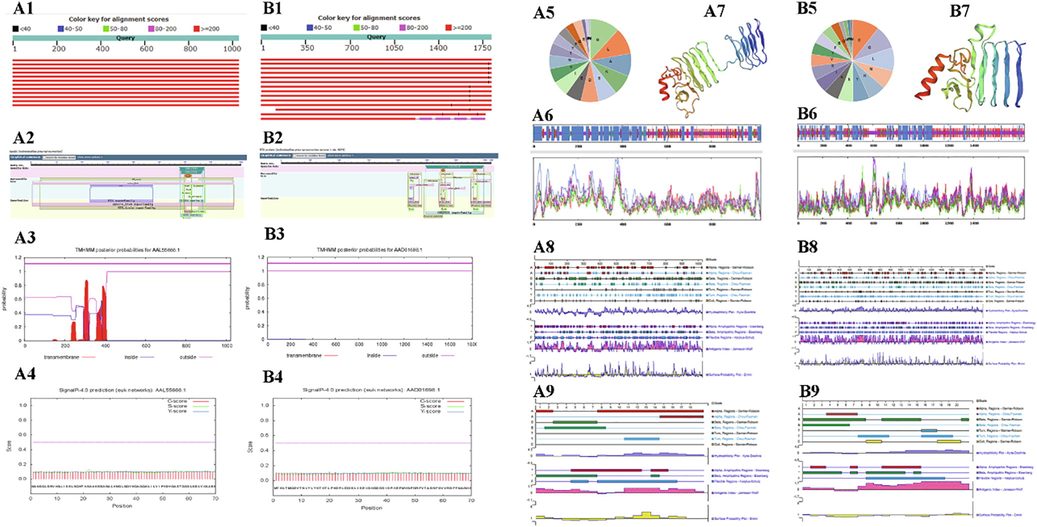
The letters A and B show the results of bioinformatics analysis of the amino acid sequences of ApxIVA and ApxIVA. A1 and B1: ApxIA and ApxIVA amino acid sequence specificity analysis. Red means. that the score is greater than or equal to 200. A2 and B2: CD-Search results for the protein sequence of ApxIA and ApxIVA. Functional sites are mapped to the query sequence and depicted as triangles. A3 and B3: Transmembrane domain of ApxIA and ApxIVA protein. The dark purple line represents the ApxIA and ApxIVA protein, the purple line below represents the outer cell membrane, and the blue line represents the inner cell membrane. A4 and B4: Signal peptide of ApxIA and ApxIVA protein. The red line represents the C score, the green line represents the S score, and the blue line represents the Y score. A5 and B5: Amino acid composition of ApxIA and ApxIVA protein. A6 and B6: The secondary structure of ApxIA and ApxIVA protein. H stands for α-helix and c for disordered crimped state. Blue represents α-helix; purple represents irregular crimping; red represents extension chain; green represents β-rotation. A7 and B7: Tertiary structure of ApxIA and ApxIVA protein. Red indicates α-helix, yellow indicates β-turning angle, green indicates Random coil, and blue indicates transmembrane region. A8 and B8: The predicted the secondary structure and physicochemical properties of the ApxIA and ApxIVA protein. A9 and B9: The predicted secondary structure and physicochemical properties of ApxIA and ApxIVA polypeptide fragments.
