Translate this page into:
Prediction of RNA editing sites and genome-wide characterization of PERK gene family in maize (Zea mays L.) in response to drought stress
⁎Corresponding authors. ali.sher@mnsuam.edu.pk (Muhammad Ali Sher), Zulfiqar_ali@uaf.edu.pk (Zulfiqar Ali)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Objectives
Inadvertent climate changes continuously threating the crops production and thus affecting the livelihood of peoples across the world. The maize production at world level has severely been hampered by the drought stress. Proline-rich extensin-like receptor kinases (PERKs) are considered among the sub-class of plants larger protein family, receptor kinases. Member of PERK gene family play significant role in both abiotic and biotic stress and in various plant metabolic activities and pathways.
Methods
As of now, no comprehensive research is reported for PERK genes in maize. We have performed a genome wide in-silico analysis and identify twenty-three PERK genes in maize. We performed phylogenetic analysis, sequence logos, motif analysis, promoter analysis, chromosomal and subcellular localization, synteny and expression analysis using RNA seq data under drought stress. We also predict RNA editing sites in mitochondrial and chloroplast genome.
Results
Phylogenetic study of PERK genes from eight different plant species divided into four distinct clades. Four subclasses group of ZmPERKs were observed based on domain organization, motif pattern, and phylogenetic analysis. The exon–intron arrangement of the ZmPERK were conserved among members of the same subclasses. In the promoter region different cis-elements were found those were involved in the growth and development, as well as light and stress response. Through gene duplication analysis it was observed that segmental duplications in ZmPERKs played major role in maize evolution. The Ka/Ks ratios indicated that most ZmPERK genes during the evolution have experienced strong purifying selection. The conversion of cytosine (C) to uracil (U) was observed in all predicted editing sites (U). These transitions were mostly based on changes in the first and second codon bases. The in-silico expression analysis of transcriptome data revealed the differential expression of ZmPERK genes in response to drought stress and oil content accumulation.
Conclusion
The current study provides base information on the PERK gene family in maize. Our findings can serve as a reference for further functional analysis of ZmPERKs. These genes can be further explored and used in breeding program to develop cultivars resilient to drought stress.
Keywords
PERK
Genome wide analysis
RNA editing sites drought
Oil content
1 Introduction
Maize is considered as important crop which is grown for food and feed around the globe. Abiotic stress especially drought stress severely affected the maize production at world level. Plant breeders are considering the drought stress as one of the most important abiotic stress that causing the hindrance in getting the higher grain yield in different crops especially in maize (Liu and Qin, 2021). Hence it is dire need of the time that breeders should tailor their modern varieties with novel traits that have capacity to buffer the drastic effects of the abiotic stresses especially drought stress. The advent of modern genotyping techniques especially the next generation sequencing (NGS) has revolutionized the field of genetics, furthermore the release of genome dataset has made a substantial increment in developing the strategies to quip the plants with novel traits (Leng and Zhao, 2020). In plants the receptor-like kinases (RLKs) having similar structure are considered as large superfamily of proteins. In this group the PERKs (Proline rich extension like receptor kinases) gene family is also included. Plant Species such as Arabidopsis and rice comprises large gene family of receptor kinase. There is 600 members are reported in arabidopsis for the receptor kinase family and their analogous have been found in around 20 different species (Morris and Walker, 2003). These kinase family play have been found playing a crucial role in the growth and development phase of plants and also defensive mechanisms (Shiu and Bleecker, 2001, Morris and Walker, 2003, Shiu et al., 2004). Role of most of the members of receptor kinase is still unknown. Receptors like kinase are the protein comprising extracellular, carboxyl terminal and intercellular domain with putative amino terminal (see Fig. 1).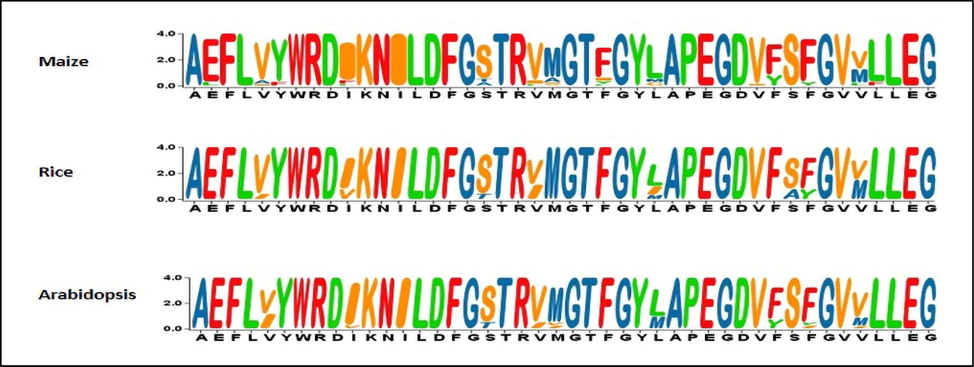
Sequence logos of PERK gene family between Maize, Rice and Arabidopsis.
Depending on their extracellular domain, receptor kinases interact to a wide range of substances for example, carbohydrates and cell wall components. This domain organization have much resemblance to the animal receptor tyrosine kinases (Shiu, 2001). Receptor kinases have specific and extracellular domains, for example, leucine-rich repeat (LRR) and proline-rich extension-like receptor kinases. Gene duplication and functional redundancy also reported among these different classes of receptor kinase (Champion et al., 2004). The CLV1 and ERECTA receptor kinase is the evidence of existence of functional redundancy (Diévart et al., 2003, Shpak et al., 2003, Shpak et al., 2004). PERK gene family of Arabidopsis have maximum sequence identity to Brassica napus and such as PERK1 of Arabidopsis is much like PERK1 of Brassica napus. Researchers have reported fifteen PERK genes in the Arabidopsis yet their functions still need to be characterize (Silva and Goring, 2002, Nakhamchik et al., 2004, Bai et al., 2009). In Arabidopsis the PERK1 is identified which do functions in response to any wound that occur in the plasma membrane (Silva and Goring, 2002). Likewise, PERK4 is predicted as key regulator for Ca2+ signaling that contributes in production of abscisic acid in root (Bai et al., 2009). It is well documented that under abiotic stresses plant accumulate more calcium contents in cells to boost the production of antioxidant enzyme activity, regulate lipid peroxidation of cell membranes and stomatal apertures to mitigate the impact of stresses on plant growth (Mansfield et al., 1990; Abadi and Sepehri, 2016). The production of reactive oxygen species (ROS) decline in the presence of PERKs whereas increasing level of ROS work as a signal for root hair development (Xing et al., 2013). In an organism the first line of defense against superoxide radicals is the production of superoxide dismutase (SODs) which catalyze the superoxide radical to hydrogen peroxide and molecular oxygen. The copper/zinc SOD (Cu/Zn SOD) is catalyzed through the MAPK cascade under high light-induction. The homologous proteins like MPK3 and MPK6 in plants are detected using the anti-PERK antibodies from animals (Samuel and Ellis, 2002; Hwang et al., 2016). Environmental stresses like heat, drought, nutrients, heavy metals, pathogens, keep threating the plants to express its fully genetic potential. It is becoming more important to scientist to reveal how plants response to internal and external stimuli. Plant sense the environmental changes through the use of cell surface receptors and initiate different signaling pathways to trigger the adaptive responses (Zhu, 2016).
Erratic climate change has become a major constraint in achieving the higher crop yield. At crop level it affects plant morphological, anatomical, and physiological attributes which ultimately results in drastic economic yield loss. Maize is a major food and feed crop grown all over the world. It rated as the world's third most significant staple grain crop (Tiwari and Yadav, 2019). Characterization of PERK family in maize can help us to understand the plant molecular mechanism of tolerance against biotic and abiotic stresses. Only a few PERK genes have been characterized, and the functions of most of them is still unknown. High-throughput genome sequencing of the maize provided an excellent opportunity for genome wide analysis of genes families. In our study, we performed in-silico genome wide analysis of PERK genes in maize. We analyzed phylogenetic relationship between 8 species and only maize separately. Furthermore, gene structure Intron/exons, motif distribution, conserved domain analysis, sequence logos, physio-chemical properties. The structural and functional importance of genes were also assessed using Ka/Ks values and synteny analysis. In-silico expression analysis were also performed for these genes to predict their role and function. The present study and their result enabled us to conclude that PERK gene family paly vital role in maize development and stress response (Fig. 2).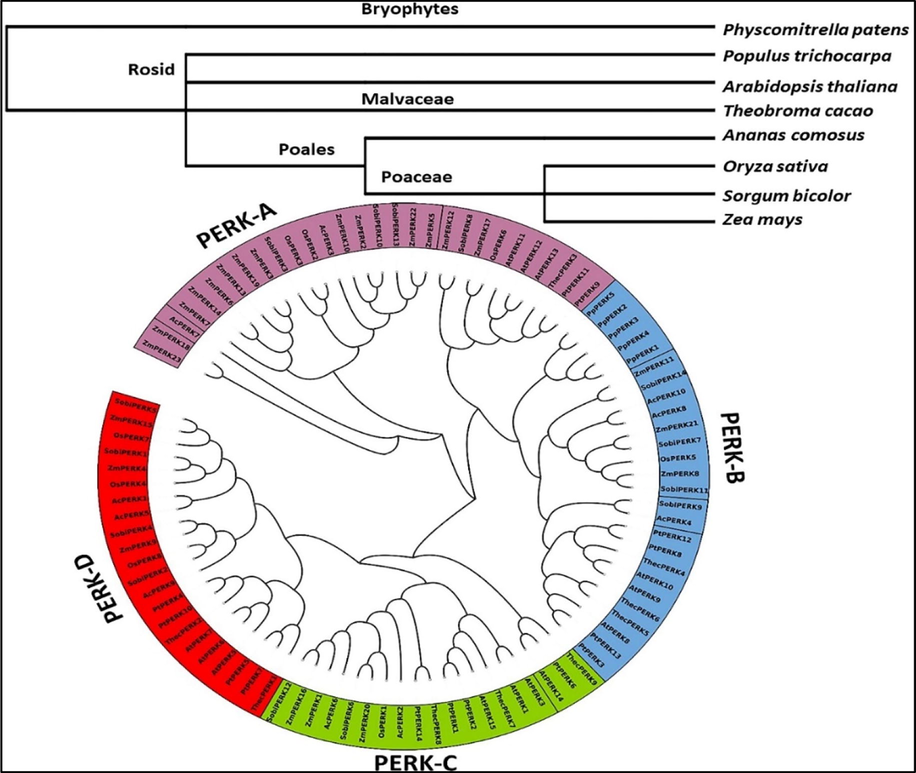
PERK gene family phylogenetic tree. The major cluster of orthologous genes is distinguished with various colours (PERKA-D).
