Prediction of protein aggregation on key proteins involved in ischemic stroke
⁎Corresponding author. alaguraj.veluchamy@kaust.edu.sa (Alaguraj Veluchamy)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Stroke is a genetic condition comprising multiple subtypes and arising from both genic and other multi factors. Genetic basis of stroke is well established through several studies. Advances in integrating sequencing methods and Genome-wide association studies have shown that genetics of stroke is manifested in several genic disorders. Many of the neurodegenerative disorders show aggravated protein aggregation through amyloid formation. Through the protein aggregation prediction, we observed a higher protein disorder in 46 stroke-associated proteins. Also, we observed a large number of aggregation residues distributed as a pattern in multiple regions of these candidate proteins. Overall, we present a study showing that there is a possible interrelationship between protein aggregation and stroke.
Keywords
Genetic disorder
Ischemic stroke
Protein aggregation
Gene ontology
KEGG pathway
1 Introduction
Stroke is a complex heterogenous condition that is one of the major causes of ailment and death in the world. Characterizing stroke involves classification into subtypes. Several classification systems have been proposed to differentiate subtypes of stroke and distinguish between ischemic and hemorrhagic stroke, subarachnoid hemorrhage, cerebral venous thrombosis, and spinal cord stroke (Amarenco et al., 2009). Studies such as the association of monozygotic twins to stroke provide the evidence of implication of genetic factors in stroke pathophysiology. Next generation sequencing (NGS), genome-wide association studies (GWAS) provided evidence that stroke is both a monogenic and a polygenic disorder. Both above classification systems and genetic studies are vital in grouping patients for therapeutic purposes.
Whole-genome sequencing studies have resulted identifying novel variants associated with stroke subtypes. A genome-wide association study through Trans-Omics for Precision Medicine (TOPMed) Program identified 5 novel loci associated with subtypes of stroke in a multi-ancestry population (Hu et al., 2022). Besides this, MEGASTROKE consortium have performed genotyping and GWAS studies that resulted in identification of stroke risk variants such as NKX2-5, ANK2, LRCH1, REEP3, JAZF1(de Vries et al., 2019; Malik et al., 2018).
Although multiple stroke specific risk loci are detected, functional characterization of these loci are challenging as these variants fall mostly on non-coding regions of the genome. Hence recent approaches use data from gene expression, DNA methylation to establish a causal relationship to stroke susceptible genes. Analysis of candidate genes involved in ischemic stroke resulted in identification of at least five susceptibility genes such as factor V Leiden Gln506, ACE I/D, MTHFR C677T, prothrombin G20210A, PAI-1 5G (Bentley et al., 2010).
Emerging evidence shows that protein aggregates formed in Ischemic stroke. Misfolded proteins tend to form fibers of aggregates (Hu et al., 2001). In particular ischemic stroke (Luo et al., 2013); stroke and aggregation (Zhang et al., 2020). Protein aggregates are found to be form deposits in degenerate cells and are involved in cellular toxicity (Tutar et al., 2013). Several neurological disorders such as Parkinson’s Disease (PD), Huntington’s disease (HD), prion diseases, Amyotrophic lateral sclerosis (ALS) are associated with the formation of protein aggregate (Pedersen & Heegaard, 2013). Aβ-peptide (1–40/1–42) forms amyloid plaque in regions such as cortex, hippocampus and forebrain. Proteins such as Tau, α-synuclein, Ataxins, superoxide dismutase (SOD1) and RNA binding proteins TDP43, FUS, TAF15 are found to form lewy bodies, intranuclear inclusion, axonal spheroids and cytoplasmic aggregates (Kumar et al., 2016).
Efforts on analysis of protein aggregation and characterization shown significant improvement in the development of protein aggregation prediction methods. More than 20 different computational algorithms are available now for the prediction of protein aggregation based on amino acid sequences (Santos et al., 2020). Tools such as TANGO, PASTA2.0, AGGRESCAN uses either protein features or experimental data to predict protein aggregation (Conchillo-Solé et al., 2007; de Groot et al., 2005; Walsh et al., 2014). Taking advantage of the available methods of protein aggregation prediction and sequences available on the stroke dataset, here we explored the connection or common theme between the above stroke and amyloid formation.
2 Methods
2.1 Dataset of candidate genes associated with disorders related to stroke
We performed text mining and sequence database search through pubmed to obtain a base dataset of genes associated with Ischemic stroke. As stroke is linked to both monogenic and polygenic disorders, we obtained list of genes reported earlier (Ekkert et al., 2022). Each of these genes in the dataset have certain impact on stroke pathogenesis. Mutiple literature and database search resulted in a dataset of 46 genes linked to risk of stroke.