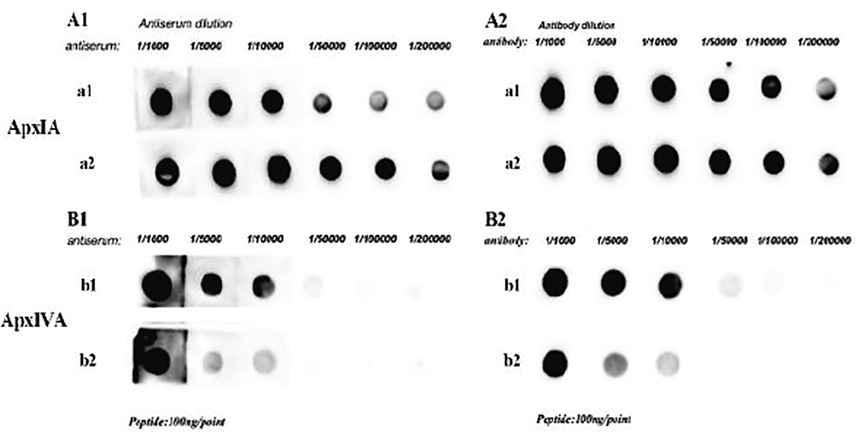
Titers of the prepared antiserum were determined by ELISA and Dot blotting. A1 and A2: ApxIA polyclonal antibodies have high titers detected by ELISA and Dot blotting, respectively. A1 and A2 are the results of ApxIA antiserum titers at concentrations 2.27 mg/mL and 2.50 mg/mL, respectively. B1 and B2: ApxIVA polyclonal antibodies have high titers detected by ELISA and Dot blotting respectively. B1 and B2 are the results of ApxIVA antiserum titers at concentrations 3.47 mg/mL and 1.80 mg/mL, respectively.
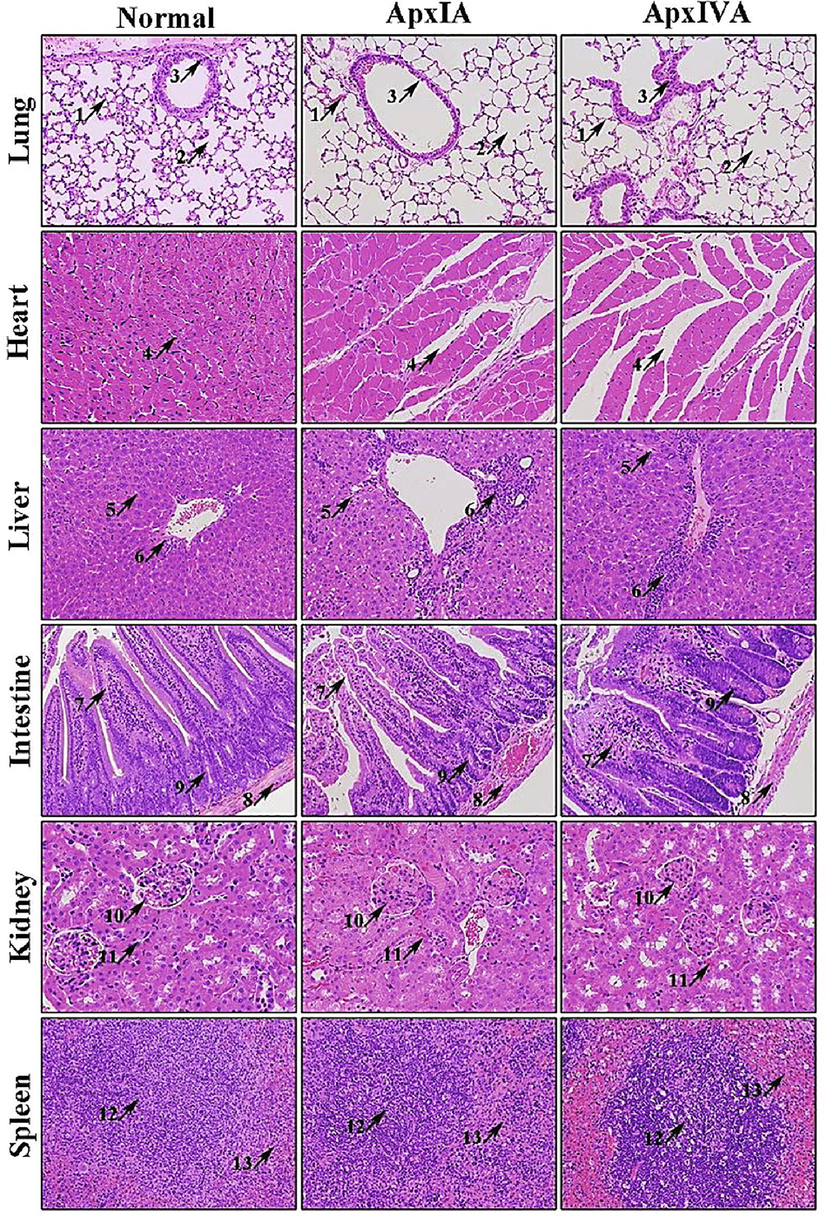
Histological sections of various organs of mice inoculated with ApxIA and ApxIVA toxins of Actinobacillus pleuropneumoniae. H & E Stain. 200x. Lungs: Thinning of alveolar wall and the perialveolar hemorrhage (Arrow 1), pulmonary emphysema (Arrow 2), and sloughing of bronchiole epithelium. Heart: Intercellular space increased (Arrow 4). Liver: Vacuolation (Arrow 5), and chronic inflammatory cells infiltration (Arrow 6). Intestines: Intestinal villi atrophied and sloughing of villi epithelium (Arrow 7), congestion of intestinal muscles (Arrow 8), and intestinal glands atrophied, and detached from the lamina propria (Arrow 9). Kidneys: Increased urinary space in glomeruli (Arrow 10), congestion in the proximal convoluted tubules (black arrow 11). Spleen: Number of lymphoid follicles increased, the germinal center enlarged (Arrow 12), and congestion in surrounding lymphoid tissue (Arrow 13).