2 Materials and methods
2.1 PERK gene family identification and characterization in maize genome
Whole maize genome sequence, as well as the general feature format file (GFF3), was downloaded from the Maize Genetic and Genomic Database (Maize GDB)(https://gamma.maizegdb.org). For the purposes of finding the probable candidates of PERK family in maize, the online Pfam database (https://www.sanger.ac.uk/Software/Pfam/) was used to download the PERK domain HMM profile and then subjected as a query into Blastp (Finn et al., 2014). For all of the retrieved protein sequences, the SMART tools (https://smart.embl-heidelberg.de/) were used to verify the presence of the PERK domain (Letunic et al., 2015). Maize PERK gene family sequence were downloaded from maize genome database. TAIR 10 (http: /https://www.Arabidopsis.org) was used to retrieved the arabidopsis sequences while all other sequences of studied organisms were retrieved from online plant database Phytozome version 11 (https://phytozome.jgi.doe.gov/pz/portal.html). ExPASyProtParam, (https://us.expasy.org/tools/protparam.html) an online web tools, were used to retrieve the physiochemical properties.
2.2 Sequence logos and phylogenetic/evolutionary analysis
The MEGA 7 software was used to find out the conserved sequences for amino acids. Sequence are aligned using ClustalW and the structure was constructed using TBtool (https.//github.com/CJ-Chen/TBtools). Furthermore, using this software the Neighbor-Joining method was used to get the phylogenetic tree to deduce the evolutionary history (Chothia et al., 2003). The distances of the number of amino acid sites in units were measured using the poisson correction parameters (Yang et al., 2008). The Bootstrap algorithm employed with 1000 repetitions to estimate the stability of the nodes in the phylogenetic tree. Total 98 amino acid sequences were used for this analysis.
2.3 Predicted protein motifs, structure of exon/intron and conserved domain analysis
To find preserved motif of PERK protein online web server Multiple Em for Motif Elicitation (MEME) is used (https://meme-suite.org/tools/meme). The TBtool was used to construct the motif structure using the MEME.xml file which is obtained through MEME suite. The default parameters were as follows: motif recurrence was set to 1 per sequence; frequency of motifs was set to 10; motif width was set to 5–50 residues; and the minimum number of motif sites was set to 5. Arrangement of Exon and Intron of PERK genes was investigated by using gff3 file downloaded from maize GDB. Structure is constructed using TBtool software. Afterward, the NCBI CDD tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) was used to perform the conserved domain analysis.
2.4 Chromosomal localization, gene duplication events and synteny analysis
The start and end position for each identified ZmPERK gene were procure from the Maize Genetic and Genomic Database (Maize GDB) and validated against the GFF3 file. Total chromosomal length is retrieved through using FASTA stat in TBtool. Finally, using MapChart v2.32 (Voorrips, 2002), the ZmPERK genes were spatially mapped onto the maize chromosomes. The phylogenetic tree was used to identify the putative paralogous PERK gene pairs. The resulting pairs were subjected to TBtool software to determine synonymous and non-synonymous substitution rates (Ka). To determine the nature of codon selection the Ka/Ks ratio was also calculated that allegedly occurred during evolution. Further, using the formula T = Ks/2 and assuming a clock rate of 6.05 X10 9 substitutions/synonymous site/year for maize, the approximate period of duplication event was calculated (Kong et al., 2013). The genome sequence files, and gene annotation files (GFF3) of sorghum, rice and maize were used for the collinearity analysis. Required files generated using one step MC scan. TBtool software was used to visualize the results, and the parameter filtering genes in the collinearity block was set to 40.
2.5 RNA editing sites prediction, subcellular localization, and promoter analysis
RNA editing is a method in which certain cytidines in mitochondrial and chloroplast transcripts of plants are converted to uridines. The online web server like PREP-Cp (for chloroplast genes) and PREP-Mt (for mitochondrial genes) software (https://prep.unl.edu/) with the cutoff value to 0.8 were used in predicting the RNA editing sites (Mower, 2009). Location of genes at cellular level were also predicted using online web server softberry (https://www.softberry.com). In order to perform promoter analysis, the 5′ upstream region of each gene of the ZmPERK was downloaded from NCBI (https://www.ncbi.nlm.nih.gov/) and the resulting file was submitted to the online database plantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) for a cis-element scan.
2.6 In-silco expression analysis
The NCBI Geodataset (https://www.ncbi.nlm.nih.gov/gds) was used to obtain transcriptome data (GSE136087, GSE40070). RNA-sequencing analysis was performed on seed embryos from 15 days after pollination (DAP) to 40 days after pollination (DAP) (Sekhon et al., 2013). We also retrieved drought stress expression data. Inbred line B73 maize (Zea mays) plants grown in the green house under well-watered and drought stress circumstances until they reached the reproductive stage (at the onset of silk emergence). Plants were hand pollinated for drought stress two to three days after irrigation was stopped, and measurements and samples were collected 24 h later for transcriptome analysis. This gene expression data was used to construct heat map.
3 Results
3.1 Identification and characterization of PERK gene family in maize genome
A systematic approach was followed to identify the PERK protein encoding genes. Taking the advantage of publically available genome of maize, after removing redundant genes we excavated 23 PERK genes in maize genome and named them as ZmPERK1 to ZmPERK23. The PERK gene family comprises 15 Arabidopsis thaliana genes, 8 Oryza sativa genes, 14 Populus trichocarpa genes, 14 Sorghum bicolor genes, 9 Theobroma cacao genes, 23 Z. mays genes, 10 Ananas comosus genes, and 5 Physcomiterlla patens genes. The biophysical properties of the ZmPERK family genes were then determined, including genes ID, start and end positions of genes on chromosomes, polarity of strand, length of CDS sequence (bp), length of protein sequence (aa), protein molecular weights (MW), isoelectric points (pl), and predicted subcellular localization of ZmPERK genes. Table 1 presents all additional estimated biophysical properties. ZmPERK proteins had a peptide length ranged from 432 to 958 amino acids, with an average of 695 A.A (Table 1). The PI (Isoelectric point) of maize PERKs varied between 6.2 and 9.134, while the molecular weight ranged between 36.45 and 100.93 kDa, with an average of 68.69 kDa. The length of nucleotide, amino acid sequences varied greatly, indicating that the ZmPERK genes are highly complex, implying a high level of complexity.
