Translate this page into:
Prefoldin proteins 2/6, and HMG20B are regulated by HDAC1, HDAC3 and are novel therapeutic and prognostic biomarkers in hepatocellular carcinoma
⁎Corresponding author. fmuhammad@ksu.edu.sa (Muhammad Farooq Khan)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Epigenetic mechanisms, such as histone deacetylases (HDACs), play an important role in the commencement and development of hepatocellular carcinoma (HCC). Previously, we have identified proteins with binds with HDAC1 and HDAC3 in liver cancer cells and also have shown that depletion of either HDAC1 or HDAC3 suppressed the expression of HDAC1/3 interacting proteins, including the prefoldin protein 2/6 (PFDN2/6), CR4-NOT transcription complex subunit 1 (CNOT1), and high mobility group 20B (HMG20b). In this study, online databases were utilized to analyze the expression of HDAC1/3, PFDN2/6, CNOT1, and HMG20b in a large panel of liver cancer cell lines, cancer tissues, and human normal and tumor liver tissues. These databases are “RNA Expression Atlas (https://www.ebi.ac.uk/gxa/home), Cancer Genome Atlas (TCGA), gene expression profiling interactive analysis (GEPIA), integrative molecular databases of hepatocellular carcinoma (HCCDB), human protein atlas, and Kaplan-Meier Plotter for RNA sequences in liver cancer (http://kmplot.com/analysis/index.php?p=service&cancer=liver_rnaseq#). The expression of these genes was verified experimentally in human HepG2 cells via semi-quantitative RT PCR. The results showed that all these genes are expressed in twenty-three human liver cancer cell lines, higher expression in human liver cancer than normal tissues. However, HDAC1 and PFDN2 are expressed at higher levels than other genes analyzed in this study. The analysis of these six genes in 364 human liver cancer patients by Kaplan Meier plotter predicted HDAC1 and PFDN2 as poor, while HMG20b as a favorable prognostic biomarker in hepatocellular carcinoma. PFDN2 and HMG20b are novel prognostic markers of hepatocellular carcinoma, identified first in this study. Further clinical studies are needed to verify the expression and patient survival concerning the expresion of PFDN2 and HMG20b in human hepatocellular carcinoma patients.
Keywords
Histone deacetylases
Prefolding proteins
Hepatocellular carcinoma
Prognostic value
1 Introduction
The management of any cancer relies on validating biomarkers to predict cancer patients' recurrence and survival. Suitable treatment approaches may be adopted for an individual patient based on the expression of certain prognostic factors (Baylot et al., 2020). These prognostic factors are essential for predicting patients' survival outcomes, and they provide a guideline for deciding therapeutic applications according to risk. Hepatocellular carcinoma (HCC) is the predominant liver cancer, accounting for around 6% of newly diagnosed cancers worldwide. Clinically, many advances have been made to treat HCC; however, its prognosis remains poor (Petrizzo et al., 2018; Qin and Tang, 2002).
Alpha-fetoprotein (AFP) was primarily used as a serum biomarker to diagnose HCC in clinical practice (Zhu et al., 2019). However, the specificity of AFP is undetermined, and the elevation of AFP has been observed in other tumors and conditions (Wong et al., 2015). Following these discoveries, serum osteopontin (OPN) is another serum biomarker that has been proposed for the diagnosis of HCC. Moreover, while OPN is associated with poor prognosis and dedifferentiated HCC (Pan et al., 2003), the accuracy of OPN at early tumor stages of HCC detection is not certain. Insulin growth factor-1 (IGF-1) has also been proposed to detect patients with an advanced HCC diagnosis but has not been implemented in daily practice (Pinero et al., 2020). It should be noted that elevated Glypican-3 (GPC3) levels have been reported in HCC, specifically (Tremosini et al., 2012), and GPC3 is considered a poor prognostic biomarker for HCC. However, the prognostic values of GPC3 were significantly varied when evaluating the overall survival and disease-free survival status of HCC patients (Liu et al., 2018). More specifically, compared to AFP, GPC3 showed a medium level of accuracy (Hippo et al., 2004). Besides these, other biomarkers, such as vascular endothelial growth factor (VEGF), angiopoietin 2 (ANG-2), Golgi protein 73 (Gp-73), hepatic growth factor (HGF), and c-MET, have also been proposed as serum biomarkers to diagnose HCC or as prognostic biomarkers for HCC. Most of these biomarkers have shown poor prognostic potential, either in early or advanced HCC, and most of the suggested biomarkers are not used in clinical practice. Therefore, the real challenge is developing novel biomarkers that can be used for early diagnosis, adequate treatment, and post-treatment prognosis.