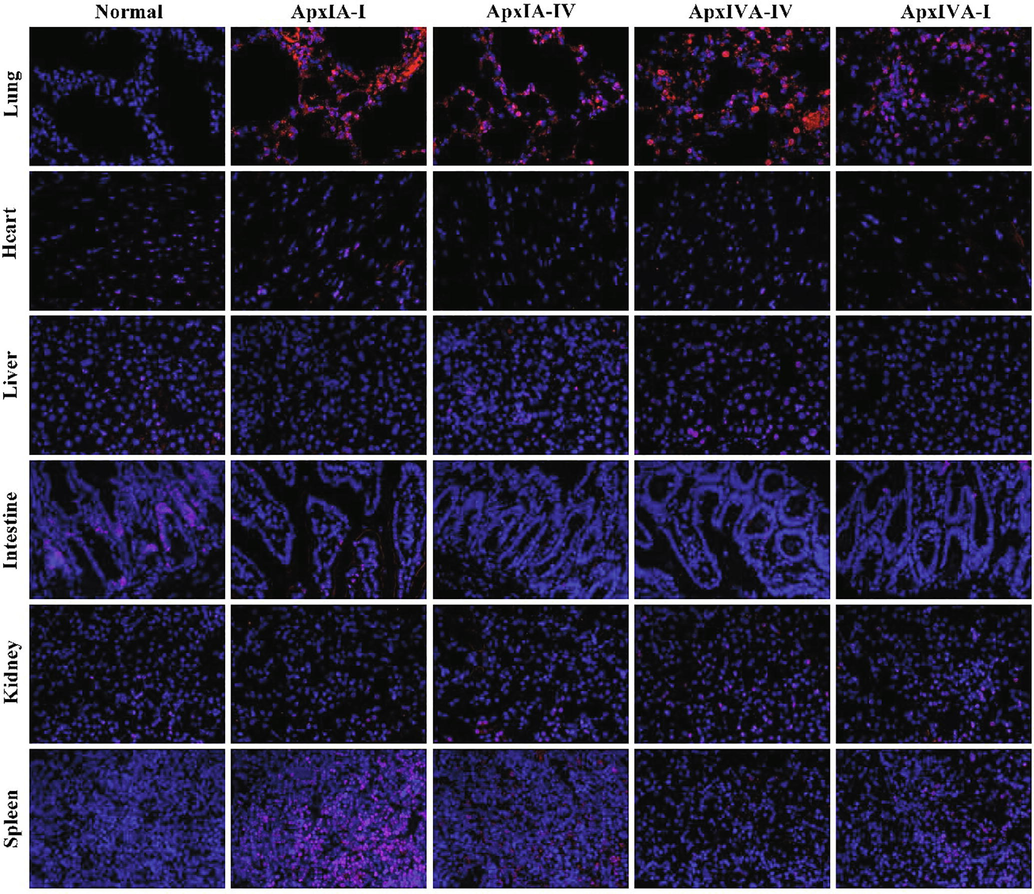
Immunofluorescence (IFC) was used to detect the distribution of ApxIA and ApxIVA in the visceral tissues of mice infected with the toxin. (×400). Normal: the lung, heart, liver, intestine, kidney, and spleen of normal mice were negative by immunohistochemical staining. Line ApxIA-I: visceral tissue sections of mice infected with ApxIA have incubated with ApxIA polyclonal antibody. and the red fluorescence pointed to ApxIA IFC positive cells. Line ApxIA-IV: visceral tissue sections of mice infected with ApxIA have incubated with ApxIVA polyclonal antibody, and the red fluorescence pointed to ApxIVA IFC positive cells. Line ApxIVA-IV: visceral tissue sections of mice infected with ApxIVA have incubated with ApxIVA polyclonal antibody, and the red fluorescence pointed to ApxIVA IFC positive cells. Line ApxIVA-I: visceral tissue sections of mice infected with ApxIVA have incubated with ApxIA polyclonal antibody, and the red fluorescence pointed to ApxIA IFC positive cells.
3.2 The titer of the prepared anti-ApxIA and anti-ApxIVA protein antiserum
The ApxIA and ApxIVA antiserum determined by ELISA and Dot blotting showed that the titer could be up to 1: 200000 and 1: 50000, respectively after immunizing five times (Fig. 2). The ApxIA and ApxIVA antiserum were isolated from blood of rabbit 1 and 2, respectively, and were purified to a1 (2.27 mg/mL), a2 (3.47 mg/mL), b1 (2.50 mg/mL), and b2 (1.80 mg/mL).
3.3 Histopathology