Transcripts ID
Gene name
chromosome
Position
Strand
CDS (bp)
Protein
length A.AProtein Molecular Weight KDA
PI
Grand
average
of hydropathicity (GRAVY)Subcelluar
lcalization
Start End
Zm00001d037811
ZmPERK1
6
138,545,311
138,550,589
-
1299
432
59.97726
6.47
−0.483
Cell membrane/plasma membrane
Zm00001d043480
ZmPERK2
3
201,473,204
201,476,633
–
2208
735
76.90377
8.51
−0.41
Cell membrane/plasma membrane
Zm00001d011908
ZmPERK3
8
164,313,713
164,318,410
–
1479
792
53.98962
9.3
−0.633
Cell membrane/plasma membrane
Zm00001d034257
ZmPERK4
1
288,176,999
288,179,853
–
1761
588
61.80629
9.13
−0.475
Cell membrane/plasma membrane
Zm00001d030218
ZmPERK5
1
113,763,279
113,766,247
+
1488
495
53.51192
6.41
0.37
Cell membrane/plasma membrane
Zm00001d041476
ZmPERK6
3
122,414,822
122,416,911
–
1524
507
56.60236
9.12
−0.039
Cell membrane/plasma membrane
Zm00001d020148
ZmPERK7
7
95,798,411
95,800,410
–
1797
598
63.45431
8.31
−0.228
Cell membrane/plasma membrane
Zm00001d035774
ZmPERK8
6
47,428,910
47,432,469
–
1788
595
63.40166
8.91
−0.311
Cell membrane/plasma membrane
Zm00001d028337
ZmPERK9
1
31,103,531
31,114,482
–
1752
583
61.77299
6.5
−0.461
Cell membrane/plasma membrane
Zm00001d012743
ZmPERK10
8
179,562,397
179,566,026
+
1233
410
45.3461
9.17
−0.463
Cell membrane/plasma membrane
Zm00001d037464
ZmPERK11
6
126,133,881
126,136,630
+
1671
556
58.84482
5.68
−0.401
Cell membrane/plasma membrane
Zm00001d011450
ZmPERK12
8
150,848,228
150,851,704
–
2052
683
71.52957
8.67
−0.488
Cell membrane/plasma membrane
Zm00001d026668
ZmPERK13
10
149,652,422
149,656,130
–
2877
958
100.93417
5.24
−0.048
Cell membrane/plasma membrane
Zm00001d049391
ZmPERK14
4
28,932,486
28,936,031
+
1002
333
36.4539
8.97
−0.07
Cell membrane/plasma membrane
Zm00001d007848
ZmPERK15
2
240,835,869
240,838,408
–
1614
535
56.9581
9.15
−0.383
Cell membrane/plasma membrane
Zm00001d010421
ZmPERK16
8
114,332,739
114,337,964
+
1989
662
69.62817
8.72
−0.523
Cell membrane/plasma membrane
Zm00001d037066
ZmPERK17
6
111,296,289
111,300,781
+
1789
662
75.50927
6.3
−0.515
Cell membrane/plasma membrane
Zm00001d031482
ZmPERK18
1
191,554,665
191,555,908
+
1401
466
51.97133
6.2
0.159
Cytoplasm
Zm00001d042185
ZmPERK19
3
155,343,261
155,349,083
–
1476
491
53.59911
9.15
−0.608
Cell membrane/plasma membrane
Zm00001d039311
ZmPERK20
3
1,451,510
1,456,285
–
1125
374
57.67767
8.41
−0.488
Cell membrane/plasma membrane
Zm00001d040127
ZmPERK21
3
28,623,779
28,627,767
–
2076
691
72.32502
8.44
−0.278
Cell membrane/plasma membrane
Zm00001d038708
ZmPERK22
6
163,090,768
163,092,715
–
2691
896
93.90373
8.61
−0.3
Cell membrane/plasma membrane
Zm00001d039176
ZmPERK23
6
171,824,226
171,828,823
+
1401
466
51.97133
6.2
0.159
Cytoplasm
3.2 Sequence logos and phylogenetic/evolutionary analysis
Sequence logos analysis provide more comprehensive information for sequence similarities, significant alignment aspects, and sequence conservation patterns. To check PERK family evolution, we generate sequence logos and results showed that this family remained conserved throughout evolution. For comparison study, the protein sequences of maize, rice and Arabidopsis were used. The results reveal that consensus sequence residues were highly conserved, and there was no compositional bias seen across any specie. These results help in discover and analyze and evaluate PERK gene family protein sequence across the species.
Phylogenetic tree serves an important way to understand the evolutionary relationships pathways. In our study we created phylogenetic tree of PERK genes to depict the evolutionary relationships. The phylogenetic or evolutionary analysis revealed the oldest plant lineage, of the PERK gene family as its members were found in Ananas comosus (angiosperm), Physcomitrella patens (bryophytes), dicots (Arabidopsis thaliana, Theobroma cacao, Populus trichocarpa), and monocots (Oryza sativa, Sorghum bicolor and Z. mays). These findings suggested that these genes evolved in ancient land plants, and that probable orthologous genes can be found across the plant kingdom. The PERK genes were characterized by 29 members in the PERK-A clade, 26 members in PERK-B, 19 members in PERK-C, and 22 members in PERK-D in the phylogenetic study. PERK genes were randomly distributed in all four clades from dicot, monocot, and bryophytes plant species, indicating that these genes evolved after the split of bryophytes. This finding showed that PERK genes possibly expanded and diversified after the radiation of these different species. These evolutionary linkages can facilitate the identification of orthologous genes and help to accelerate their functional characterization.
3.3 Predicted protein motifs, structure of exon/intron and conserved domain analysis
The 23 ZmPERK protein sequences were classified into four subfamilies using a rectangular phylogenetic tree (subfamily I, II, III, IV). 10 members were found in the Subfamily I, followed by subfamily II (5), subfamily III (4), and subfamily IV (4) (Fig. 3A). In addition, we examined the conserved motifs using MEME software to further investigate the diversity of ZmPERK protein family (Fig. 3). In this study, total 10 motifs were found (Table S3). All the gene exhibits same motif pattern. The type, order, and number of motifs were consistent within a subfamily, but varied across subfamilies. The patterns of ZmPERK protein motif distribution revealed that conserved distribution patterns existed for similar motifs. Domains 1, 2 and 3 represent the distinctive protein kinase-binding domain that is found in all 23 ZmPERK proteins (Fig. 3C). Similarly, Fig. 3D depicts the relative lengths of introns and exon sequence conservation within each ZmPERK gene in maize. A gene's biological function is linked to the distribution of exons and introns. All these genes contain exons ranged between 2 and 10. The findings demonstrated obvious conservation, laying the groundwork for functional conservatism and guiding future functional research.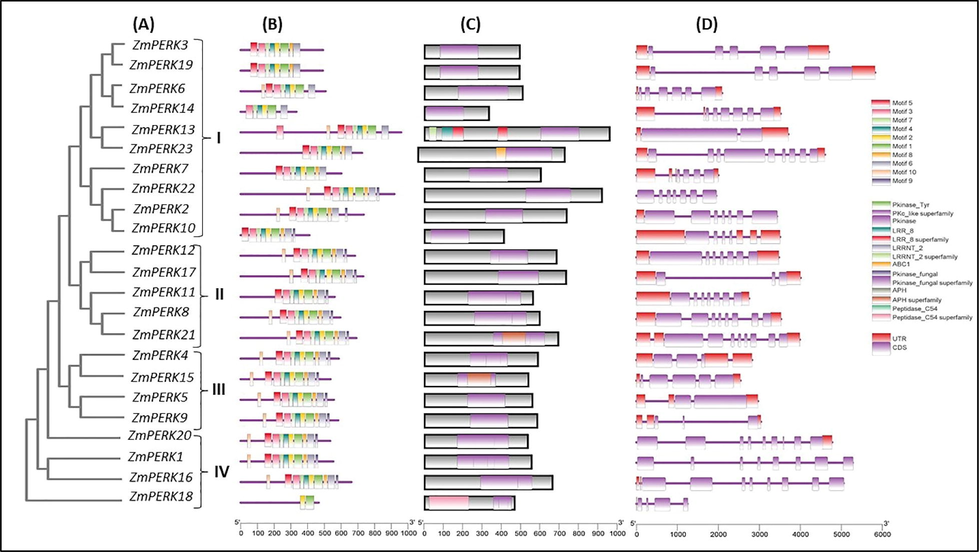
A: Phylogenetic tree-based categorization of ZmPERK genes. An un-rooted phylogenetic tree an un-rooted phylogenetic tree based on full-length peptide sequences (ZmPERK) was generated. Classification is shown based on a phylogenetic tree using differences into groups. 3B: Motif pattern of ZmPERK genes 3C: Conserved domains of maize PERK protein 3D: Exon–intron structure analyses of ZmPERK genes. The purple line represents introns, while the purple boxes represent exons.
3.4 Chromosomal localization, gene duplication events and synteny analysis
Each ZmPERK gene's genomic DNA sequence was analyzed in the maize genome database using BLASTn to establish its location, and MapChart was used to visualize the position of identified ZmPERK members on their respective chromosomes. The chromosome map revealed that 23 PERK genes were dispersed out over 8 of the 10 chromosomes. (Fig. 4A). The most ZmPERK genes were found on chr06, which had six members, followed by chr01, 03, and 08, which had 4, 5, and 4 members, respectively. While chromosomes 2, 4, 7, and 10 each had only one gene. Zmchr06 had the most PERK genes (26.08%), followed by Zmchr3 (21.73%), Zmchr1, and Zmchr8 (17.39%), while Zmchr02, Zmchr04, Zmchr06, Zmchr07, and Zmchr10 had the lowest percentage (4.34%) (Fig. 4B).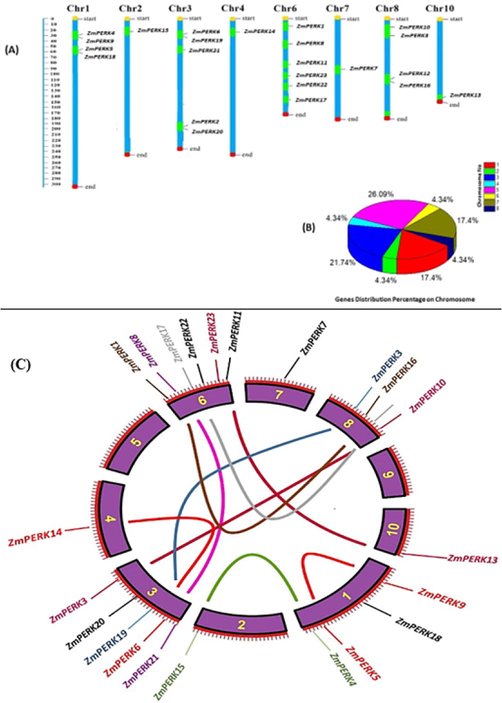
A: Distribution of 23 ZmPERK genes on their respective chromosomes.4B. Pie chart representing percentage of genes present on chromosome.4C: Pictorial representation of paralog gene pairs on chromosome indicating the type of duplication either tandem or segmental.