With new advancements in molecular biology and fields related to cancer biology, differential gene expression in HCC has been meticulously studied. For instance, the molecular factors related to cancer cell invasion, metastasis, recurrence, and survival factors related to HCC have been explored. Post-translational modification of histones facilitates the number of biological processes via chromatin modification, resulting in the expression or repression of target genes. Their misregulation in cancer can result in the inappropriate activation of oncogenes or, otherwise, inactivation of tumor suppressors. Next, histone acetylation, which broadly influences chromatin's compaction state, is largely associated with active transcription, whereas histone deacetylation (HDAC) is associated with an inactive transcription. (Di Cerbo and Schneider, 2013). Eleven (11) human zinc-dependent HDACs are known so far. They are grouped into different classes based on their general sequence homology to yeast; HDAC1, 2, 3, 8 are grouped in class I; class IIa comprises HDAC4, 5, 7, 9; and class IIb includes HDAC6, 10. In contrast, class III comprises non-zinc-dependent HDACs – so-called sirtuins. HDAC11 is categorized as class IV.
Many cancers, including HCC, have been linked to histone deacetylases (HDACs). (Eckschlager et al., 2017). The upregulation of HDAC1 and HDAC2 was believed to be the reason for high mortalities of hepatocellular carcinoma patients (Ler et al., 2015). Moreover, the upregulation of HDACs 1–3 has been reported by (Quint et al., 2011), and “HDAC2″ was proposed as an independent prognostic marker of survival in HCC. It was additionally reported that HCC patients with high mortality rates had higher expression of various HDAC isoforms, especially HDAC1 and HDAC3, and this hyperexpression was also related to poor prognosis (Liu et al., 2014; Lu et al., 2018). Bearing in mind the important role ”HDAC1/3″ plays in liver carcinogenesis, HDAC1/3 interacting proteins were previously identified in HepG2 cells (Farooq et al., 2013; Farooq and Wadaan, 2013).
The main objective of this study was to analyze the expression of HDAC1/3, PFDN2/6, CNOT1, and HMG20b in a large panel of liver cancer cell lines, to check the differential expression in cancer vs. normal human liver tissues by utilizing online databases which are designed for such purposes. In addition, the prognostic values of these five genes were analyzed using a Kaplan-Meier plotter.
2 Material and methods
2.1 Cell culture
Human HCC cell line (HepG2) was obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained in Dulbecco's Modified Eagle's Medium (DMEM) (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, MA, USA) in a humidified atmosphere of 5% (v/v) CO2 and 95% air at 37 °C.
2.2 Reverse transcriptase-polymerase chain reaction (RT PCR)
Total RNA was isolated using a total RNA purification kit (Jena Bioscience, Jena Germany; Cat.No. PP-210S). The RNA was reverse transcribed into first-strand cDNA using a two-step cDNA Synthesis Kit from Roche Cat.No.11 483 188 001 following the instruction manual. The sequences of primers and PCR conditions were the same as described previously (Al-Yhya et al., 2021)
2.3 Identification of differential gene expression (IDEG)
The online and freely available Gene Expression Profiling Interactive Analysis database (GEPIA: http://gepia.cancer-pku.cn/index.html) (Tang et al., 2017) was used to analyze selected genes differentially expressed in Hepatocellular carcinoma (LIHC) and normal liver cells.
2.4 Analyzing HCC tumor grade-related expression of the gene
UALCAN is a comprehensive, user-friendly, and interactive web resource for analyzing cancer OMICS data (Chandrashekar et al., 2017). UALCAN was searched to analyze the differential expression of genes at various stages of the tumor.
2.5 mRNA expression in a panel of liver cancer cell lines
The mRNA expression levels of the selected genes across 23 hepatocellular carcinoma and hepatoblastoma cancer cell lines were analyzed using the Expression Atlas from The European Bioinformatics Institute (EMBL-EBI) online database (Petryszak et al., 2016).
2.6 Protein expression analysis
The publicly available Human Protein Atlas database (http://www.proteinatlas.org/) was used to validate the expression of candidate genes on transcriptional and translational levels.
2.7 Survival analysis of HDAC1/3-interacting proteins using Kaplan–Meier (KM) analysis
The association of patients' overall survival (OS) with mRNA expression of HDAC1, HDAC3, CNOT1, PFDN2, PFDN6, and HMG20b in human liver cancer was analyzed using a KM plot, as described previously (Menyhart et al., 2018).