The pathological sections of mice infected with ApxIA and ApxIVA toxins showed various lesions (Fig. 3). In the lungs, alveolar epithelial cells decreased, the alveolar wall became thinner, the perialveolar hemorrhages (arrow 1), the alveolar cavity was highly dilated, the alveolar septum was ruptured, the adjacent alveolar cavity fused to form a large cystic cavity, i.e., pulmonary emphysema (arrow 2), and sloughing of bronchiolar epithelium (arrow 3). In the heart, the number of cardiomyocytes in mice infected with ApxIA and ApxIVA toxins decreased, and the intercellular space increased (arrow 4).
In the liver, vacuolation of hepatocytes of mice infected with ApxIA and ApxIVA toxins increased (arrow 5), and chronic inflammatory cell infiltration in the hepatic portal area (arrow 6). In the intestine, the intestinal villi of mice infected with ApxIA and ApxIVA toxins atrophied and sloughed of villi epithelium (arrow 7). The intestinal muscle layer was congested and decreased muscle cells (arrow 8). The intestinal glands at the root of the intestinal villi were atrophied and detached from the lamina propria (arrow 9).
In the kidneys of mice infected with ApxIA and ApxIVA toxins, increased urinary space in glomeruli (arrow 10), there was multiple congestion in the proximal convoluted tubules, and the wall cells decreased (arrow 11). In the spleen, the number of lymphoid follicles increased, the germinal center enlarged (arrow 12), and congestion of surrounding lymphoid tissue (arrow 13).