Gene duplications, either whole-genome or segmental, as well as tandem duplications, are critical for gene family evolution. Although it has been proven that segmental and tandem duplications play a key role in gene family evolution in all plants specie (Cannon et al., 2004).To investigate ZmPERK gene duplications and evolutionary processes in maize, we identified 09 pairs of probable paralogous genes using the maize PERK phylogenetic tree. It is well documented facts that tandem duplication observed when paralogous genes are present on the same chromosome, whereas segmental duplication arise when paralogous genes are located on distinct chromosomes (Panchy et al., 2016). All the paralogous gene pair appeared to have evolved by segmental duplication except one (ZmPERK5-ZmPERK9) which evolved through tandem duplication indicating that the evolution of PERK genes appears to have been dominated by segmental duplications in maize. (Fig. 4C) Segmental duplication is the primary force that drives the evolution of a gene family. The estimated time of divergence for paralogous gene pairs was determined using synonymous (Ks) and non-synonymous (Ka) substitution rates. The Ka/Ks ratios for all paralog ZmPERK varied between 0.10 and 0.65 (Table 2). It indicates that purifying selection may have been performed on codons in the development and proliferation of parallel PERK genes in maize. Non-synonymous and synonymous substitutions are designated by Ka and Ks, respectively.
Gene I
Gene II
Ka
Ks
Ka/Ks
Type of Duplication
T = Ks/2λ
ZmPERK3
ZmPERK19
0.022323415
0.212693
0.104956101
Segmental
6.97
ZmPERK6
ZmPERK14
0.241933478
0.521139
0.658737178
Segmental
4.21
ZmPERK13
ZmPERK23
0.05503852
0.043242
0.120303727
Segmental
1.31
ZmPERK2
ZmPERK10
0.146633906
0.370521
0.395750292
Segmental
1.21
ZmPERK12
ZmPERK17
0.45049383
1.189433
0.378746589
Segmental
3.901
ZmPERK8
ZmPERK21
0.222577445
2.176048
0.102285191
Segmental
7.13
ZmPERK4
ZmPERK15
0.291936478
0.54189
0.538737178
Segmental
1.77
ZmPERK5
ZmPERK9
0.070311852
0.062761
0.160303727
Tandem
2.05
ZmPERK1
ZmPERK16
0.028134859
0.206977
0.135932012
Segmental
6.788
Multiple collinearity scan tool was used to find orthologous genes among genomes of maize, Sorghum, and rice to further understand the Synteny links of ZmPERK genes with these plant species. (Fig. 5). 18 pairs of collinearity genes of PERK gene family between maize and rice whereas sixteen pairs in maize and sorghum were observed in the synteny analysis (Table S2). Gene IDs of all collinear genes is given supplemental file. According to these findings, the collinearity between maize and sorghum is significant as compared to the collinearity values between maize and rice furthermore, these PERK genes in maize derived from a common ancestor.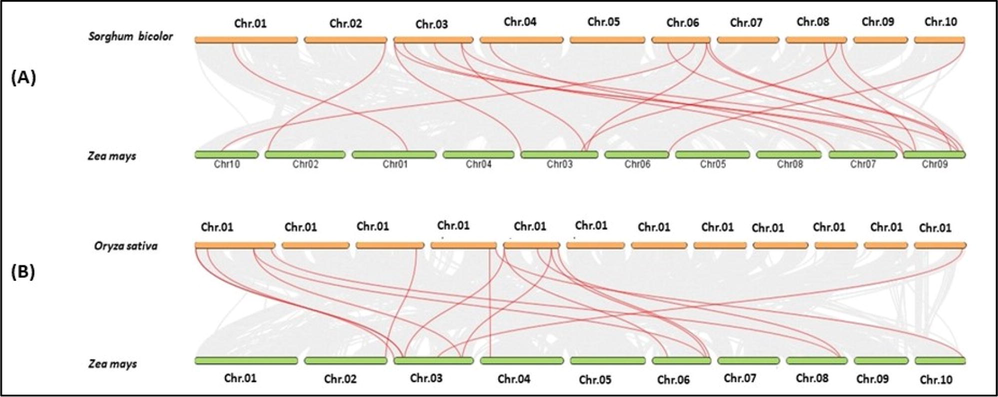
Collinearity analysis of maize, rice, and sorghum. (A) Collinearity analysis of all chromosomes reveals duplicated PERK genes in maize and sorghum. The lines connect the pairs of duplicated genes. (B) The collinearity study of maize and rice chromosomes. The PERK genes are represented by the red flags on distinct chromosomes.
3.5 Prediction of RNA editing sites, subcellular localization, and promoter analysis
The Prep-CP and Prep-Mt prediction tools were used to find the RNA editing sites of ZmPERK chloroplast and mitochondrial genes, respectively. In chloroplast genes, 196 RNA editing sites were predicted (Table 3A) and 268 in mitochondrial genes (Table 3B). All predicted editing sites in the chloroplast and mitochondrial genomes were distributed among 23 genes, with an average of 8.5 and 11.26 editing sites per gene, respectively. The chloroplast gene ZmPERK1 contains maximum RNA editing sites (12) while minimum editing sites (5) were predicted in ZmPERK14. Similarly, mitochondrial gene ZmPERK12 contain maximum editing sites (21) while minimum sites (7) were predicted in ZmPERK14. The position of RNA editing sites was further explored, and it was observed that all the predicted sites were based on first and second codon base changes. At the third codon base, we couldn't find any site for RNA editing. The transition of cytosine → uracil (C-U) seems to be present in all editing sites, resulting in amino acid substitutions. Eleven type of amino acid change found in chloroplast and mitochondrial genes (Fig. 6A). Amino acid conservation caused by RNA editing including A (Alanine) → V (Valine), T (Threonine) → I (Isoleucine), H (Histidine) → Y (Tyrosine), P (Proline) → S (Serine, P (Proline) → L (Leucine), R (Arginine) → C (Cysteine), S (Serine) → F (Phenylalanine), R (Arginine) → W (Tryptophan), P (Proline) → F (Phenylalanine), S (Serine) → L (Leucine), T (Threonine) → M (Methionine), L (Leucine)F (Phenylalanine).
Genes
Nucleotide Position
Amino acid Position
Amino acid Conservation
Genes
Nucleotide Position
Amino acid Position
Amino acid Conservation
Genes
Nucleotide Position
Amino acid Position
Amino acid Conservation
Genes
Nucleotide Position
Amino acid Position
Amino acid Conservation
ZmPERK1
14
5
TCC (S) => CTC (F)
ZmPERK7
134
45
CCG (P) => CTG (L)
ZmPERK13
319
107
CTC (L) => TTC (F)
ZmPERK19
4
2
CCC (P) => TCC (S)
175
59
CCG (P) => TCG (S)
485
162
TCT (S) => TTT (F)
850
284
CCG (P) => TCG (S)
65
22
TCG (S) => TTG (L)
203
68
CCG (P) => CTG (L)
514
172
CCG (P) => TCG (S)
853
285
CCC (P) => TCC (S)
77
26
ACG (T) => ATG (M)
280
94
CCA (P) => TCA (S)
521
174
CCA (P) => CTA (L)
1187
396
ACC (T) => ATC (I)
