Translate this page into:
Polymorphisms of TP53 gene and its association with colorectal cancer: A case-control investigation
⁎Corresponding author. 437106985@ksu.edu.sa (Abdullah M Alhadheq)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The tumor suppressor gene (TP53) is crucial for DNA repair mechanism, apoptosis, and cell cycle regulation and progression. In human cancer, TP53 is mutated and highly polymorphic. In the current case-control research investigation, we investigated TP53 gene SNPs, in exonic and intronic regions, as potential risk factors for colorectal cancer (CRC). This study comprised of 192 patients and 192 control. Obtained data illustrated that only the G allele; rs1042522 (Pro72Arg (C > G), demonstrated a statistically significant association, almost 1.5-fold induction promotes the risk of CRC development in contrast to individuals with the C allele (OR = 1.5, χ2 = 7.28, p = 0.00696). The homozygous variant GG genotype of rs1042522 was also a significant risk factor to CRC development (OR = 2.1, χ2 = 6.41, p = 0.01136). SNP rs1042522 polymorphism established a considerably elevated odds of CRC among male patients aged < 57 years and in patients’ with tumors situated in colon region. In silico analysis exhibited that proline to arginine amino acid substitution affects the protein structure. Both rs1642785 and rs9894946 SNPs did not demonstrate any significant statistical association with CRC. In conclusion, this study confirmed that rs1042522 SNP within TP53 gene is correlated with possibility of developing CRC in the Saudi population. This finding highlights those polymorphisms within TP53 gene could act as a diagnostic indicator for CRC.
Keywords
TP53
Genotyping
Cancer
Colorectal
SNP
Polymorphism
1 Introduction
Colorectal cancer (CRC) is deemed among major malignancies worldwide. It occurs as a result of accumulation of set of genetic and epigenetic modifications over time in different pathways that are proven to drive and transform the colonic epithelial cells into tumors (Houlston and Tomlinson 2001; Sung et al., 2021). It may take 10 to 15 years for the development and progression of carcinogenesis, which involves concurrent histological and nuclear changes (Fearon and Vogelstein 1990; Katerji and Duerksen-Hughes, 2021). Generally, variations in several biochemical pathways play vital roles in the development and transition from adenoma to carcinoma (Jesionek-Kupnicka et al., 2017). In colon cancer, approximately 90 % of the cases are reported to have mutations in APC and in TP53 pathways (Michor et al., 2005; Kanth and Inadomi, 2021; Vuik et al., 2019; Chittleborough et al., 2020). In this regard, the role of TP53 gene in suppression of tumor cannot be undermined. The human TP53 gene is positioned on chromosome 17 which has 11 exons and 10 introns (Lamb and Crawford 1986), and encodes TP53 protein. In over 50 % of human cancers, TP53 undergoes mutation, while the remaining cases exhibit changes in its regulators or targets (Hu et al., 2021). In 1979, the TP53 protein was initially identified as an oncogene by different groups (Hernandez Borrero and El-Deiry 2021). Simultaneously, it was found to complex with the SV40 virus T antigen in cells undergoing 8–11 tumor transformation. Subsequently, other studies have identified interactions between this 53 kDa protein and adenovirus and human papillomavirus proteins (Werness et al., 1990). The “tumor antigen” is upregulated in tumor cells and seems to collaborate with other oncogenes like HRAS in converting primary cells into cultured cells (Miret et al., 2003). TP53 plays a crucial part in various biological processes, including DNA repair pathways, cell cycle regulation, and apoptosis (Levine and Oren 2009). TP53 is often altered in various cancers and it is a polymorphic gene. Many studies showed an enormous number of single nucleotide polymorphisms (SNPs) in the exon, promoter, and intron regions of the TP53 gene. Some of these studies showed association between SNPs in TP53 and cancer (Whibley et al., 2009, Jesionek-Kupnicka et al., 2017, Sobieszkoda et al., 2017). This gene is critical to tumor progression and therapeutic response. Therefore, the current study was intended to study the potential association of SNPs in TP53 gene with CRC in the Saudi population. We analyzed the association of TP53 (rs1042522, rs1642785, and rs9894946) polymorphism with CRC development among Saudi population.