3.4 Immunohistochemical detection
The results of immunofluorescence detection showed that red fluorescence was detected only in the lungs of mice infected with ApxIA and ApxIVA toxins, and no fluorescence was found in other organs of the negative control group and the experimental group (Fig. 4).
4 Discussion
The preparation of high titer antibodies against ApxIA and ApxIVA is very important in the clinical diagnosis of A. pleuropneumoniae. We synthesized ApxIA and ApxIVA polypeptides as antigens by chemical synthesis, and successfully prepared ApxIA and ApxIVA polyclonal antibodies with titers as high as 1: 200000 and 1: 50000, respectively. In the visceral localization detection of mice infected with ApxI and ApxIV toxins, it was found that these two antibodies were highly expressed in the lungs.
The preparation of high titer antibody is closely related to the method of obtaining immunogens. The common immunogens are recombinant proteins and synthetic peptides, in which recombinant proteins are obtained from tissues and specific recombinant protein molecules are obtained by gene recombination technology, and this method is complex and costly (Dreyfus et al., 2004; Sevinc et al., 2015a; Sevinc et al., 2015b; Li et al., 2020; Kamel and El-Sayed, 2019). The synthetic peptides are synthesized chemically by selecting a suitable amino acid sequence, and then coupled with the carrier can be used as an immune source to prepare antibodies (Trier et al., 2019; Peng et al., 2020). It can be stated that synthetic peptides were used in this direction in the present study.
Polypeptide antibodies usually have high specificity and affinity and can be used in various analyses in the field of diagnosis to help diagnose infections and diseases, to make accurate quantification, or to identify or locate the presence of a given substance (Ripoll et al., 2017). In addition, it has been reported that Apx is thermostable, and it is difficult to obtain bioactive natural Apx after purification (Chang et al., 2014). Therefore, it is hard to synthesis ApxIA and ApxIVA proteins by gene recombination to prepare antibodies, and a better choice is to prepare antibodies with synthetic peptides. Our work also proved that the polypeptides synthesized by this method can produce high titer polyclonal antibodies against ApxIA and ApxIVA, and the synthetic polypeptides have a good purification effect on the prepared polyclonal antibodies.
The high titer and specificity of peptide antibodies are often determined by the characteristics of peptide sequences. Generally speaking, the most ideal peptide sequence should have the characteristics of hydrophilicity, protein surface and structural deformability, and so on (Chiang et al., 2009; To et al., 2016; Wu et al., 2018). In our study, the polypeptide C-RVKGLIDSLNQHTKSAAK was synthesized from the 7-25aa of the ApxIA amino acid sequence, and the polypeptide C-RNSVIDAGAGNDTVNGGNGDD was synthesized from the 1367-1387aa of the ApxIVA amino acid sequence. These two sequences have a large number of linear epitopes and have good specificity and hydrophilicity. The linear antigen epitope is the structure that binds specifically to the antibody, which determines the specificity of the antigen. As we all know, the specificity of antigen is the most prominent feature of immune response and the theoretical basis of immunological diagnosis and prevention (Saravanan and Gautham, 2015). Our study supports and considerably data showing the peptide sequence has a large number of linear epitopes and strong specificity, which increases the probability of antigen–antibody binding, which is conducive for the preparation of more polyclonal antibodies. Hydrophilicity is often considered in the design of peptide antigens because hydrophilic sequences are easier to dissolve and easier to synthesize (Trier et al., 2019).