3 Results
3.1 The HDAC1/3 interacting genes overexpressed in twenty-three human hepatocellular carcinoma cell lines.
The online RNA seq expression database, containing the expression of RNA-seq of the entire human genome in 934 human cancer cell lines (https://www.ebi.ac.uk/gxa/experiments/E-MTAB-2770/Results), was searched to analyze the expression of these six genes in human liver cancer cell lines. The database contains a panel of 23 human cancer cell lines of hepatoblastoma and hepatocellular carcinoma origin. The search results indicated that all these cell lines express HDA1, HDAC3, CNOT1, PFDN2, PFDN6, and HGM20B, albeit the expression level of genes varies from cell line to cell line. As shown in Fig. 1A, HDAC1 and PFDN2 are expressed at a higher level than other genes. PFDN2 shows the highest expression between 60 and 180 fragments per kilobase of transcript per million mapped reads (FPKM), while HDAC1 expression is between 40 and 90 FPKM. The third highly expressed gene is CNOT 1, with an expression between 20 and 50 FPKM.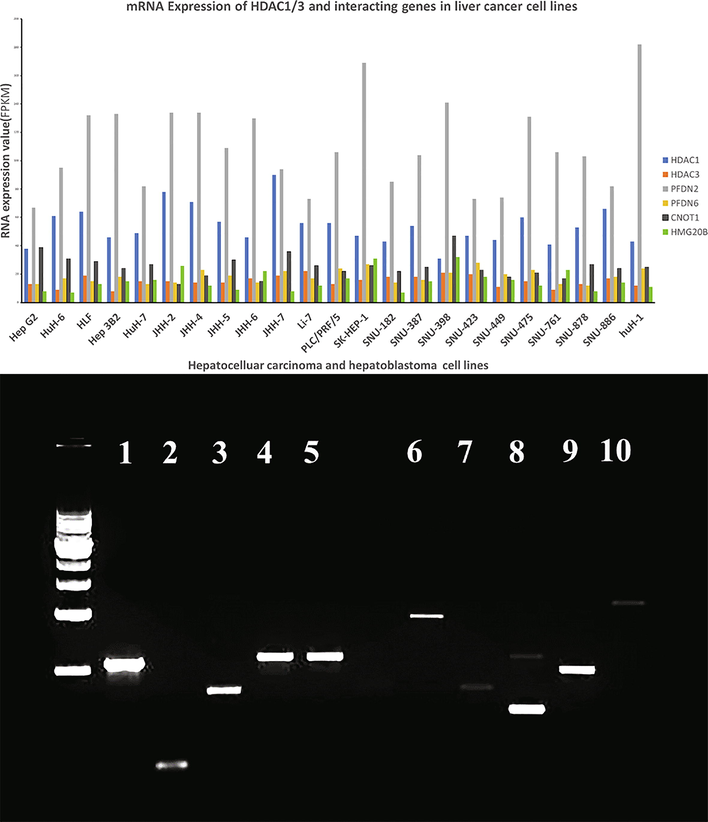
A: Differential expression of HDAC1/3 interacting proteins in liver cancer cell lines. The mRNA expression levels of the selected genes across 23 hepatocellular carcinoma and hepatoblastoma cancer cell lines. The data was analyzed using the Expression Atlas from The European Bioinformatics Institute (EMBL-EBI) online database. B: Reverse transcriptase-polymerase chain reaction (RT PCR) to validate the expression of HDAC1/3 interacting genes in HepG2 cells. 1 GAPDH 5 CNOT1 9 PFDN6. 2 Actin Beta 6 TBL1X 10 HMG20b. 3 HDAC1 7 CDKA1. 4 HDAC3 8 PFDN2 1 KB DNA ladder.
Semi-quantitative RT-PCR was used to check the expression of genes in the HepG2 cell line. The PCR analysis shows higher HDAC1, HDAC3, CNOT1, PFDN2, PFDN6 and lowers CDKA1 and HMG20b RNA expression in HepG2 cells. The expression of these six genes is presented in Fig. 1B.
3.2 Differential gene expression (DEG) analysis of six selected genes in hepatocellular carcinoma vs. adjacent normal tissues
The Gene Expression Profiling Interactive Analysis (GEPIA http://gepia.cancer-pku.cn/index.html) database was searched to check the expression of these genes in hepatocellular carcinoma (LIHC) tumor samples (n = 369) and adjacent normal tissues (n = 50). The differential gene expression analysis in HCC was conducted with a criterion of P < 0.05 and |log2FC| > 2. The GEPIA revealed higher expression of the candidate six genes in HCC tumor samples than in normal tissues. However, the tumor tissues showed a higher expression of HDAC1 compared to HDAC3, CNOT1, PFDN6, and HMG20b (Fig. 2).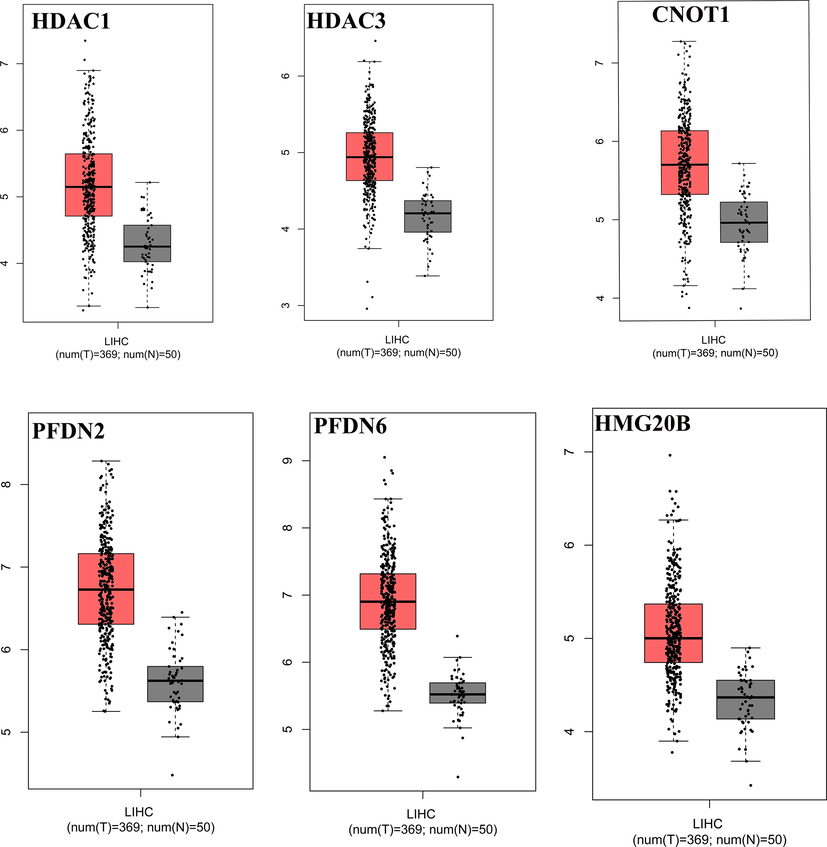