2 Material and methods
2.1 Patient samples
Samples from Saudi Arabia of CRC patients (n = 192) and matched controls (n = 192) were obtained from collaborators and clinicians according to the guidelines of 12/3352/IRB. Patients visiting the Endoscopy Department at King Khalid University Hospital (KKUH) were examined by the oncologist alongside a routine examination. There were no restrictions in patient group in terms of age and CRC stages. For genotyping studies, each patient donated blood volume of 5 ml. Clinical and demographic traits such as tumor location, sex, age, and ethnicity, family history of cancer, lymph node status, and smoking habit were recorded for all the study participants (both cases and controls). Informed consent statements were collected from all the study participants in accordance with the rules of the ethical review committee at King Saud Medical City, King Saud University.
2.2 Nucleic acid Isolation
Blood samples were utilized in the extraction process for Genomic DNA, by QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA) as per manufacturer’s guide. Briefly, 20 µl protease was inserted in 200 µl of blood samples in 1.5 ml tubes and mixed. Next, AL buffer was incubated for 10 min at temperature of 56 °C. Mixed was then centrifugated and then transferred to spin columns. Wash buffers 1 & 2 were inserted to the column, and centrifugated after each wash. The elution was performed in 50 µl AE buffer. The yielded DNA was measured for volume and purity using the NanoDrop8000 spectrophotometer (Thermo Scientific).
2.3 Genotyping
Both CRC and normal DNA samples were genotyped and amplified for TP53 SNPs by real-time polymerase chain reaction (PCR) using a TaqMan SNP genotyping assays as described (Alanazi et al., 2013, Angelopoulou et al., 2017, Ozdemirkiran et al., 2017). Each well containing 20 ng of genomic DNA, 5.6 μl of TaqMan® Genotyping Master Mix (Applied Biosystems, Foster City, CA, USA), 0.2 μl of 40 × TaqMan® Genotyping SNP Assay (Applied Biosystems). QuantStudio™ Real-Time PCR (Applied Biosystems) was used for each genotyping run with and an endpoint reaction reading. The setting of the PCR run are as follows; 1) pre-read stage for 30 s at 60 °C temperature, 2) hold stage 10 min at 95 °C, 3) 40 cycles at PCR stage 15 s for denaturation at 95 °C and annealing for 1 min at 60 °C, and 4) post-read stage 30 s at 60 °C.
2.4 Statistical analysis
The data analysis was conducted by calculating the allele frequencies. Genotype evaluated the differences between the samples, and the calculation was executed as per Pearson’s goodness-of-fit chi-square. The allelic variations were calculated on the basis of wild type which was treated as a reference for the present investigation. Chi-square, odds ratios, p-values, and confidence intervals were computed by using IBM SPSS version 23. Haploview software was used to plot the Linkage disequilibrium (Barrett et al., 2004). In silico study done by using the online tool (https://www3.cmbi.umcn.nl/hope/input).
3 Results
The present study included 192 patients’ samples diagnosed with CRC and 192 CRC patients, with earlier assent from every person. Clinical and demographic details are presented in (Table 1). Genetic polymorphisms that were identified, i.e., rs1042522 (Pro72Arg (C > G)) from the exonic region, rs1642785 (C > G) in the intronic region, and rs9894946 (A > G) in the intronic region, in TP53 gene variants, were examined in Saudi cohort diagnosed with CRC, to assess the risk of susceptibility to develop CRC. The genotype distributions are demonstrated in Table 2. The genotype distributions for all the SNPs were in agreement with Hardy-Weinberg Equilibrium (HWE). In the overall analysis, a significant link was observed only with the G allele of rs1042522 (Pro72Arg (C > G)), which showed a nearly 1.5-fold increase in odds of CRC development in comparison to the individuals with the C allele (OR = 1.5, χ2 = 7.28) (Table 1). The p-value for this association was 0.00696, which is statistically significant at the 0.05 level. Moreover, the homozygous variant GG genotype of rs1042522 also had a statistically significant association with risk of CRC (OR = 2.1, χ2 = 6.41, p = 0.01136). This suggest that individuals with the GG genotype were 2.1 times more likely to develop CRC than individuals with the CC genotype.