187
63
CAC (H) => TAC (Y)
331
111
CCT (P) => TCT (S)
856
286
CAC (H) => TAC (Y)
1246
416
CTC (L) => TTC (F)
209
70
CCG (P) => CTG (L)
338
113
CCG (P) => CTG (L)
949
317
CAC (H) => TAC (Y)
1379
460
CCA (P) => CTA (L)
250
84
CCG (P) => TCG (S)
340
114
CCG (P) => TCG (S)
1034
345
GCA (A) => GTA (V)
1556
519
TCC (S) => TTC (F)
292
98
CAT (H) => TAT (Y)
347
116
CCA (P) => CTA (L)
1225
409
CCC (P) => TTC (F)
1744
582
CAC (H) => TAC (Y)
349
117
CTT (L) => TTT (F)
544
182
CAC (H) => TAC (Y)
1226
409
CCC (P) => TTC (F)
1877
626
ACC (T) => ATC (I)
353
118
CCG (P) => CTG (L)
655
219
CAT (H) => TAT (Y)
355
119
CCG (P) => TCG (S)
ZmPERK8
236
79
CCC (P) => CTC (L)
ZmPERK14
379
127
CGG (R) => TGG (W)
362
121
CCG (P) => CTG (L)
268
90
CCG (P) => TCG (S)
424
142
CTT (L) => TTT (F)
415
139
CCC (P) => TTC (F)
313
105
CCT (P) => TCT (S)
442
148
CCA (P) => TCA (S)
ZmPERK20
211
71
CCA (P) => TCA (S)
ZmPERK2
416
139
CCC (P) => TTC (F)
812
271
TCG (S) => TTG (L)
881
294
GCC (A) => GTC (V)
236
79
CCT (P) => CTT (L)
461
154
TCA (S) => TTA (L)
1229
410
GCT (A) => GTT (V)
896
299
CCG (P) => CTG (L)
277
93
CCA (P) => TCA (S)
577
193
CCC (P) => TCC (S)
1277
426
GCT (A) => GTT (V)
322
108
CCG (P) => TCG (S)
742
248
CCG (P) => TCG (S)
1307
436
ACT (T) => ATT (I)
ZmPERK15
326
109
CCG (P) => CTG (L)
373
125
CCT (P) => TCT (S)
778
260
CAC (H) => TAC (Y)
421
141
CCG (P) => TCG (S)
389
130
CCC (P) => CTC (L)
841
281
CAC (H) => TAC (Y)
ZmPERK9
604
202
CCG (P) => TCG (S)
676
226
CAC (H) => TAC (Y)
406
136
CCA (P) => TCA (S)
979
327
CAT (H) => TAT (Y)
865
289
CAC (H) => TAC (Y)
1034
345
GCG (A) => GTG (V)
475
159
CCT (P) => TCT (S)
1093
365
CAC (H) => TAC (Y)
958
320
CAC (H) => TAC (Y)
1187
396
GCC (A) => GTC (V)
512
171
ACA (T) => ATA (I)
997
333
CCG (P) => TCG (S)
1241
414
CCG (P) => CTG (L)
863
288
ACC (T) => ATC (I)
1063
355
CCC (P) => TCC (S)
1244
415
ACG (T) => ATG (M)
ZmPERK21
826
276
CCG (P) => TCG (S)
ZmPERK3
1036
346
CCT (P) => TCT (S)
1457
486
TCC (S) => TTC (F)
1306
436
CCC (P) => TTC (F)
851
284
ACG (T) => ATG (M)
1103
368
CCT (P) => CTT (L)
1307
436
CCC (P) => TTC (F)
1000
334
CCG (P) => TCG (S)
1151
384
CCT (P) => CTT (L)
ZmPERK10
19
7
CTT (L) => TTT (F)
1520
507
CCC (P) => CTC (L)
1003
335
CCT (P) => TCT (S)
1187
396
TCG (S) => TTG (L)
241
81
CAC (H) => TAC (Y)
1040
347
CCG (P) => CTG (L)
1195
399
CAT (H) => TAT (Y)
556
186
CCT (P) => TCT (S)
ZmPERK16
529
177
CCG (P) => TCG (S)
1072
358
CCG (P) => TCG (S)
1214
405
CCG (P) => CTG (L)
737
246
GCT (A) => GTT (V)
586
196
CCA (P) => TCA (S)
1117
373
CCA (P) => TCA (S)
842
281
GCG (A) => GTG (V)
628
210
CCT (P) => TCT (S)
1148
383
TCC (S) => TTC (F)
289
97
CCC (P) => TCC (S)
883
295
CCG (P) => TCG (S)
1021
341
CAC (H) => TAC (Y)
1312
438
CCT (P) => TCT (S)
ZmPERK4
482
161
CCG (P) => CTG (L)
1132
378
CTC (L) => TTC (F)
1327
443
CAT (H) => TAT (Y)
491
164
CCG (P) => CTG (L)
1172
391
CCG (P) => CTG (L)
1379
460
GCA (A) => GTA (V)
ZmPERK22
205
69
CCC (P) => TCC (S)
565
189
CAT (H) => TAT (Y)
1435
479
CTT (L) => TTT (F)
286
96
CAC (H) => TAC (Y)
577
193
CCC (P) => TCC (S)
ZmPERK11
178
60
CCG (P) => TCG (S)
1441
481
CTT (L) => TTT (F)
611
204
CCA (P) => CTA (L)
619
207
CCG (P) => TCG (S)
181
61
CCA (P) => TCA (S)
715
239
CCT (P) => TCT (S)
626
209
ACA (T) => ATA (I)
211
71
CCG (P) => TCG (S)
ZmPERK17
106
36
CCG (P) => TCG (S)
965
322
GCA (A) => GTA (V)
859
287
CAC (H) => TAC (Y)
259
87
CCT (P) => TTT (F)
121
41
CCT (P) => TTT (F)
1307
436
CCA (P) => CTA (L)
260
87
CCT (P) => TTT (F)
122
41
CCT (P) => TTT (F)
1319
440
ACC (T) => ATC (I)
143
48
TCG (S) => TTG (L)
302
101
TCT (S) => TTT (F)
167
56
TCA (S) => TTA (L)
1358
453
CCA (P) => CTA (L)
ZmPERK5
238
80
CCG (P) => TCG (S)
352
118
CCG (P) => TCG (S)
329
110
CCG (P) => CTG (L)
1385
462
CCA (P) => CTA (L)
752
251
TCA (S) => TTA (L)
359
120
CCG (P) => CTG (L)
338
113
CCA (P) => CTA (L)
1010
337
GCC (A) => GTC (V)
376
126
CCC (P) => TCC (S)
382
128
CCT (P) => TTT (F)
ZmPERK23
649
217
CCT (P) => TCT (S)
1031
344
CCG (P) => CTG (L)
388
130
CCG (P) => TCG (S)
383
128
CCT (P) => TTT (F)
652
218
CCC (P) => TCC (S)
1226
409
GCC (A) => GTC (V)
425
142
TCT (S) => TTT (F)
793
265
CTT (L) => TTT (F)
1367
456
TCC (S) => TTC (F)
ZmPERK12
389
130
CCG (P) => CTG (L)
821
274
TCG (S) => TTG (L)
1394
465
GCG (A) => GTG (V)
394
132
CCG (P) => TCG (S)
ZmPERK18
863
288
ACT (T) => ATT (I)
842
281
TCC (S) => TTC (F)
430
144
CCC (P) => TCC (S)
1036
346
CCT (P) => TCT (S)
847
283
CCA (P) => TCA (S)
275
92
ACT (T) => ATT (I)
535
179
CCA (P) => TCA (S)
1150
384
CCT (P) => TCT (S)
859
287
CCG (P) => TCG (S)
ZmPERK6
400
134
CCT (P) => TTT (F)
542
181
CCG (P) => CTG (L)
1190
397
CCT (P) => CTT (L)
916
306
CCA (P) => TCA (S)
401
134
CCT (P) => TTT (F)
571
191
CCG (P) => TCG (S)
1198
400
CAT (H) => TAT (Y)
1252
418
CTC (L) => TTC (F)
512
171
ACA (T) => ATA (I)
920
307
TCG (S) => TTG (L)
1274
425
ACC (T) => ATC (I)
940
314
CCC (P) => TCC (S)
1105
369
CTC (L) => TTC (F)
1319
440
ACC (T) => ATC (I)
1319
440
ACG (T) => ATG (M)
1369
457
CCT (P) => TCT (S)
1343
448
GCG (A) => GTG (V)
1439
480
TCC (S) => TTC (F)
1490
497
ACG (T) => ATG (M)
Genes
Nucleotide Position
Amino acid Position
Amino acid Conservation
Genes
Nucleotide Position
Amino acid Position
Amino acid Conservation
Genes
Nucleotide Position
Amino acid Position
Amino acid Conservation
Genes
Nucleotide Position
Amino acid Position
Amino acid Conservation
ZmPERK1
5
2
ACG (T) => ATG (M)
ZmPERK8
8
3
TCC (S) => TTC (F)
ZmPERK13
319
107
CTC (L) => TTC (F)
ZmPERK20
104
35