2.2 Functional annotation of genes linked to stroke disorders
Curated canonical protein sequences are obtained from UniProtKB/Swiss-Prot protein sequence database (The UniProt Consortium, 2021). Only full-length protein sequences are used for each of these genes. Alternative sequences for each ids are avoided to remove redundancy and only unique sequences are further analyzed. For functional annotation of gene list, DAVID knowledgebase which is a webserver for bioinformatics resource providing functional enrichment analysis is utilized (Sherman et al., 2022).
2.3 Prediction of protein aggregation in stroke disorder associated protein sequences
To evaluate the tendency of proteins associated with stroke to form protein aggregate, we performed analysis whether these sequences forms β-sheet enriched secondary structure conformation. Using a pairwise energy potential, intrinsic disorder and secondary structure, protein aggregation calculation for the candidate protein sequences were performed in PASTA2.0 webserver (Walsh et al., 2014).
2.4 Amyloidogenic region in the protein sequences
Smaller fragments of regions in the protein sequences responsible for the amylodogenesis (Ivanova et al., 2004). These regions are composed of aminoacids which are unique and distinct from other non-aggregating regions or peptides. Using expected contact of the residues in the protein sequences, amyloidogenic regions are predicted (Garbuzynskiy et al., 2010).
3 Results
We have shown here that the protein aggregation might occur among the candidate proteins involved in stroke associated disorders. The pathophysiology between the neurodegenerative disorders and protein aggregation are shown to be shared. Our approach has shown that there could be a significant overlap between the pathophysiology of amyloid formation and ischemic stroke.
3.1 Genes involved in monogenic and polygenic disorders associated with stroke
Around 46 genes related to stroke associated disorders are retrieved from different databases. These genes are found to have around 687 splice variants in the UniprotKB. We used full length canonical protein sequences for further analysis. Annotation of genes retrieved through DAVID shows that most genes are related to signaling function including receptors (Table1). Multiple candidate genes functions have stroke phenotypic manifestation.
| SI | From | Species | David Gene Name |
|---|---|---|---|
| 1 | NOTCH3 | Homo sapiens | notch receptor 3(NOTCH3) |
| 2 | FOXC1 | Homo sapiens | forkhead box C1(FOXC1) |
| 3 | CASZ1 | Homo sapiens | castor zinc finger 1(CASZ1) |
| 4 | WNT2B | Homo sapiens | Wnt family member 2B(WNT2B) |
| 5 | LINC01492 | Homo sapiens | long intergenic non-protein coding RNA 1492(LINC01492) |
| 6 | HTRA1 | Homo sapiens | HtrA serine peptidase 1(HTRA1) |
| 7 | ADCY2 | Homo sapiens | adenylate cyclase 2(ADCY2) |
| 8 | PRPF8 | Homo sapiens | pre-mRNA processing factor 8(PRPF8) |
| 9 | HDAC9 | Homo sapiens | histone deacetylase 9(HDAC9) |
| 10 | ABO | Homo sapiens | ABO, alpha 1–3-N-acetylgalactosaminyltransferase and alpha 1–3-galactosyltransferase(ABO) |
| 11 | ZCCHC14 | Homo sapiens | zinc finger CCHC-type containing 14(ZCCHC14) |
| 12 | EDNRA | Homo sapiens | endothelin receptor type A(EDNRA) |
| 13 | SH3PXD2A | Homo sapiens | SH3 and PX domains 2A(SH3PXD2A) |
| 14 | CBS | Homo sapiens | cystathionine beta-synthase(CBS) |