Clinical characteristics
Cases
Controls
Age
Median age, years
57
58
Range, years
23–79
21–76
≤57 years
91
114
>57 years
101
78
Gender
Male
116
108
Female
76
80
Tumor location
Colon
109
–
Rectum
70
–
Family history of cancer
32
–
SNP
Variant
Cases (Freq)
Controls
OR
P- Value
rs1042522
CC
0.40
0.49
Ref
CG
0.39
0.37
1.28
0.27
GG
0.22
0.13
2.10
0.011
C
226 (0.59)
0.68
Ref
G
158 (0.41)
0.31
1.50
0.007
rs1642785
CC
0.15
0.09
Ref
CG
0.43
0.49
0.52
0.057
GG
0.42
0.42
0.61
0.16
C
0.36
0.34)
Ref
G
0.64
0.66
0.89
0.46
rs9894946
AA
0.44
0.52
Ref
AG
0.40
0.37
1.28
0.27
GG
0.16
0.11
1.66
0.11
A
0.64
0.70
Ref
G
0.36
0.30
1.33
0.065
The genotype frequencies of rs1642785 and rs9894946 did not show statistically significant associations in an overall comparison between CRC cases and controls (Table 2). Furthermore, we divided the samples into two subgroups stratified based on the median age of patients, i.e., below or above 57 years to study the influence of TP53 SNPs rs1042522, rs1642785 and rs9894946 on risk of CRC. As detailed in Table 3, a notable correlation was observed in the case of the rs1042522 allele among individuals with CRC who were younger than 57 years old (Specifically, the CG genotype at p = 0.008, the GG genotype at p = 0.014, and the presence of the G allele at p = 0.001). However, this significant association did not evident in the older patient group. Both SNPs rs1642785 and rs9894946 did not display any association on age stratification among patients. We also studied the effect of TP53 SNPs rs1042522, rs1642785, and rs9894946 on risk of CRC development in subgroups stratified by sex (male and female). As shown in Table 4, only rs1042522 polymorphism showed significant risk for developing CRC among male patients (Specifically, the GG genotype at p = 0.041, and the presence of the G allele at p = 0.017); SNPs rs1642785 and rs9894946 not showing any association with CRC among patients stratified by the male gender. Among female, none of these SNPs confirmed statistical significance (Table 4).