Identification of differential gene expression (IDEG). The online and freely available Gene Expression Profiling Interactive Analysis database (GEPIA: http://gepia.cancer-pku.cn/index.html) was used to analyze selected genes differentially expressed in Hepatocellular carcinoma (LIHC) and normal liver cells.
3.3 HCC tumor grade-related differential expression of genes
UALCAN is an interactive web resource for analyzing cancer OMICS data, allowing users to identify biomarkers or perform in silico validation of potential genes of interest. This analysis can be very helpful to identify biomarkers expressed at the very early stages of tumors to design better strategies for the treatment of HCC. To analyze the differential expression of six genes at various tumor grades of HCC in this present research, the UALCAN database was searched, and the results are presented in Fig. 3. It was revealed that all the genes exhibited varying expression levels in all tumor grades. The HDAC1 was expressed at a weaker level 75 transcript per million (TPM) at grade 1 of HCC, and its expression enhanced with the progression of the tumor. The highest level of HDAC1 was detected at grade 4 (130 TPM), and a higher level of HDAC3 was detected at the early stage of HCC (45 TPM grade 1). Interestingly, the expression of HDAC3 remained stable until grade 3, after which a downregulation was detected at grade 4 of HCC.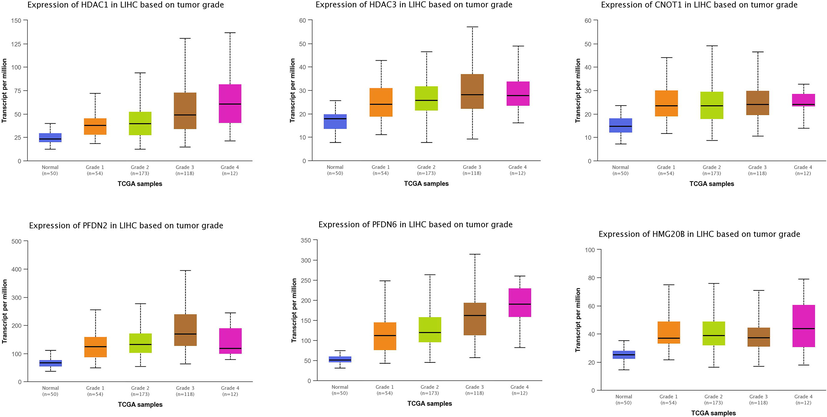
Analyzing tumor grade-related expression of the genes in HCC. University of Alabama Cancer Database (UALCAN) UALCAN is a comprehensive, user-friendly, and interactive web resource for analyzing cancer OMICS data. UALCAN (http://ualcan.path.uab.edu/) was searched to analyze the differential expression of genes at various stages of HCC.
Similarly, the CNOT1 was also expressed at the early stages of HCC with varying expression levels at 45, 50, and 45 TPM at stages 1, 2, and 3, respectively, while its expression dropped significantly at grade 4 HCC. As shown in Fig. 3, PFDN2 was expressed at a higher level at grade 3 HCC, with an expression of almost 400 TPM. However, the expression of PFDN2 significantly decreased at grade 4, and 250 TPM was detected at this grade HCC. A similar trend in the expression of PFDN6 in the development of HCC was also detected. The PFDN6 was detected at a higher level at grade 1 with 250 TPM, while the highest expression of PFDN6 was detected at grade 3 of HCC with 300 TPM. The expression of PFDN6 was reduced at grade 4 of HCC. The HMG20b was detected at all grades of HCC; its expression remained steady at grades 1, 2, and 3, and grade 4 showed a higher expression.