| 15 | PITX2 | Homo sapiens | paired like homeodomain 2(PITX2) |
| 16 | ZNF566 | Homo sapiens | zinc finger protein 566(ZNF566) |
| 17 | NKX2-5 | Homo sapiens | NK2 homeobox 5(NKX2-5) |
| 18 | SH2B3 | Homo sapiens | SH2B adaptor protein 3(SH2B3) |
| 19 | HABP2 | Homo sapiens | hyaluronan binding protein 2(HABP2) |
| 20 | RGS7 | Homo sapiens | regulator of G protein signaling 7(RGS7) |
| 21 | FGA | Homo sapiens | fibrinogen alpha chain(FGA) |
| 22 | ZFHX3 | Homo sapiens | zinc finger homeobox 3(ZFHX3) |
| 23 | FOXF2 | Homo sapiens | forkhead box F2(FOXF2) |
| 24 | TREX1 | Homo sapiens | three prime repair exonuclease 1(TREX1) |
| 25 | ABCC6 | Homo sapiens | ATP binding cassette subfamily C member 6(ABCC6) |
| 26 | ANK2 | Homo sapiens | ankyrin 2(ANK2) |
| 27 | PDZK1IP1 | Homo sapiens | PDZK1 interacting protein 1(PDZK1IP1) |
| 28 | TBX3 | Homo sapiens | T-box transcription factor 3(TBX3) |
| 29 | MMP12 | Homo sapiens | matrix metallopeptidase 12(MMP12) |
| 30 | COL3A1 | Homo sapiens | collagen type III alpha 1 chain(COL3A1) |
| 31 | LRCH1 | Homo sapiens | leucine rich repeats and calponin homology domain containing 1(LRCH1) |
| 32 | CDK6 | Homo sapiens | cyclin dependent kinase 6(CDK6) |
| 33 | GAL | Homo sapiens | galanin and GMAP prepropeptide(GAL) |
| 34 | COL4A2 | Homo sapiens | collagen type IV alpha 2 chain(COL4A2) |
| 35 | COL4A1 | Homo sapiens | collagen type IV alpha 1 chain(COL4A1) |
| 36 | PDE3A | Homo sapiens | phosphodiesterase 3A(PDE3A) |
| 37 | KCNK3 | Homo sapiens | potassium two pore domain channel subfamily K member 3(KCNK3) |
| 38 | LOC100505841 | Homo sapiens | zinc finger protein 474-like(LOC100505841) |
| 39 | FBN1 | Homo sapiens | fibrillin 1(FBN1) |
| 42 | ILF3 | Homo sapiens | interleukin enhancer binding factor 3(ILF3) |
| 43 | CDKN2A | Homo sapiens | cyclin dependent kinase inhibitor 2A(CDKN2A) |
| 44 | ZNF318 | Homo sapiens | zinc finger protein 318(ZNF318) |
| 45 | FURIN | Homo sapiens | furin, paired basic amino acid cleaving enzyme(FURIN) |
| 46 | TM4SF4 | Homo sapiens | transmembrane 4 L six family member 4(TM4SF4) |
| 47 | PMF1 | Homo sapiens | polyamine modulated factor 1(PMF1) |
| 48 | SMARCA4 | Homo sapiens | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4(SMARCA4) |
3.2 Prediction of protein aggregation
Formation of amyloid aggregates is implicated in several neurodegenerative disorders. We use protein disorder as a scale for predicting protein aggregation. Propensity of aggregation remains relatively similar across multiple methods for candidate stroke related genes (Fig. 1). We further used PASTA2.0 to determine the percentage of protein disorder, number of amyloid within the protein sequence, percentage of α-helix, percentage of β-strand etc. Percentage disorder of proteins vary ranging from 1 to upto 80 for stroke associated genes. For a random dataset of non-stroke related genes this range from 1.5 to 63. Median value differs significantly between these two groups of proteins (Fig. 2). Statistical test (t-test) using R between the above two groups of proteins was performed. This test reveals a significantly lower p-value (p-value = 0.007744). This is highly significant and percentage disorder is higher for stroke associated genes.

- Consensus methods predicting same amyloid regions.
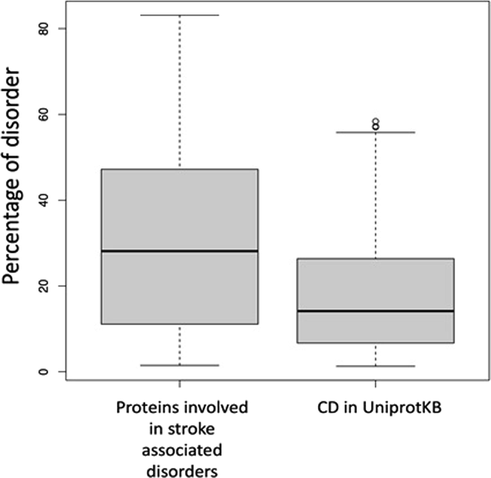
- Boxplot showing the differences in the disorder among two groups of proteins.