SNP
Variant
Cases (Freq)
Controls
(Freq)OR
P- Value
rs1042522
(age: below 57)
CC
0.36
0.57
Ref
CG
0.46
0.32
2.27
0.008
GG
0.18
0.09
2.90
0.014
C
0.59
0.74
Ref
G
0.41
0.25
1.96
0.001
(age: above 57)
CC
0.43
0.37
Ref
CG
0.32
0.44
0.62
0.157
GG
0.26
0.17
1.25
0.582
C
0.58
0.59
Ref
G
0.42
0.40
1.05
0.819
rs1642785
(age: below 57)
CC
0.16
0.07
Ref
CG
0.37
0.46
0.36
0.036
GG
0.48
0.47
0.45
0.101
C
0.34
0.30
Ref
G
0.66
0.70
0.83
0.380
(age: above 57)
CC
10.14
0.12
Ref
CG
0.49
0.55
0.73
0.512
GG
0.38
0.33
0.94
0.900
C
0.38
0.39
Ref
G
0.62
0.61
1.04
0.849
rs9894946
(age: below 57)
AA
0.46
0.49
Ref
AG
0.40
0.43
0.98
0.945
GG
0.14
0.08
1.92
0.167
A
0.66
0.71
Ref
G
0.34
0.29
1.24
0.310
(age: above 57)
AA
0.42
0.55
Ref
AG
0.41
0.28
1.91
0.057
GG
0.18
0.17
1.418
0.409
A
0.62
0.69
Ref
G
0.38
0.31
1.38
0.148
SNP
Variant
Cases
(Freq)Controls
(Freq)OR
P- Value
rs1042522
Male
CC
0.44
0.56
Ref
CG
0.35
0.31
1.44
0.220
GG
0.21
0.12
2.21
0.041
C
0.62
156 (0.72)
Ref
G
0.38
0.27
1.62
0.017
Female
CC
0.33
0.37
Ref
CG
0.43
0.47
1.04
0.909
GG
0.24
0.15
1.80
0.199
C
0.55
0.61
Ref
G
0.45
0.38
1.31
0.234
rs1642785
Male
CC
0.14
0.10
Ref
CG
0.43
0.48
0.65
0.321
GG
0.43
0.42
0.76
0.542
C
0.35
0.34
Ref
G
0.65
0.66
0.96
0.831
Female
CC
0.16
0.08
Ref
CG
0.43
0.51
0.40
0.093
GG
0.41
0.41
0.47
0.171
C
0.38
0.33
Ref
G
0.63
0.67
0.82
0.418
rs9894946
Male
AA
0.45
0.54
Ref
AG
0.37
0.34
1.30
0.378
GG
0.18
0.12
1.80
0.139
A
0.63
153 (0.71)
Ref
G
0.37
0.29
1.40
0.093
Female
AA
0.42
0.48
Ref
AG
0.45
0.41
1.22
0.555
GG
0.13
0.11
1.32
0.592
A
0.64
0.68
Ref
G
0.36
0.32
1.18
0.495
The analysis extended to consider the location of tumors within the colorectal region. Among the investigated variants, rs1042522 exhibited a significantly higher risk of developing CRC in the colonic region for individuals with the homozygous GG allele (χ2: 7.00; CI: 1.24–4.66; p = 0.008), showing a 2.41-fold increase in risk. There was also an elevated risk association for the G allele (χ2: 7.78; CI: 1.15–2.29; p = 0.005) with a 1.63-fold increase in risk, but this association was not observed in the rectal region. Notably, there was no discernible evidence indicating any association between the SNPs rs1642785 and rs9894946 and tumors in either the colon or rectal regions.
The pairwise linkage disequilibrium (LD) values (D’ and r2) are listed in Fig. 1. For the SNPs, LD analysis revealed weak LD, forming one haplotype block, suggesting that haplotype evaluation may be beneficial. SNP rs9894946 showed disequilibrium (D’= 0.428, r2 = 0.168) in cases. The remaining two SNPs showed > 0.1 D’ and r2 values in cases and controls. Fig. 2 and Fig. 3 illustrate the regional LD plot for the TP53 SNPs and the effect of amino acid replacement in SNP rs1042522 (Pro72Arg).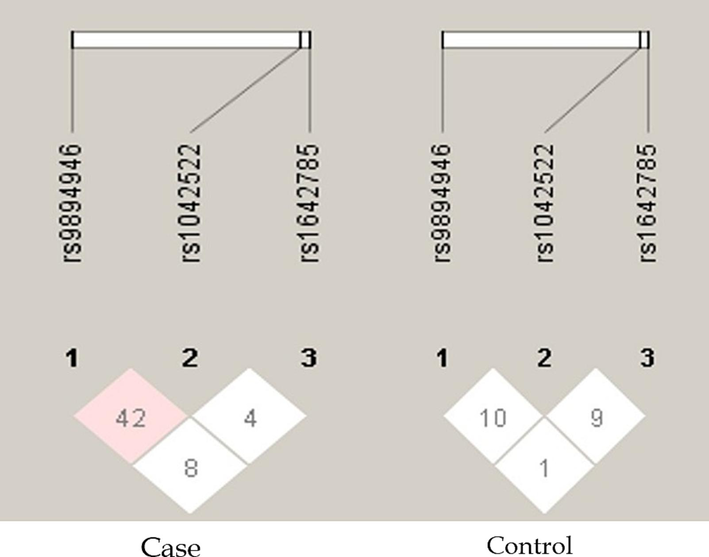
Linkage Disequilibrium (LD) of studied TP53 loci in colorectal cancer cases and controls.