3.4 Protein expression in tissues with immunohistochemistry
To analyze protein expression patterns in normal human tissues and HCC tissues, the Human Protein Atlas database (https://www.proteinatlas.org) was analyzed. The relative expression of six genes at the protein level is shown in Fig. 4. Immunohistochemistry analysis here reveals higher protein expression of HDAC1, HDAC3, CNOT1, and PFDN2 in HCC tissues than normal liver tissues. HDAC1, HDAC3 were expressed in the nucleus of positive liver cells, while cytoplasmic staining was observed for CNOT1 and PFDN2. As shown in Fig. 4, a weaker to moderate staining of PFDN6 and HMG20b was detected in HCC tissues than normal liver tissues. The subcellular localization staining shows cytoplasmic staining of PFDN6 and nuclear staining of HMG20b.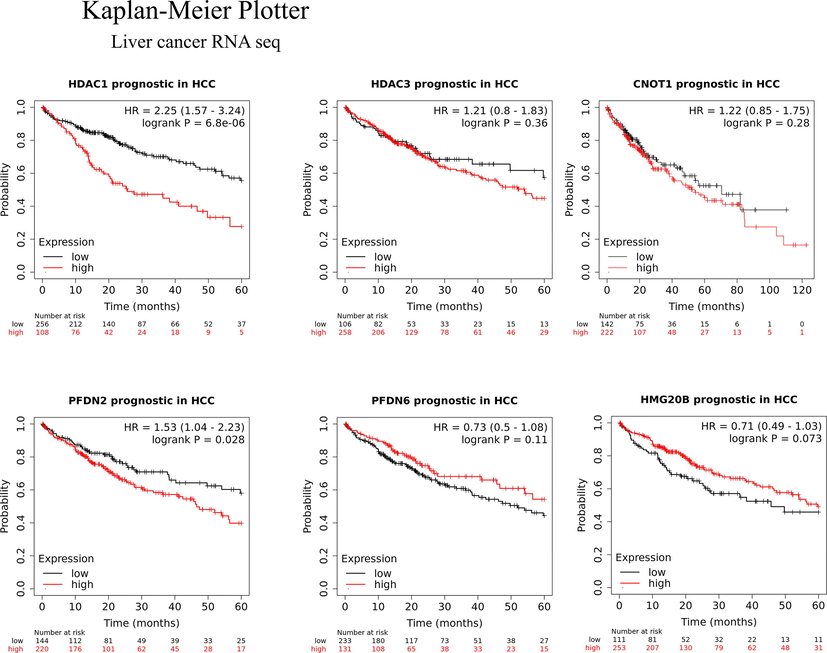
Kaplan–Meier plot showing the prognostic value and overall survival (OS) of selected HDAC1/3 interacting genes. The expression data showed that expression of HDAC1, HDAC3, CNOT1, and PFDN2 are unfavorable prognostic markers of HC C, while expression of HMG20b and PFSDN6 favorable prognostic markers in hepatocellular carcinoma.
3.5 Determining the prognostic value of HDA1/3 interacting proteins in liver cancer
To investigate the prognostic values of HDAC1/3, CNOT-1, PFDN2/6, and HMGG20 in hepatocellular carcinoma, an online Kaplan–Meier plotter analysis tool (http://kmplot.com/analysis/index.php?p=service&cancer=liver_rnaseq#) was utilized. The 5-year overall survival (OS) rate of 364 HCC patients (250 males and 121 females) expressing the mRNA of HDAC1, HDAC3, CNOT1, PFDN2/6, and HMG20b was investigated. As shown in Fig. 5, the high mRNA expression of HDAC1 (HR = 2.25, 95% CI: 1.57–3.24, p = 6.8e − 06), PFDN2 (HR = 1.53, 95% CI: 1.04–2.23, p = 0.28), HDAC3 (HR = 1.21, 95% CI: 0.8–1.83, p = 0.36), and CNOT1 (HR = 1.22, 95% CI: 0.85–1.75, p = 0.028), and low mRNA expression of HMG20b (HR = 0.71, 95% CI: 0.49–1.03, p = 0.073) and PFDN6 (HR = 0.73, 95% CI: 0.5–1.08, p = 0.11) was observed in HCC tissues. High mRNA expression of HDAC1 and PFDN2 showed an association with significantly shorter OS and unfavorable outcomes. HDAC3 and CNOT1 also showed associations with an unfavorable outcome, but this was not statistically significant. HMG20b expression was associated with significantly longer OS with a favorable outcome. PFDN6 was also associated with a favorable outcome, but this was not significant. The expression data showed that HDAC1, HDAC3, CNOT1, and PFDN2 are oncogenes, while HMG20b and PFSDN6 are tumor suppressor genes in hepatocellular carcinoma.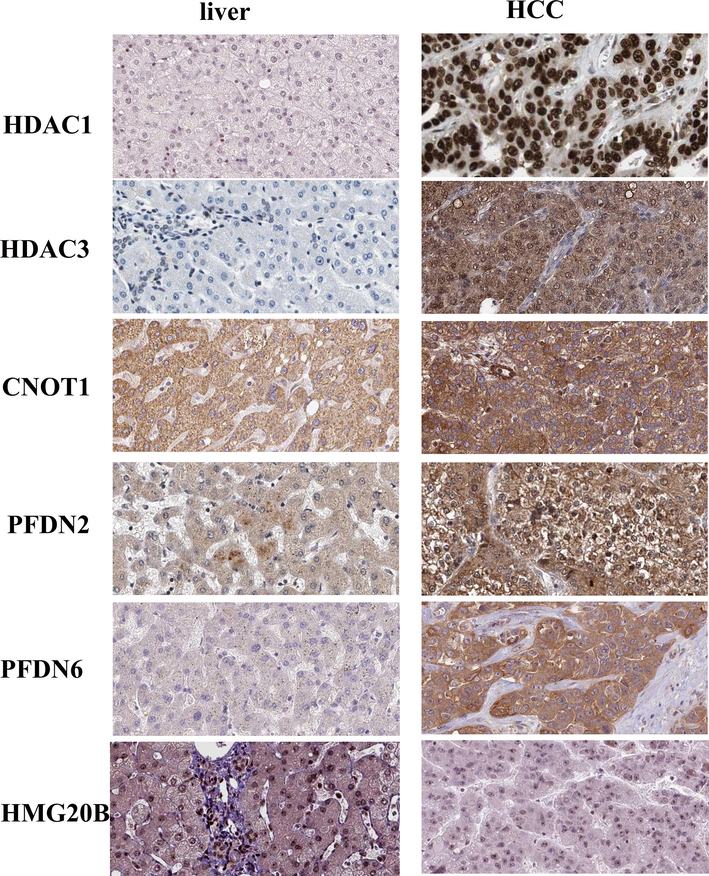
Protein expression analysis. Immunohistochemistry analysis here reveals higher protein expression of HDAC1, HDAC3, CNOT1, PFDN2, and PFDN6 in HCC tissues than normal liver tissues, whereas higher protein expression of HMG20b in normal liver tissues as compared to HCC. The data has been analyzed using Human Protein Atlas database (http://www.proteinatlas.org/).
4 Discussion