3.3 Residue based prediction of aggregation specific protein region
Each protein sequences are predicted to have atleast 20 poteintial amyloid forming short amino acid sequence pattern (Table 2). Consensus pattern derived from multiple amyloid predicting tools such as TANGO, AGGRESCAN, WALTZ through FoldAmyloid shows that these patterns are detected in all methods (Fig. 1). For example, in protein WNTB-2B, these aggregation forming residues are distributed in 14 different sites (Fig. 3). Most of these patterns are 4–14 aminoacid length (Fig. 4).
| Protein name (from fasta header) | Length | # Amyloids | Best energy | % disorder | % α-helix | % β-strand | % coil |
|---|---|---|---|---|---|---|---|
| spQ08462 | 1091 | 20 | −39.221871 | 4.216 | 64.25 | 8.07 | 27.68 |
| spO95255 | 1503 | 20 | −19.941325 | 8.715 | 59.41 | 6.25 | 34.33 |
| spQ9Y2L9 | 728 | 20 | −19.362124 | 27.33 | 42.58 | 10.03 | 47.39 |
| spP09958 | 794 | 20 | −18.84564 | 18.26 | 15.49 | 25.06 | 59.45 |
| spQ13113 | 114 | 20 | −17.268009 | 16.66 | 48.25 | 8.77 | 42.98 |
| spO14649 | 394 | 20 | −17.165792 | 28.17 | 65.48 | 2.79 | 31.73 |
| spP25101 | 427 | 20 | −16.917126 | 7.259 | 64.64 | 6.32 | 29.04 |
| spQ9UM47 | 2321 | 20 | −15.802264 | 13.09 | 17.36 | 16.59 | 66.05 |
| spP48230 | 202 | 20 | −15.752635 | 7.92 | 54.46 | 2.97 | 42.57 |
| spQ6P2Q9 | 2335 | 20 | −14.995744 | 1.498 | 46.55 | 13.28 | 40.17 |
| spQ14432 | 1141 | 20 | −14.680867 | 33.74 | 41.89 | 7.01 | 51.1 |
| spP16442 | 354 | 20 | −14.397722 | 3.672 | 33.9 | 16.95 | 49.15 |
| spP02671 | 866 | 20 | −10.907301 | 35.33 | 16.17 | 24.02 | 59.82 |
| spQ01484 | 3957 | 20 | −10.281311 | 42.91 | 26.38 | 14.43 | 59.19 |
| spP35520 | 551 | 20 | −10.195829 | 15.06 | 37.75 | 14.7 | 47.55 |
| spQ5TCZ1 | 1133 | 20 | −9.781373 | 45.18 | 11.92 | 21.27 | 66.81 |
| spQ15911 | 3703 | 20 | −9.775551 | 52.17 | 32.19 | 8.37 | 59.44 |
| spQ93097 | 391 | 20 | −9.619822 | 10.48 | 45.78 | 14.07 | 40.15 |
| spQ8N726 | 132 | 20 | −9.546539 | 66.66 | 11.36 | 14.39 | 74.24 |
| spQ12906 | 894 | 20 | −9.535643 | 47.2 | 28.75 | 10.18 | 61.07 |
| spQ00534 | 326 | 20 | −9.230796 | 13.49 | 43.86 | 16.87 | 39.26 |
| spQ92743 | 480 | 20 | −8.959704 | 13.33 | 18.96 | 30.83 | 50.21 |
| spQ9UQQ2 | 575 | 20 | −8.630576 | 45.04 | 24.35 | 14.61 | 61.04 |
| spQ12947 | 444 | 20 | −8.290021 | 52.02 | 22.52 | 3.83 | 73.65 |
| spP51532 | 1647 | 20 | −8.254159 | 50.15 | 46.63 | 5.22 | 48.15 |
| spQ9UKV0 | 1011 | 20 | −8.19617 | 29.57 | 40.26 | 8.51 | 51.24 |
| spP35555 | 2871 | 20 | −7.913752 | 3.065 | 4.25 | 31.8 | 63.95 |
| spP39900 | 470 | 20 | −7.626874 | 4.042 | 23.83 | 23.83 | 52.34 |
| spP49802 | 495 | 11 | −7.622278 | 11.11 | 52.93 | 2.42 | 44.65 |