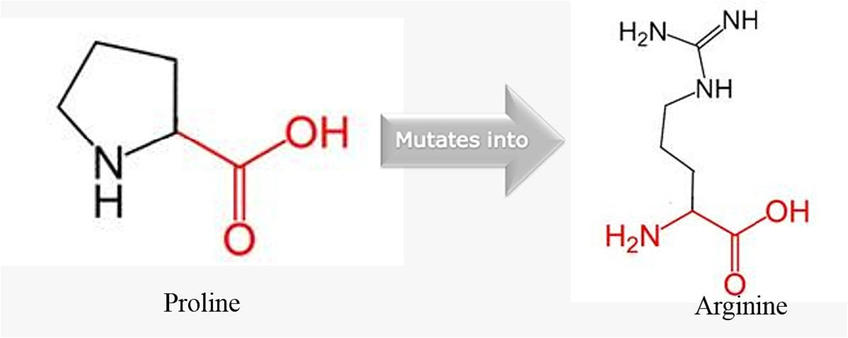
Effect of amino acid replacement in SNP rs1042522 (Pro72Arg).
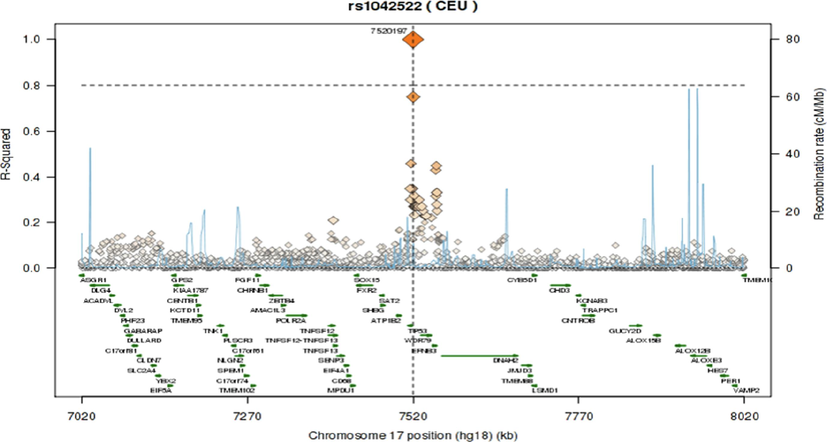
Regional LD plot for the TP53 SNPs (A) rs1042522; (B) rs1642785; (C) rs9894946.
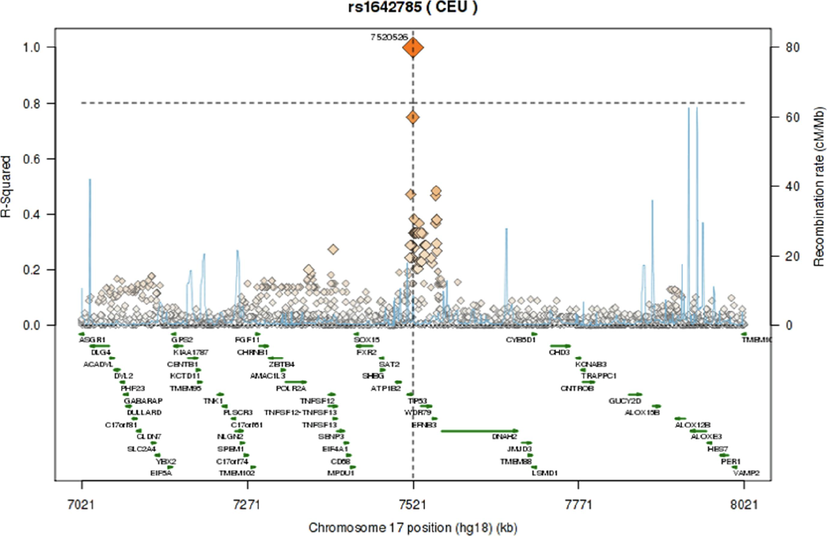
Regional LD plot for the TP53 SNPs (A) rs1042522; (B) rs1642785; (C) rs9894946.