The specific antibody can quickly and accurately capture the antigen that can react with it in the animal body. In our study, pathological observation showed that the lungs of mice infected with ApxIA and ApxIVA toxins were seriously damaged, and their alveolar walls became thinner or even ruptured, resulting in an enlarged alveolar cavity. Cytopenia and internal structural changes also occurred in other visceral tissues. However, when the prepared polyclonal antibody was used in immunofluorescence localization, we found that it was only positive in the lungs of mice infected with ApxIA and ApxIVA toxins. Studies have shown that the target organ of A. pleuropneumoniae is the lung, which rarely infects other organs, but is often accompanied by pathological phenomena such as pericarditis, hepatitis and splenomegaly (Zuo et al., 2013; Wang et al., 2016; Li et al., 2019; Nahar et al., 2021; Dos Santos et al., 2018). This is consistent with our pathological observation and shows that it is reasonable for us to localize ApxIA and ApxIVA antigens in the lungs with polyclonal antibodies, and the polyclonal antibodies ApxIA and ApxIVA prepared by us are more specific.
5 Conclusion
In conclusion, the polyclonal antibodies against ApxIA and ApxIVA with good specificity can be prepared by using synthetic ApxIA and ApxIVA peptides. This study provides a method for the preparation of polyclonal antibodies against ApxIA and ApxIVA with high titer. The results of ELISA, Dot blotting and immunofluorescence showed that the antibodies provided an effective tool for determining the localization and expression level of ApxIA and ApxIVA antigens under different pathophysiological conditions and laid a foundation for the diagnosis of A. pleuropneumoniae.
6 Novelty statement
As far as the authors know, there is no report on the preparation of ApxIA and ApxIVA polyclonal antibodies from chemically synthesized ApxIA and ApxIVA polypeptide antigens of Actinobacillus pleuropneumoniae. Although the method of preparing antibodies from chemically synthesized peptide antigens is widely used, there is little literature on the preparation of antibodies using synthetic peptides of ApxIA and ApxIVA as antigens. The preparation of ApxIA and ApxIVA antibodies from synthetic peptides is simpler and faster than that of traditional recombinant proteins as antigens to prepare ApxIA and ApxIVA antibodies. The preparation of antibodies against ApxIA and ApxIVA high valence peptides is of great significance for the rapid diagnosis and treatment of porcine pleuropneumonia.
CRediT authorship contribution statement
Qingqing Li: Software, Writing – original draft, Writing – review & editing, Validation. Sufang Cheng: Data curation, Validation. Guyue Li: Methodology, Formal analysis. Pei Liu: Writing – original draft. Zhenxing Zou: Methodology, Formal analysis. Xiaolu Hou: Writing – original draft. Vincent Latigo: Writing – original draft. Lin Li: Methodology. Xiaoquan Guo: Methodology, Visualization, Resources, Funding acquisition. Guoliang Hu: Validation, Resources, Funding acquisition. Huajun Huang: Software, Writing – review & editing. Ahrar Khan: Software, Writing – review & editing. Ping Liu: Software, Conceptualization, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Acknowledgement
This project was supported by the National Natural Science Foundation of China grant (No. 31960723, Beijing, P. R. China) awarded to PL, the Natural Science Foundation of Jiangxi Province grant (No. 20171ACB21026; 2017ACB20012) awarded to PL, the Technology R&D Program of Jiangxi Province grant (No. GJJ170243, Nanchang, P. R. China) awarded to PL, the Special funds for postgraduates' Innovation in Jiangxi Province (No. YC2019-S174) and the Innovative Entrepreneurship training Program for College students in Jiangxi Province (No. 201910410026).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Histo-Morphometric compression finding of the small intestine in rats and rabbit according to different foods. Int. J. Vet. Sci.. 2020;9:433-437.
- [CrossRef] [Google Scholar]
- Cryptosporidium spp. CP15 and CSL protein-derived synthetic peptides’ immunogenicity and in vitro seroneutralisation capability. Vaccine.. 2018;36(45):6703-6710.
- [Google Scholar]
- Ginger extract and ginger nanoparticles; characterization and applications. Int. J. Vet. Sci.. 2020;9:203-209.
- [CrossRef] [Google Scholar]
- Actinobacillus pleuropneumoniae: pathobiology and pathogenesis of infection. Microbes Infect.. 2002;4:225-235.