CCG (P) => CTG (L)
38
13
TCG (S) => TTG (L)
31
11
CCG (P) => TCG (S)
850
284
CCG (P) => TCG (S)
121
41
CCC (P) => TCC (S)
40
14
CCC (P) => TCC (S)
38
13
CCG (P) => CTG (L)
853
285
CCC (P) => TCC (S)
149
50
TCG (S) => TTG (L)
82
28
CTC (L) => TTC (F)
80
27
GCC (A) => GTC (V)
1187
396
ACC (T) => ATC (I)
182
61
GCA (A) => GTA (V)
185
62
CCT (P) => CTT (L)
103
35
CCC (P) => TCC (S)
1246
416
CTC (L) => TTC (F)
239
80
CCA (P) => CTA (L)
215
72
CCC (P) => CTC (L)
107
36
GCA (A) => GTA (V)
1379
460
CCA (P) => CTA (L)
272
91
TCT (S) => TTT (F)
284
95
CCC (P) => CTC (L)
139
47
CAC (H) => TAC (Y)
1556
519
TCC (S) => TTC (F)
277
93
CCA (P) => TCA (S)
305
102
ACC (T) => ATC (I)
215
72
CCA (P) => CTA (L)
1744
582
CAC (H) => TAC (Y)
284
95
GCC (A) => GTC (V)
314
105
GCA (A) => GTA (V)
250
84
CCG (P) => TCG (S)
1877
626
ACC (T) => ATC (I)
305
102
CCA (P) => CTA (L)
328
110
CCT (P) => TCT (S)
278
93
TCG (S) => TTG (L)
1925
642
GCC (A) => GTC (V)
320
107
GCG (A) => GTG (V)
332
111
CCT (P) => CTT (L)
302
101
GCC (A) => GTC (V)
2045
682
CCC (P) => CTC (L)
329
110
GCA (A) => GTA (V)
362
121
GCG (A) => GTG (V)
308
103
GCA (A) => GTA (V)
2054
685
GCT (A) => GTT (V)
329
110
CCT (P) => CTT (L)
ZmPERK21
535
179
CCT (P) => TCT (S)
ZmPERK2
389
130
CCT (P) => CTT (L)
341
114
GCA (A) => GTA (V)
ZmPERK14
92
31
ACG (T) => ATG (M)
545
182
CCG (P) => CTG (L)
391
131
CCA (P) => TCA (S)
428
143
CCG (P) => CTG (L)
170
57
TCA (S) => TTA (L)
550
184
CCA (P) => TCA (S)
406
136
CCG (P) => TCG (S)
386
129
CCT (P) => CTT (L)
566
189
TCT (S) => TTT (F)
440
147
CCA (P) => CTA (L)
ZmPERK9
164
55
CCG (P) => CTG (L)
394
132
CAT (H) => TAT (Y)
641
214
CCT (P) => CTT (L)
469
157
CCG (P) => TCG (S)
166
56
CCA (P) => TCA (S)
752
251
GCA (A) => GTA (V)
749
250
GCG (A) => GTG (V)
512
171
TCA (S) => TTA (L)
230
77
TCG (S) => TTG (L)
923
308
GCA (A) => GTA (V)
815
272
GCG (A) => GTG (V)
566
189
ACG (T) => ATG (M)
254
85
GCT (A) => GTT (V)
950
317
GCC (A) => GTC (V)
829
277
CCG (P) => TCG (S)
572
191
TCA (S) => TTA (L)
389
130
GCA (A) => GTA (V)
836
279
CCT (P) => CTT (L)
578
193
CCC (P) => CTC (L)
457
153
CCG (P) => TCG (S)
ZmPERK15
32
11
CCG (P) => CTG (L)
857
286
CCG (P) => CTG (L)
808
270
CCG (P) => TCG (S)
671
224
TCG (S) => TTG (L)
106
36
CCC (P) => TCC (S)
935
312
CCG (P) => CTG (L)
877
293
CCT (P) => TCT (S)
803
268
GCC (A) => GTC (V)
230
77
GCG (A) => GTG (V)
881
294
TCG (S) => TTG (L)
922
308
CTT (L) => TTT (F)
233
78
GCG (A) => GTG (V)
ZmPERK22
34
12
CAC (H) => TAC (Y)
958
320
CAC (H) => TAC (Y)
281
94
GCC (A) => GTC (V)
76
26
CCA (P) => TCA (S)
ZmPERK3
223
75
CTT (L) => TTT (F)
1358
453
CCC (P) => CTC (L)
326
109
CCG (P) => CTG (L)
205
69
CCC (P) => TCC (S)
317
106
GCT (A) => GTT (V)
1741
581
CAA (Q) => TAA (X)
404
135
GCG (A) => GTG (V)
667
223
CCT (P) => TCT (S)
931
311
CCT (P) => TCT (S)
428
143
CCG (P) => CTG (L)
836
279
CCA (P) => CTA (L)
1198
400
CAC (H) => TAC (Y)
ZmPERK10
5
2
ACG (T) => ATG (M)
430
144
CCG (P) => TCG (S)
890
297
CCA (P) => CTA (L)
1277
426
GCC (A) => GTC (V)
8
3
CCG (P) => CTG (L)
439
147
CCG (P) => TCG (S)
1000
334
CAT (H) => TAT (Y)
1322
441
ACC (T) => ATC (I)
10
4
CCG (P) => TCG (S)
488
163
GCG (A) => GTG (V)
1033
345
CCC (P) => TCC (S)
1373
458
CCA (P) => CTA (L)
31
11
CCG (P) => TCG (S)
571
191
CCC (P) => TCC (S)
1040
347
GCT (A) => GTT (V)
1382
461
ACC (T) => ATC (I)
98
33
GCG (A) => GTG (V)
614
205
GCC (A) => GTC (V)
1181
394
TCG (S) => TTG (L)
476
159
GCG (A) => GTG (V)
659
220
ACC (T) => ATC (I)
ZmPERK4
8
3
TCT (S) => TTT (F)
608
203
GCT (A) => GTT (V)
748
250
CCC (P) => TCC (S)
ZmPERK23
125
42
CCG (P) => CTG (L)
31
11
CCA (P) => TCA (S)
614
205
ACA (T) => ATA (I)
155
52
CCG (P) => CTG (L)
38
13
CCG (P) => CTG (L)
884
295
CCG (P) => CTG (L)
ZmPERK16
235
79
CCC (P) => TTC (F)
191
64
TCG (S) => TTG (L)
41
14
TCT (S) => TTT (F)
236
79
CCC (P) => TTC (F)
218
73
GCA (A) => GTA (V)
74
25
TCT (S) => TTT (F)
ZmPERK11
34
12
CCT (P) => TCT (S)
302
101
CCA (P) => CTA (L)
395
132
GCC (A) => GTC (V)
89
30
ACT (T) => ATT (I)
77
26
ACG (T) => ATG (M)
331
111
CCG (P) => TCG (S)
944
315
CCA (P) => CTA (L)
107
36
GCG (A) => GTG (V)
176
59
ACT (T) => ATT (I)
425
142
ACG (T) => ATG (M)
1165
389
CCT (P) => TTT (F)
137
46
CCC (P) => CTC (L)
194
65
CCG (P) => CTG (L)
571
191
CCG (P) => TCG (S)
1166
389
CCT (P) => TTT (F)
164
55
GCG (A) => GTG (V)
224
75
CCC (P) => CTC (L)
626
209
CCT (P) => CTT (L)
1405
469
CCG (P) => TCG (S)
182
61
GCT (A) => GTT (V)
232
78
CCA (P) => TCA (S)
632
211
GCT (A) => GTT (V)
1739
580
GCT (A) => GTT (V)
197
66
CCA (P) => CTA (L)
239
80
GCT (A) => GTT (V)
815
272
TCG (S) => TTG (L)
272
91
CCC (P) => CTC (L)
254
85
CCT (P) => CTT (L)
1021
341
CAC (H) => TAC (Y)
260
87
CCT (P) => CTT (L)
1124
375
GCG (A) => GTG (V)
ZmPERK5
208
70
CCC (P) => TCC (S)
289
97
CCT (P) => TTT (F)
1228
410
CAT (H) => TAT (Y)
236
79
CCG (P) => CTG (L)
290
97
CCT (P) => TTT (F)
1277
426
GCT (A) => GTT (V)
257
86
ACC (T) => ATC (I)
340
114
CCA (P) => TCA (S)
577
193
CCG (P) => TCG (S)
347
116
GCA (A) => GTA (V)
ZmPERK17
34
12
CAC (H) => TAC (Y)
686
229
GCG (A) => GTG (V)
371
124
CCG (P) => CTG (L)
76
26
CCA (P) => TCA (S)
943
315
CCA (P) => TCA (S)
383
128
GCC (A) => GTC (V)
205
69
CCC (P) => TCC (S)
1010
337
GCC (A) => GTC (V)
395
132
CCT (P) => CTT (L)
667
223
CCT (P) => TCT (S)
1094
365
GCG (A) => GTG (V)
836
279
CCA (P) => CTA (L)
ZmPERK12
266
89
TCG (S) => TTG (L)
890
297
CCA (P) => CTA (L)
ZmPERK6
49
17
CCA (P) => TCA (S)
268
90
CCG (P) => TCG (S)
1000
334
CAT (H) => TAT (Y)
64
22