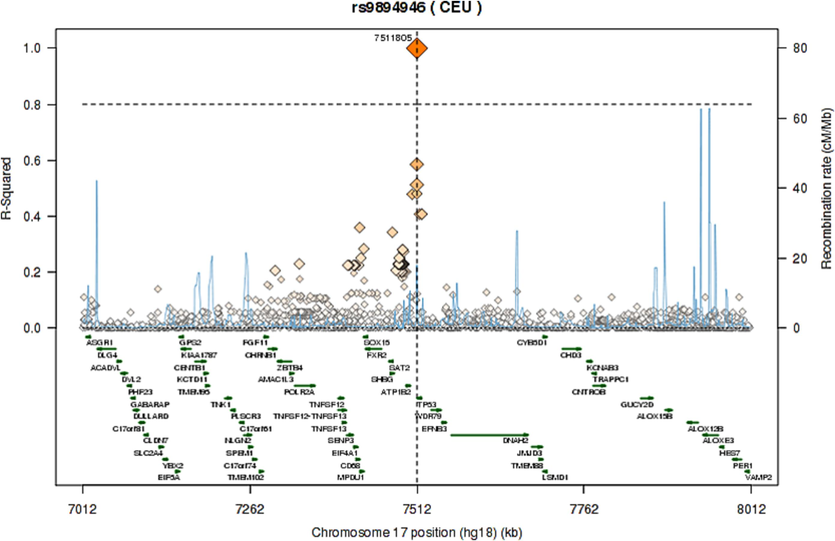
Regional LD plot for the TP53 SNPs (A) rs1042522; (B) rs1642785; (C) rs9894946.
4 Discussion
TP53 protein is a crucial component in maintaining genomic integrity which prevents the cells from oncogenic transformation. Inactivation of TP53 is common occurrence in majority of the cancers. TP53 serves to regulate DNA repair, cell cycle, and cell development. Any changes in TP53 function will abrupt these functions and culminate in loss of genome integrity. Polymorphisms of the TP53 gene are widely established to take part in progression of CRC. If the TP53 functioning is normal, it is a fundamental obstacle for carcinogenesis. Few SNPs of TP53 coding region are strongly linked with the process of carcinogenesis. They are commonplace in a large number of cancers, contributing to severe aberration in TP53 function. Till now, approximately > 200 SNPs are documented to be found commonly in TP53 which are expected to cause aberrations in TP53 functioning. The purpose of current study was to perceive any associations of TP53 polymorphisms with development of CRC in Saudi Arabian population. This study investigated the role of exonic and intronic SNPs (exonic rs1042522, Pro 72 Arg (C > G), intronic rs1642785 (C > G), and intronic rs9894946 (A > G) of the TP53 gene as potential CRC risk factors in a case-control study involving 192 CRC cases and 192 matched normal control samples.