- [CrossRef] [Google Scholar]
- Elucidating the role of ApxI in hemolysis and cellular damage by using a novel apxIA mutant of Actinobacillus pleuropneumoniae serotype 10. J. Vet. Sci.. 2014;15:81.
- [CrossRef] [Google Scholar]
- Expression of canine kynurenine 3-monooxygenase by baculovirus for canine mammary tumor diagnosis. Pak. Vet. J.. 2020;40:145-150.
- [Google Scholar]
- Immunogenicity and protective efficacy of ApxIA and ApxIIA DNA vaccine against Actinobacillus pleuropneumoniae lethal challenge in murine model. Vaccine.. 2009;27:4565-4570.
- [CrossRef] [Google Scholar]
- Dos Santos, L.F., Costa Polveiro, R., Scatamburlo Moreira, T., Pereira Vidigal, P.M., Chang, Y.F., Scatamburlo Moreira, M.A., 2018. Polymorphism analysis of the apxIA gene of Actinobacillus pleuropneumoniae serovar 5 isolated in swine herds from Brazil. PloS One. 13 12 2018 10.1371/journal.pone.0208789 e0208789 e0208789.
- Use of recombinant ApxIV in serodiagnosis of Actinobacillus pleuropneumoniae infections, development and prevalidation of the ApxIV ELISA. Vet. Microbiol.. 2004;99(3–4):227-238.
- [CrossRef] [Google Scholar]
- RTX toxins of animal pathogens and their role as antigens in vaccines and diagnostics. Toxins.. 2019;11:719.
- [CrossRef] [Google Scholar]
- Pathogenicity of feed-borne Bacillus cereus and its implication on food safety. Agrobiol. Rec.. 2021;3:1-16.
- [Google Scholar]
- Development of a sybr green real-time PCR assay with melting curve analysis for simultaneous detection of Actinobacillus pleuropneumoniae and Haemophilus parasuis. Kafkas Univ. Vet. Fakult. Dergisi.. 2020;26:665-670.
- [CrossRef] [Google Scholar]
- Utilization of herpesviridae as recombinant viral vectors in vaccine development against animal pathogens. Virus Res.. 2019;270:197648
- [CrossRef] [Google Scholar]
- Endobronchial inoculation with Apx toxins of Actinobacillus pleuropneumoniae leads to pleuropneumonia in pigs. Infect. Immun.. 1997;65(10):4350-4354.
- [CrossRef] [Google Scholar]
- Global gene networks in 3D4/31 porcine alveolar macrophages treated with antigenic epitopes of Actinobacillus pleuropneumoniae ApxIA, IIA, and IVA. Sci. Rep.. 2019;9(1):5269.
- [CrossRef] [Google Scholar]
- Antibody production with synthetic peptides. Meth. Mol. Biol. (Clifton, N.J.).. 2016;1474:25-47.
- [CrossRef] [Google Scholar]
- Activation of porcine alveolar macrophages by Actinobacillus pleuropneumoniae lipopolysaccharide via the toll-like receptor 4/NF-κB-mediated pathway. Infect. Immun.. 2018;86:e00642-e717.
- [CrossRef] [Google Scholar]
- Exploring the potential STAT3 gene in broiler with ascites syndrome by bioinformatics analysis. Agrobiol. Rec.. 2020;2:24-30.
- [CrossRef] [Google Scholar]
- A requirement of TolC1 for effective survival, colonization and pathogenicity of Actinobacillus pleuropneumoniae. Microb. Pathog.. 2019;134:103596
- [CrossRef] [Google Scholar]
- Detection of Actinobacillus pleuropneumoniae through duplex PCR based on ApxIA and ApxIVA genes. Pak. Vet. J.. 2018;38:276-280.
- [CrossRef] [Google Scholar]
- Evaluation of immunogenicity and protective efficacy of Eimeria maxima immune mapped protein 1 with EDA adjuvant in chicken. Pak. Vet. J.. 2021;41:209-213.