CCT (P) => TTT (F)
296
99
CCT (P) => CTT (L)
1033
345
CCC (P) => TCC (S)
65
22
CCT (P) => TTT (F)
299
100
CCT (P) => CTT (L)
1040
347
GCT (A) => GTT (V)
92
31
ACA (T) => ATA (I)
304
102
CCG (P) => TCG (S)
1181
394
TCG (S) => TTG (L)
775
259
CTT (L) => TTT (F)
332
111
CCG (P) => CTG (L)
1316
439
TCA (S) => TTA (L)
356
119
GCG (A) => GTG (V)
ZmPERK18
55
19
CCC (P) => TTC (F)
1412
471
ACA (T) => ATA (I)
359
120
GCG (A) => GTG (V)
56
19
CCC (P) => TTC (F)
1420
474
CCG (P) => TCG (S)
386
129
GCA (A) => GTA (V)
62
21
GCC (A) => GTC (V)
1441
481
CCG (P) => TCG (S)
395
132
CCG (P) => CTG (L)
83
28
GCC (A) => GTC (V)
440
147
GCC (A) => GTC (V)
223
75
CTC (L) => TTC (F)
ZmPERK7
65
22
CCG (P) => CTG (L)
449
150
ACG (T) => ATG (M)
317
106
GCT (A) => GTT (V)
109
37
CTT (L) => TTT (F)
461
154
TCA (S) => TTA (L)
931
311
CCT (P) => TCT (S)
137
46
GCC (A) => GTC (V)
476
159
GCG (A) => GTG (V)
1127
376
TCA (S) => TTA (L)
143
48
GCG (A) => GTG (V)
521
174
ACC (T) => ATC (I)
1226
409
GCT (A) => GTT (V)
257
86
GCT (A) => GTT (V)
524
175
GCC (A) => GTC (V)
1370
457
CCA (P) => CTA (L)
260
87
CCA (P) => CTA (L)
535
179
CCA (P) => TCA (S)
1379
460
ACC (T) => ATC (I)
275
92
GCG (A) => GTG (V)
616
206
CCA (P) => TCA (S)
290
97
TCA (S) => TTA (L)
620
207
CCT (P) => CTT (L)
ZmPERK19
4
2
CCC (P) => TTC (F)
311
104
GCG (A) => GTG (V)
644
215
CCG (P) => CTG (L)
5
2
CCC (P) => TTC (F)
320
107
GCT (A) => GTT (V)
674
225
TCG (S) => TTG (L)
26
9
CCG (P) => CTG (L)
323
108
GCT (A) => GTT (V)
38
13
TCG (S) => TTG (L)
524
175
ACA (T) => ATA (I)
43
15
CCG (P) => TCG (S)
614
205
CCC (P) => CTC (L)
59
20
TCT (S) => TTT (F)
770
257
GCG (A) => GTG (V)
70
24
CTT (L) => TTT (F)
98
33
GCG (A) => GTG (V)
194
65
CCG (P) => CTG (L)
200
67
CCG (P) => CTG (L)
299
100
GCC (A) => GTC (V)
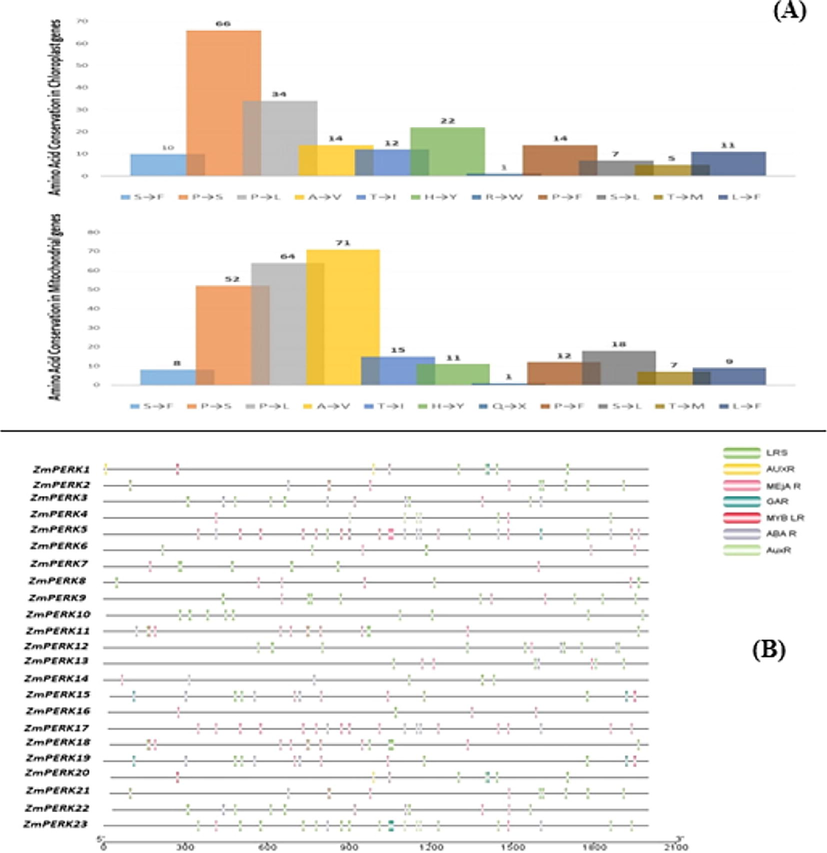
(A) RNA editing of the PERK genes results in amino acid conservation. (B) Identified cis-acting elements in ZmPERK gene family promoters.
Location of gene at cellular level was also determined. Results indicated that 21 of the 23 ZmPERK proteins were localized to the plasma membrane, while two (ZmPERK18 and ZmPERK23) were localized to the cytoplasm, Table1 contains the details of these parameters. The promoter region, which is located upstream of the start codon area, controls gene transcription. Understanding gene regulation and function requires a thorough examination of cis-elements (Higo et al., 1999). We discovered and classified cis-acting factors in the upstream region of the ZmPERK genes. (See Table S4) The cis-elements were categorized based on their roles in growth and development, as well as light and stress actions. The upstream region of ZmPERK genes contained cis-acting factors like MeJA responsive, MYB-binding sites associated with light responsive elements, ABA responsive elements, defense, stress, low temperature, gibberellin acid (GA), and salicylic acid (SA)responsive elements, as per the promoter analysis results (Fig. 6B). The promoters of the ZmPERK gene have the most MeJA responsiveness elements. where they were found in 296 promoters. There were 166 light responsive elements, 88 ABA responsive elements, 27 GA responsive elements, 25 MYB light responsive elements, and 4 auxin responsive elements. The cis-element analysis showed that during abiotic stress and plant development phase the ZmPERK genes could respond.
3.6 In-silico expressions analysis
Expression patterns give information regarding the biological activities of genes because gene expression is required for optimal regulation of plant growth and development. We looked examined the expression patterns of the ZmPERK under drought stress and at different stages of seed embryo development from 15 days after pollination (DAP) to 40 days after pollination (DAP) in two distinct varieties (High oil content and low oil content) (Fig. 7A).Under drought stress 9 genes were upregulated in both tissues leave and cob (ZmPERK2, ZmPERK 3, ZmPERK 8, ZmPERK 10, ZmPERK 14, ZmPERK 16, ZmPERK 19, ZmPERK 20) 4 genes (ZmPERK 1, ZmPERK 7, ZmPERK 18, ZmPERK 21) only upregulated in leave tissue and 1 gene ZmPERK 18 upregulated in cob (Fig. 7B). While Seven genes downregulated (ZmPERK 4, ZmPERK 5, ZmPERK 11, ZmPERK 12, ZmPERK 15, ZmPERK 17, ZmPERK 22) under drought stress.For oil content accumulation in embryo 9 genes shows upregulated expression pattern (ZmPERK 1, ZmPERK 3, ZmPERK 6, ZmPERK 8, ZmPERK 13, ZmPERK 14, ZmPERK 16, ZmPERK 19 ZmPERK,20) while 12 genes show downregualted trend (ZmPERK 2, ZmPERK 5, ZmPERK 6, ZmPERK 7, ZmPERK 9, ZmPERK 10, ZmPERK 11, ZmPERK 12, ZmPERK 17, ZmPERK 18, ZmPERK 22, ZmPERK 23). Interestingly, some genes show similar expression pattern under both conditions. For example, ZmPERK 3 ZmPERK 8 ZmPERK 14 ZmPERK 16 ZmPERK 19 ZmPERK 20 regardless of the tissues or stresses applied, they were always upregulated. We may conclude from these findings that ZmPERK gene expression is involved in drought stress and oil content accumulation in the embryo.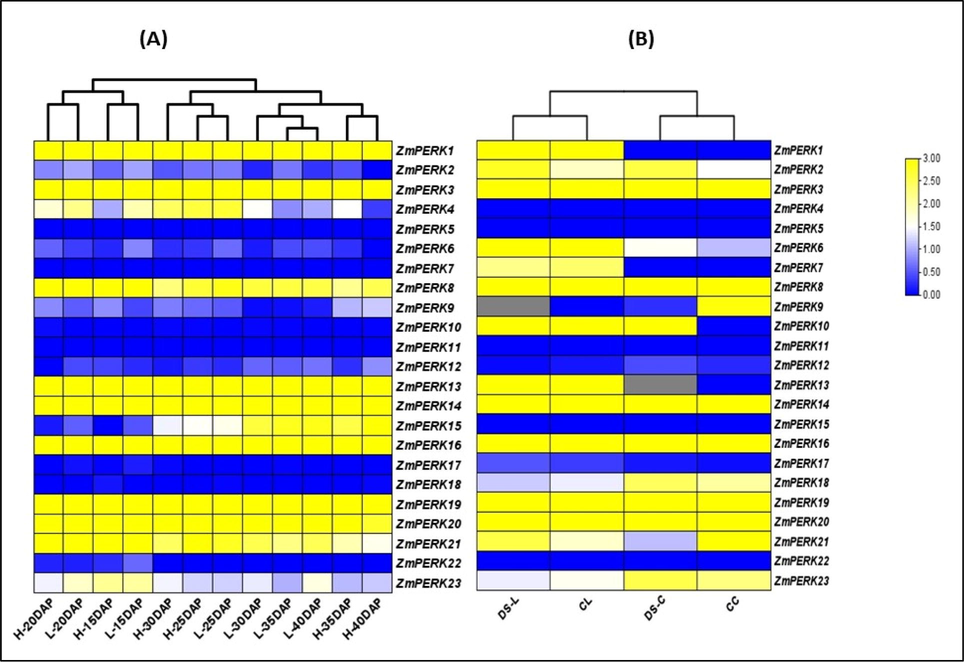
A: ZmPERK gene expression patterns at different developmental stages of seed embryos from 15 days after pollination (DAP) to 40 days after pollination (DAP). H-represent expression in high oil content varaiety while L represent low oil content varaiety B: Expression pattern of ZmPERKgene under drought stress DS-L(Drought stress leave sample) DS-C (Drought stress cob sample) CL(Controled leave sample) CC (Control Cob sample.