Prognosis remains an integral part of the patient assessment to improve patient survival, especially in liver cancer patients, and make good therapeutic and disease management decisions. Prognostic factors, when applied appropriately, allow for the stratification of patients' risks to categorize therapy based on the possible outcome. Unfortunately, studies investigating prognostic factors in HCC have been characterized with heterogenous subjects having different underlying factors such as the presence or absence of liver diseases such as cirrhosis (Tandon and Garcia-Tsao, 2009), making results from such studies inapplicable to a wider population. Conventionally, studies investigating the prognostic value of specific genes do so by examining one gene at a time, which is time-consuming and labor-intensive, not to talk of the cost implication (Huang da et al., 2009). On the contrary, through the application of bioinformatics tools, we were able to analyze multiple differentially expressed genes (listed above), which were further investigated by enrichment analysis. Findings showed that these genes are implicated in the pathophysiology of HCC. A search of online TCGA cancer tissue data (https://www.proteinatlas.org/ENSG00000204220-PFDN6/pathology) suggested a moderate to high-level expression of PFDN6 in liver cancer tissues than the normal human liver.
Eukaryotic prefoldins (PFDN) are hexameric chaperone complexes consisting of six different subunits (26), and their main job is to keep freshly generated peptides stable and promote proper cytoskeletal protein folding, preventing protein misfolding and aggregation (27). PFDN2 is a 154-amino-acid protein with a 16,648-dalton molecular mass. PFDN2 is linked to malignancies and can be used as a biological marker for various disorders. For bladder tumor classification and clinical outcome, PFDN2 is a predictive biomarker. The expression of PFDN2 in diverse subtypes of gastric cancer is a biological indicator of poor prognosis and shows poor overall survival. PFDN6 is a hydrophilic protein in the major histocompatibility complex with an unknown function (Yesseyeva et al., 2020a). However, expression of PFDN6 was associated with overall poor survival in gastric and chemotherapy in acute lymphocytic leukemia (Dehghan-Nayeri et al., 2017; Mo et al., 2020). It is reported that κ-actin is the major component of the actin cytoskeleton in hepatocellular carcinoma tissues. The κ-actin reduced the interaction between actin and PFDN2, causing poor cytoskeletal organization and promoting abnormal cells' growth, proliferation, or metastasis in liver cancer (Riester et al., 2014).
We previously have discovered that several PFDN subunits have a significant affinity for HDAC1 in hepatocellular carcinoma HepG2 cells (Farooq et al., 2013), strengthening the notion that PFDN subunits are involved in hepatocellular carcinoma progression and metastasis. The oncogenic behavior of PFDN2 in hepatocellular cancer (HCC) patients suggests that it could be used as a novel therapeutic target to treat liver cancer. Although PFDN2 and PFDN6 affect liver cancer, the underlying mechanism has yet to be discovered.