- [Google Scholar]
- An anti-propionibacterium acnes antibody shows heterologous resistance to an Actinobacillus pleuropneumoniae infection independent of neutrophils in mice. Immunol. Res.. 2017;65:1124-1129.
- [CrossRef] [Google Scholar]
- Comparative serological, histopathological and immunohistochemical evaluation of immune status of broiler chickens experimentally infected with velogenic Newcastle disease virus in different ages. Int. J. Vet. Sci.. 2019;8:143-150.
- [Google Scholar]
- Allethrin induced toxicopathological alterations in adult male albino rats. Agrobiol. Rec.. 2021;5:8-14.
- [CrossRef] [Google Scholar]
- Actinobacillus pleuropneumoniae: The molecular determinants of virulence and pathogenesis. Adv. Microb. Physiol.. 2021;78:179-216.
- [CrossRef] [Google Scholar]
- Serovars, antimicrobial susceptibility and molecular characteristics of Haemophilus parasuis isolates in southern China. Kafkas Univ. Vet. Fakultesi Dergisi. 2020;26:491-497.
- [CrossRef] [Google Scholar]
- The RPSP: Web server for prediction of signal peptides. Polymer.. 2007;48(19):5493-5496.
- [CrossRef] [Google Scholar]
- Combinatorial peptide-based epitope mapping from Ebola virus DNA vaccines and infections reveals residue-level determinants of antibody binding. Hum. Vacc. Immunother.. 2017;13:2953-2966.
- [CrossRef] [Google Scholar]
- Immunogenicity of synthetic peptide constructs based on PvMSP9E795-A808, a linear B-cell epitope of the P. vivax Merozoite Surface Protein-9. Vaccine.. 2019;37(2):306-313.
- [Google Scholar]
- Harnessing computational biology for exact linear b-cell epitope prediction: A novel amino acid composition-based feature descriptor. Omics.. 2015;19:648-658.
- [CrossRef] [Google Scholar]
- Characterization of apxIVA, a new RTX determinant of Actinobacillus pleuropneumoniae. Microbiology (Reading, England).. 1999;145(Pt 8):2105-2116.
- [CrossRef] [Google Scholar]
- Identification and expression of Babesia ovis secreted antigen 1 and evaluation of its diagnostic potential in an enzyme-linked immunosorbent assay. J Clin. Microbiol.. 2015;53(5):1531-1536.
- [Google Scholar]
- A new immunoreactive recombinant protein designated as rBoSA2 from Babesia ovis: Its molecular characterization, subcellular localization and antibody recognition by infected sheep. Vet. Parasitol.. 2015;214:213-218.
- [CrossRef] [Google Scholar]
- An immunosorbent assay based on the recombinant ApxIA, ApxIIA, and ApxIIIA toxins of Actinobacillus pleuropneumoniae and its application to field sera. J. Vet. Diagn. Invest.. 2011;23:736-742.
- [CrossRef] [Google Scholar]
- Genetic and antigenic characteristics of ApxIIA and ApxIIIA from Actinobacillus pleuropneumoniae serovars 2, 3, 4, 6, 8 and 15. Microbiol. Immunol.. 2016;60:447-458.
- [CrossRef] [Google Scholar]
- Peptides, antibodies, peptide antibodies and more. Int. J. Mol. Sci.. 2019;20:6289.
- [CrossRef] [Google Scholar]
- Recombinant ApxIV protein enhances protective efficacy against Actinobacillus pleuropneumoniae in mice and pigs. J. Appl. Microbiol.. 2018;124:1366-1376.
- [CrossRef] [Google Scholar]
- Transcriptional profiling of swine lung tissue after experimental infection with Actinobacillus pleuropneumoniae. Int. J. Mol. Sci.. 2013;14:10626-10660.
- [CrossRef] [Google Scholar]