| spP02462 | 1669 | 20 | −7.454205 | 83.1 | 2.64 | 8.63 | 88.74 |
| spQ14520 | 560 | 20 | −7.387258 | 6.607 | 14.46 | 26.43 | 59.11 |
| spQ9NSU2 | 314 | 20 | −6.854083 | 27.7 | 40.45 | 6.69 | 52.87 |
| spP08572 | 1712 | 20 | −6.646477 | 62.44 | 2.69 | 9.35 | 87.97 |
| spO15119 | 743 | 9 | −6.556453 | 32.03 | 28.4 | 11.84 | 59.76 |
| spQ12948 | 553 | 6 | −6.366574 | 65.82 | 22.78 | 6.69 | 70.52 |
| spQ5VUA4 | 2279 | 11 | −6.276438 | 50.89 | 27.29 | 10.79 | 61.91 |
| spQ969W8 | 418 | 1 | −5.915617 | 3.588 | 22.73 | 18.66 | 58.61 |
| spP42771 | 156 | 7 | −5.761273 | 28.2 | 50 | 0 | 50 |
| spQ86V15 | 1759 | 20 | −5.735543 | 44.11 | 19.56 | 17.45 | 62.99 |
| spQ8N6F7 | 178 | 4 | −5.629332 | 28.08 | 21.91 | 13.48 | 64.61 |
| spP22466 | 123 | 3 | −5.60644 | 80.48 | 52.03 | 0 | 47.97 |
| spQ99697 | 317 | 3 | −5.452123 | 34.7 | 37.54 | 3.79 | 58.68 |
| spQ8WYQ9 | 949 | 1 | −5.372969 | 50.36 | 18.02 | 16.97 | 65.02 |
| spP02461 | 1466 | 2 | −5.361421 | 77.96 | 3.96 | 6.48 | 89.56 |
| spQ6P1K2 | 205 | 1 | −5.021246 | 22.43 | 76.59 | 0 | 23.41 |
| spP52952 | 324 | 0 | −4.668274 | 15.12 | 30.56 | 11.11 | 58.33 |
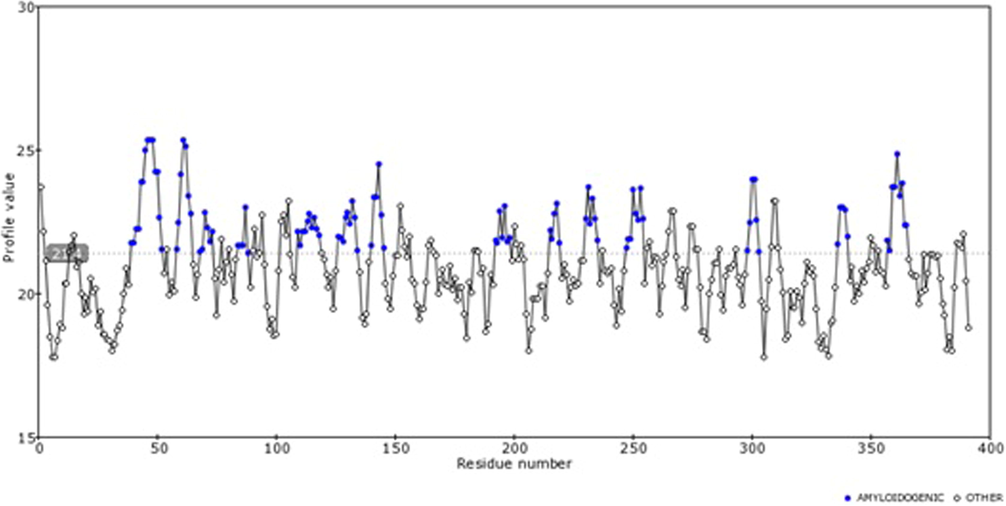
- Contact frequency predicted for different residues across the protein sequence is shown here.

- Amyloidogenic residues predicted for human Wnt-2b protein.
4 Discussion
Analysis of protein aggregation from 46 protein sequences implicated in stroke manifestation shows that large number of proteins form amyloids with varying degree of protein disorder. The role of protein aggregation and amyloid formation in the neurodegenerative diseases are well established (Pedersen & Heegaard, 2013; Tutar et al., 2013). Also, earlier studies have shown that there is higher levels or induction of protein aggregation after cerebral Ischemia (Wu & Du, 2021). Compared to the random datasets of proteins from UniprotKB, these proteins are found to have higher percentage of protein disorder. This could arise from the β-strand composition of these sequences. Furthermore, structural variation including SNP and small indels in these genes could possibly contribute to the amyloid formation. Our work contributes to the evidence that protein aggregation could be implicated in stroke disorder. Further research following our findings would improve our knowledge on the molecular level overlap between these two related process and disorder. More experimental evidences are needed for implicating the above listed genes (their products) in the protein aggregation. Establishing mouse models for stroke and investigating the aggregation through electron microscopy, laser-scanning confocal microscopy, and Western blotting could help further.