The 5-year survival of liver cancer patients expressing HDAC1, HDAC3, CNOT1, PFDN2/6, and HMG20b was calculated via Kaplan–Meier analysis. The results have indicated that high expression of HDAC1, HDAC3, CNOT1, and PDNF2 reflects short or medium-term survival, suggesting an unfavorable prognosis for HCC patients. Another study has also reported the overexpression of HDAC1, HDAC3, and PFDN2 as unfavorable prognostic biomarkers and reduced recurrence-free survival in liver cancer patients (Freese et al., 2019; Wu et al., 2010). HDACs 1 and 3 are highly expressed in the nucleus, where they are involved in numerous functions by regulating different pathophysiological processes, such as mitosis, apoptosis, and tumorigenesis (Ji et al., 2019). The oncogenic behavior of PFDN2 in hepatocellular cancer (HCC) patients suggests that it could be used as a novel therapeutic target to treat liver cancer (Yesseyeva et al., 2020a). PFDN6, on the other hand, was associated with a favorable outcome, but this was not statistically significant. PFDN2 was also previously reported as a poor prognostic biomarker in gastric cancer (Yesseyeva et al., 2020b).
HMG20b is a non-sequence-specific DNA binding protein that belongs to the high mobility group (HMG) family (Sumoy et al., 2000). HMG20b is necessary for cytokinesis during cell division, and its mutation was reported to result in the failure of cytokinesis in human epithelial cancer (Lee et al., 2011; Lee and Venkitaraman, 2014). The role of HMG20b in liver cancer is unknown. In a recent study, the suppression of liver tumor growth in mice has been reported by inhibition of HMGB1. (Li et al., 2018). The Kaplan–Meier analysis of liver cancer patients expressing HMG20b showed increased five years overall survival (OS) than patients with low expression, suggesting it as a favorable prognostic biomarker in liver cancer. The expression and prognostic value of HMG20b in liver cancer have never been reported before; thus, we consider it a novel findings in this study.
PFDN6, HMG20b are the important component of the CoREST complex and regulate many biological processes along with HDAC. The exact mechanism of how PFDN6 regulates HMG20b is not known. It is well established that the CoREST complex generally silent the gene expression and contributes to cancer and other diseases. The role of each member of the CoREST complex should be investigated; however, in the present scenario, the regulation of PFDN6 and HMG20b by HDAC1 would be the most feasible biochemical pathway involved for liver cancer progression and metastasis. Human HMG20b is a rare tumor suppressor gene that may promote carcinogenesis through heterozygous mutations caused by missense mutagenesis. A study has demonstrated that the carboxyl (C)-terminal region of HMG20b, is essential for cytokinesis and reported that this region is inactivated by missense mutation causing breast cancer (Lee and Venkitaraman, 2014).
5 Conclusion
Our recent and previous work has revealed that CNOT1, PFDN2/6, and HMG20b. overexpressed in most cell lines originated from human liver carcinoma. The analysis of human protein atlas, cancer atlas, and similar online databases has shown the higher expression of these genes in human cancer tissues compared to normal liver. The computational analysis done in this study has identified PFDN6 and HMG20b as novel prognostic biomarkers in liver cancer. Further clinical studies are needed to verify the relationship between the expression of PFDN2 and HMG20b and patient survival in human hepatocellular carcinoma patients.
Funding
This study was funded by the Deanship of Scientific Research, King Saud University, through the Vice Deanship of Scientific Research Chairs. The funding body had no role in the design, collection, analysis, interpretation of data, or manuscript writing.
Acknowledgements
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through the Research chair program.
Conflicts of interest
The authors declare that they have no competing interests.
References
- Pharmacological inhibition of HDAC1/3-interacting proteins induced morphological changes, and hindered the cell proliferation and migration of hepatocellular carcinoma cells. Environ. Sci. Pollut. Res. Int.. 2021;28:49000-49013.
- [Google Scholar]
- Prognostic factors for cancer patient admitted to a medical intensive care unit. Acta Oncol.. 2020;59:458-461.
- [Google Scholar]
- UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649-658.
- [Google Scholar]
- Identification of potential predictive markers of dexamethasone resistance in childhood acute lymphoblastic leukemia. J. Cell Commun. Signal.. 2017;11:137-145.
- [Google Scholar]
- Cancers with wrong HATs: the impact of acetylation. Brief Funct. Genomics. 2013;12:231-243.
- [Google Scholar]
- Identification of histone deacetylase 1 protein complexes in liver cancer cells. Asian Pac. J. Cancer Prev.. 2013;14:915-921.
- [Google Scholar]
- Epigenetic targets in hepatocellular carcinoma cells: identification of chaperone protein complexes with histone deacetylases. Epigenomics. 2013;5:501-512.