4 Discussion
Many ancient land plants evolved over the time also possess PERKs genes (Nakhamchik et al., 2004, Qanmber et al., 2019, Chen et al., 2020) which depict that these gene families are present from centuries in the plants. Now a day’s modern techniques of DNA sequences have revolutionized the field of DNA sequencing especially the advent of next-generation sequencing technologies, have shorten the time of sequencing with more accuracy in the results. The availability of maize genome assemblies has open the new avenues for studying various functions of genes at genome-wide level. There has been no systematic study of maize to date. However, in our study we discovered 23 ZmPERK genes in the maize genome. In phylogenetic analysis, we divided ZmPERK genes into four groups. The findings demonstrated that PERK genes were initially originated in ancient land plants and their orthologous genes may be found throughout the plant kingdom. Plant PERK genes from dicot, monocot, lycophytes, and chlorophytes were assigned to each of the four clades randomly. Our study showed that ZmPERK genes remained evolutionarily conserved, as these were found in each of the species which was used in this study. Furthermore, the expansion of these gene into higher plants was occurred with the passage of time. According to sequence logos for PERK genes, the protein sequence residues were highly conserved, and no compositional bias was seen across the studied species. To study the evolutionary history of multiple gene families (Ohta, 2010). It is essential to know the structure of the genes. The length of nucleotide and amino acid sequences varied considerably, indicating that the ZmPERK genes are diverse. It is quite worth to study the exon–intron structure as insertion/deletion events play important role in determine the structure of exon–intron. The introns gain or lose have been witnessed throughout eukaryotic diversification. The exon–intron pattern of duplicated genes is similar, whereas more diversification observed in the intron length suggesting that the intron length may be significant in ZmPERK functional diversification. In ZmPERK proteins, different combinations of conserved motifs were identified. Protein motif analysis revealed that protein from same species were tend to fall in the same cluster together. Arrangements of motif were comparable among members of the same subfamily.
The study of gene duplication events is critical as these play crucial role in the genome expansions and alignments (Tamura et al., 2011). The gene duplication events have been witnessed in various transcription factor families of plants (Liu et al., 2011; Shan et al., 2013). To differentiate whether the gene duplication was the result of tandem or segmental if the duplications are the result of the presence two or more genes the same chromosome, it will be considered as tandem duplication whereas segmental or WGD duplications are when two or more genes are duplicated on different chromosomes. The intron expansion is mainly the result of the tandem duplications and thus give rise to the formation of the new genes (Yang et al., 2008), but we only find evidence of one tandem and eight segmental duplications in this study. In Plants the environmental and selection factors have expanded multiple gene families more than other eukaryotes organisms. The Ka/Ks ratios demonstrated that maize PERK genes have been subjected to extensive selection, with relatively minimal functional variations due to whole genome and segmental duplication.
ZmPERK genes possess cis-elements associated with stress responses in their promoter regions. ZmPERK contains cis-elements such as MeJA responsive, MYB-binding sites associated with light responsiveness elements, ABA responsive, defense, and stress responsive, low temperature and gibberellin acid (GA) responsive elements. The presence of these cis elements with specified characteristics demonstrated the putative role in plant growth, development, as well as in biotic and abiotic stress response. Synteny is a framework for assessing homologous gene and gene order conservation across genomes of different species. The collinearity between maize and sorghum was shown to be more significant than the collinearity between maize and rice.
Plant growth and development are assisted by RNA editing, which is an effective strategy for regulating gene expression at the post-transcriptional level in higher plant organelle genomes. The discovery and identification of RNA editing sites is critical for a better knowledge of their biological activities and establishing the framework for future research and comprehension of their molecular processes. In this work, the RNA editing sites of chloroplast and mitochondrial genes in maize were predicted. Table 3A lists 196 RNA editing sites predicted in chloroplast genes and 268 in mitochondrial genes (Table 3B). In the chloroplast and mitochondrial genomes, these sites were detected on 23 genes, with an average of 8.5 and 11.26 editing sites per gene, respectively. The transition and conservation of cytosine (C) to uracil (U) was observed in all of the editing sites. Changes in the first and second codon nucleotides were mostly involved for these transitions. The current study laid the groundwork for future research into the biological functions of chloroplast and mitochondrial RNA editing in maize. The expression patterns of genes are closely related to their biological functions. According to expression analyses ZmPERK genes were shown to be important in drought tolerance and oil content accumulation in embryos.
5 Conclusion
The current study found 23 non-redundant ZmPERK encoding genes in maize. The PERK gene family is conserved among the analyzed plant species, according to their classification, characterization in terms of gene structure, motif, conserved domains, and comparative phylogenetic analyses. Furthermore, gene duplication analysis and syntenic relationship studies reveal that the maize paralogous genes proliferate through segmental duplications, whereas codons went under purifying selection, resulting in a significant expansion of the ZmPERK gene family. The existence of putative cis-elements in the ZmPERK gene promoter regions suggests that they have a functional role in growth, development, and stress resilience. Most of the genes were found to be up regulated in response to stress and oil content, accumulation showing that they may play a role in stress modulation and development process in maize. Overall, these findings will assist in the functional characterization of maize PERK genes. The candidate ZmPERK genes can be employed in a breeding program.
Acknowledgement
This project was supported by Researchers Supporting Project Number (RSP-2023R7) King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Effect of Piriformospora indica and Azotobacter chroococcum on mitigation of zinc deficiency stress in wheat (Triticum aestivum L.) Symbiosis. 2016;69(1):9-19.
- [Google Scholar]
- Plasma membrane-associated proline-rich extensin-like receptor kinase 4, a novel regulator of Ca2+ signalling, is required for abscisic acid responses in Arabidopsis thaliana. Plant J.. 2009;60(2):314-327.
- [Google Scholar]
- The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol.. 2004;4(1):1-21.
- [Google Scholar]
- Genome-wide analysis of proline-rich extension-like receptor protein kinase (PERK) in Brassica rapa and its association with the pollen development. BMC Genomics. 2020;21(1):1-13.
- [Google Scholar]
- CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell. 2003;15(5):1198-1211.
- [Google Scholar]
- Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res.. 1999;27(1):297-300.
- [Google Scholar]
- Cell wall-associated ROOT HAIR SPECIFIC 10, a proline-rich receptor-like kinase, is a negative modulator of Arabidopsis root hair growth. J. Exp. Bot.. 2016;67(6):2007-2022.
- [Google Scholar]
- Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genomics. 2013;14(1):1-15.
- [Google Scholar]
- Transcription factors as molecular switches to regulate drought adaptation in maize. Theor. Appl. Genet.. 2020;133(5):1455-1465.
- [Google Scholar]
- SMART: recent updates, new developments and status in 2015. Nucleic Acids Res.. 2015;43(D1):D257-D260.
- [Google Scholar]
- Genome-wide analysis of the auxin response factor (ARF) gene family in maize (Zea mays) Plant Growth Regul.. 2011;63(3):225-234.
- [Google Scholar]
- Genetic dissection of maize drought tolerance for trait improvement. Mol. Breed.. 2021;41(2):1-13.
- [Google Scholar]
- Some current aspects of stomatal physiology. Ann. Rev. Plant Physiol. Plant Mol. Biol.. 1990;41:55-75.
- [Google Scholar]
- Receptor-like protein kinases: the keys to response. Curr. Opin. Plant Biol.. 2003;6(4):339-342.
- [Google Scholar]
- The PREP suite: predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res.. 2009;37(suppl_2):W253-W259.
- [Google Scholar]
- A comprehensive expression analysis of the Arabidopsis proline-rich extensin-like receptor kinase gene family using bioinformatic and experimental approaches. Plant Cell Physiol.. 2004;45(12):1875-1881.
- [Google Scholar]
- Gene conversion and evolution of gene families: an overview. Genes.. 2010;1(3):349-356.
- [Google Scholar]
- Genome-wide identification and characterization of the PERK gene family in Gossypium hirsutum reveals gene duplication and functional divergence. Int. J. Mol. Sci.. 2019;20(7):1750.
- [Google Scholar]
- Double jeopardy: both overexpression and suppression of a redox-activated plant mitogen-activated protein kinase render tobacco plants ozone sensitive. Plant Cell. 2002;14(9):2059-2069.
- [Google Scholar]
- Maize gene atlas developed by RNA sequencing and comparative evaluation of transcriptomes based on RNA sequencing and microarrays. PLoS ONE. 2013;8(4):e61005.
- [Google Scholar]
- Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol.. 2013;31(8):686-688.
- [Google Scholar]
- Plant receptor-like kinase gene family: diversity, functions, and signaling. Sci. STKE.. 2001;18:113-122.
- [Google Scholar]
- Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci.. 2001;98(19):10763-10768.
- [Google Scholar]
- Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16(5):1220-1234.
- [Google Scholar]
- Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Develop. 2004;131(7):1491-1501.
- [Google Scholar]
- Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA leucine-rich repeat receptor–like kinase signaling pathway that regulates organ shape. Plant Cell. 2003;15(5):1095-1110.
- [Google Scholar]
- The proline-rich, extensin-like receptor kinase-1 (PERK1) gene is rapidly induced by wounding. Plant Mol. Biol.. 2002;50(4):667-685.
- [Google Scholar]
- MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol.. 2011;28(10):2731-2739.
- [Google Scholar]
- High temperature stress tolerance in maize (Zea mays L.): Physiological and molecular mechanisms. Journal of. Plant Biology.. 2019;62(2):93-102.
- [Google Scholar]
- MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered.. 2002;93(1):77-78.
- [Google Scholar]
- MKK5 regulates high light-induced gene expression of Cu/Zn superoxide dismutase 1 and 2 in Arabidopsis. Plant Cell Physiol.. 2013;54(7):1217-1227.
- [Google Scholar]
- Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol. Genet. Genomics. 2008;280(3):187-198.
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2022.102293.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