5 Conclusion
In the present study, we analyzed the aggregation properties of stroke related proteins. We selected a set of stroke-associated candidate proteins and a set of random control dataset. Overall, we observed that most proteins associated with stroke have higher protein disorder compared to a random dataset of protein sequences. These amyloids forming aggregating residues are distributed anywhere between the N-terminal and C-terminal part of the sequence of these candidate proteins. We found the contact frequency profile value of multiple residues are higher than average expected value and is part of disordered region related to protein conformation. Our study suggests that there is an overlap in pathophysiology of protein aggregation, neurological disorders and stroke related disorders.
Acknowledgment
The authors extend their appreciations to the deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number (lFP-2020-38).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Classification of stroke subtypes. Cerebrovasc. Dis. (Basel, Switzerland). 2009;27(5):493-501.
- [CrossRef] [Google Scholar]
- Causal relationship of susceptibility genes to ischemic stroke: comparison to ischemic heart disease and biochemical determinants. PLOS ONE. 2010;5(2)
- [CrossRef] [Google Scholar]
- AGGRESCAN: A server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinf.. 2007;8(1):65.
- [CrossRef] [Google Scholar]
- Prediction of “hot spots” of aggregation in disease-linked polypeptides. BMC Struct. Biol.. 2005;5(1):18.
- [CrossRef] [Google Scholar]
- A genome-wide association study identifies new loci for factor VII and implicates factor VII in ischemic stroke etiology. Blood. 2019;133(9):967-977.
- [CrossRef] [Google Scholar]
- Ischemic stroke genetics: what is new and how to apply it in clinical practice? Genes. 2022;13(1):48.
- [CrossRef] [Google Scholar]
- FoldAmyloid: a method of prediction of amyloidogenic regions from protein sequence. Bioinformatics. 2010;26(3):326-332.
- [CrossRef] [Google Scholar]
- Whole-Genome sequencing association analyses of stroke and its subtypes in ancestrally diverse populations from trans-omics for precision medicine project. Stroke. 2022;53(3):875-885.
- [Google Scholar]
- Protein aggregation after focal brain ischemia and reperfusion. J. Cereb. Blood Flow Metab.. 2001;21(7):865-875.
- [CrossRef] [Google Scholar]
- An amyloid-forming segment of β2-microglobulin suggests a molecular model for the fibril. Proc. Natl. Acad. Sci.. 2004;101(29):10584-10589.
- [CrossRef] [Google Scholar]
- Protein aggregation and neurodegenerative diseases: from theory to therapy. Eur. J. Med. Chem.. 2016;124:1105-1120.
- [CrossRef] [Google Scholar]
- Protein misfolding, aggregation, and autophagy after brain ischemia. Transl. Stroke Res.. 2013;4(6):581-588.
- [CrossRef] [Google Scholar]
- Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet.. 2018;50(4):524-537.
- [CrossRef] [Google Scholar]
- Analysis of protein aggregation in neurodegenerative disease. Anal. Chem.. 2013;85(9):4215-4227.
- [CrossRef] [Google Scholar]
- Computational prediction of protein aggregation: Advances in proteomics, conformation-specific algorithms and biotechnological applications. Comput. Struct. Biotechnol. J.. 2020;18:1403-1413.
- [CrossRef] [Google Scholar]
- DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucl. Acids Res.. 2022;gkac194
- [CrossRef] [Google Scholar]
- UniProt: The universal protein knowledgebase in 2021. Nucl. Acids Res.. 2021;49(D1):D480-D489.
- [CrossRef] [Google Scholar]
- Tutar, Y., Özgür, A., Tutar, L., 2013. Role of Protein Aggregation in Neurodegenerative Diseases. In: Neurodegenerative Diseases. IntechOpen. https://doi.org/10.5772/54487
- PASTA 2.0: An improved server for protein aggregation prediction. Nucl. Acids Res.. 2014;42(Web Server issue):W301-W307.
- [CrossRef] [Google Scholar]
- Protein aggregation in the pathogenesis of ischemic stroke. Cell. Mol. Neurobiol.. 2021;41(6):1183-1194.
- [CrossRef] [Google Scholar]
- Correlation between cellular uptake and cytotoxicity of fragmented α-synuclein amyloid fibrils suggests intracellular basis for toxicity. ACS Chem. Nerosci.. 2020;11(3):233-241.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2022.102474.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