- [Google Scholar]
- Freese, K., Seitz, T., Dietrich, P., Lee, S.M.L., Thasler, W.E., Bosserhoff, A., Hellerbrand, C., 2019. Histone deacetylase expressions in hepatocellular carcinoma and functional effects of histone deacetylase inhibitors on liver cancer cells in vitro. Cancers (Basel) (11).
- Identification of soluble NH2-terminal fragment of glypican-3 as a serological marker for early-stage hepatocellular carcinoma. Cancer Res.. 2004;64:2418-2423.
- [Google Scholar]
- Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res.. 2009;37:1-13.
- [Google Scholar]
- HDAC3 deficiency promotes liver cancer through a defect in H3K9ac/H3K9me3 transition. Cancer Res.. 2019;79:3676-3688.
- [Google Scholar]
- A cancer-associated mutation inactivates a region of the high-mobility group protein HMG20b essential for cytokinesis. Cell Cycle. 2014;13:2554-2563.
- [Google Scholar]
- A mitotic function for the high-mobility group protein HMG20b regulated by its interaction with the BRC repeats of the BRCA2 tumor suppressor. Oncogene. 2011;30:3360-3369.
- [Google Scholar]
- HDAC1 and HDAC2 independently predict mortality in hepatocellular carcinoma by a competing risk regression model in a Southeast Asian population. Oncol. Rep.. 2015;34:2238-2250.
- [Google Scholar]
- A novel albumin wrapped nanosuspension of meloxicam to improve inflammation-targeting effects. Int. J. Nanomed.. 2018;13:4711-4725.
- [Google Scholar]
- The genetic and epigenetic alterations in human hepatocellular carcinoma: a recent update. Protein Cell. 2014;5:673-691.
- [Google Scholar]
- Prognostic significance of glypican-3 expression in hepatocellular carcinoma A meta-analysis. Medicine. 2018;97
- [Google Scholar]
- Histone deacetylase 3 promotes liver regeneration and liver cancer cells proliferation through signal transducer and activator of transcription 3 signaling pathway. Cell Death Dis.. 2018;9:398.
- [Google Scholar]
- Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R. Soc. Open Sci.. 2018;5:181006
- [Google Scholar]
- The role of prefoldin and its subunits in tumors and their application prospects in nanomedicine. Cancer Manag. Res.. 2020;12:8847-8856.
- [Google Scholar]
- Overexpression of osteopontin is associated with intrahepatic metastasis, early recurrence, and poorer prognosis of surgically resected hepatocellular carcinoma. Cancer-Am Cancer Soc.. 2003;98:119-127.
- [Google Scholar]
- Cellular prognostic markers in hepatitis-related hepatocellular carcinoma. Infectious Agents Cancer. 2018;13:10.
- [Google Scholar]
- Expression Atlas update–an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res.. 2016;44:D746-D752.
- [Google Scholar]
- Biomarkers in hepatocellular carcinoma: diagnosis, prognosis and treatment response assessment. Cells-Basel. 2020;9
- [Google Scholar]
- The prognostic molecular markers in hepatocellular carcinoma. World J. Gastroenterol.. 2002;8:385-392.
- [Google Scholar]
- Clinical significance of histone deacetylases 1, 2, 3, and 7: HDAC2 is an independent predictor of survival in HCC. Virchows Arch.. 2011;459:129-139.
- [Google Scholar]
- Integrative analysis of 1q23.3 copy-number gain in metastatic urothelial carcinoma. Clin. Cancer Res.. 2014;20:1873-1883.
- [Google Scholar]
- HMG20A and HMG20b map to human chromosomes 15q24 and 19p13.3 and constitute a distinct class of HMG-box genes with ubiquitous expression. Cytogenet. Cell Genet.. 2000;88:62-67.
- [Google Scholar]
- Prognostic indicators in hepatocellular carcinoma: a systematic review of 72 studies. Liver Int.. 2009;29:502-510.
- [Google Scholar]
- GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res.. 2017;45:W98-W102.
- [Google Scholar]
- Prospective validation of an immunohistochemical panel (glypican 3, heat shock protein 70 and glutamine synthetase) in liver biopsies for diagnosis of very early hepatocellular carcinoma. Gut. 2012;61:1481-1487.
- [Google Scholar]
- Elevated alpha-fetoprotein: differential diagnosis - hepatocellular carcinoma and other disorders. Clin. Liver Dis.. 2015;19:309-323.
- [Google Scholar]
- Identification of histone deacetylase 3 as a biomarker for tumor recurrence following liver transplantation in HBV-associated hepatocellular carcinoma. PLoS ONE. 2010;5:e14460
- [Google Scholar]
- Prefoldin subunits (PFDN1-6) serve as poor prognostic markers in gastric cancer. Biosci. Rep.. 2020;40
- [Google Scholar]
- Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased alpha-fetoprotein concentrations (REACH-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol.. 2019;20:282-296.
- [Google Scholar]