Our study found a strong association between the GG genotype of SNP rs1042522 and an increased risk of CRC in the Saudi population. Additionally, we observed that the minor allele (G) of the same SNP also independently contributes to a higher susceptibility to CRC (Table 2). Our findings, consistent with previous research, demonstrate that individuals carrying the GG genotype of rs1042522 are more susceptible to CRC (Ashton et al., 2009, Ricks-Santi et al., 2010, Tian et al., 2016). This genotype is associated with a decreased ability of the TP53 protein to induce cell cycle arrest and apoptosis, which are vital mechanisms for controlling and eliminating potentially cancerous cells. Furthermore, our study has also unveiled an independent contribution of the minor allele (G) of SNP rs1042522 to an increased risk of CRC. This observation suggests that even individuals who possess a single copy of the G allele may be more predisposed to CRC than those with a different genetic makeup at this locus. Contrastingly, several other investigations have reported results inconsistent with our own findings. Notably, there is no discernible association between the rs1042522 SNP and cancer risk in their respective studies (Dahabreh et al., 2010; Kodal et al., 2017). In the current study, we sub-analyzed the possible association between three TP53 SNPs and CRC risk by age, sex, and tumor location. In a prior publication of ours (Alhadheq et al., 2016, Semlali et al., 2017), we made a noteworthy observation concerning the SNP rs8679. Specifically, we identified a statistically significant protective association between this SNP and the risk of colorectal cancer, particularly in females. The genotype distributions for TT, TC, and CC were as follows: in controls, 0.42, 0.49, and 0.09, respectively, whereas in cases, they were 0.67, 0.30, and 0.03, respectively. These findings indicated that the presence of certain alleles of rs8679 was associated with a lower susceptibility to colorectal cancer in females. It's worth noting that recent research conducted by Nazarian and Kulminski has shed new light on the potential genetic heterogeneity specific to sex in the context of colorectal cancer and lung cancer (Nazarian and Kulminski, 2021). Their work has explored the influence of sex at both the SNP (single nucleotide polymorphism) and pathway levels, revealing intriguing insights into how genetic factors may interact differently in males and females in relation to these cancers. These novel findings underscore the importance of considering sex-specific genetic factors when studying cancer susceptibility and may have significant implications for future research in this field. We noticed a noteworthy association between the rs1042522 polymorphism (CG, GG genotypes, and minor allele G) and an increased risk of CRC in individuals aged <57 years. Surprisingly, SNP rs1642785 demonstrated a significant protective effect against CRC in patients <57 years. Altogether, this finding offers valuable insights into the intricate interplay of genetic factors in colorectal cancer susceptibility, especially concerning age-related considerations. Further hold potential implications for the field of personalized medicine and refined risk assessment. The identification of individuals with distinct genetic profiles has the potential to facilitate early detection, precision screening, and proactive preventive strategies. When comparing sexes, we observed that the rs1042522 SNP was linked to a higher risk of CRC in males in contrast to females. This finding aligns with a prior study that highlighted the influence of sex in susceptibility to CRC (Micheli et al., 2009). Purim et al. reported that females are more protected against CRC development, with a lower related mortality rate when compared to males (Purim et al., 2013). The observed phenomenon may be attributed to variances in physiology, diet, and hormones between males and females (Jacobs et al., 2007). Furthermore, SNP rs1042522 exhibited a noteworthy correlation with an elevated colorectal cancer (CRC) risk, particularly among patients with tumors located in the colon. It is conceivable that this SNP influences CRC risk by heightening the susceptibility of colon cells to DNA damage. Such DNA damage events can instigate mutations that foster the growth of cancer cells. Consequently, our findings imply that the rs1042522 SNP could serve as a valuable marker for the identification of individuals at an escalated risk of CRC. This insight holds the potential to guide the development of focused strategies for prevention and treatment tailored to this specific group of patients.
Various functional studies using genetically modified mice have consistently reported a notable link between TP53 gene polymorphisms and the vulnerability of patients to tumor progression. These studies have consistently shown that mice with a silenced mutation in one TP53 allele exhibit a lower incidence of tumors compared to mice with the mutation (Donehower et al., 1992). Moreover, in a study conducted by Tina et al. (Tian et al., 2016) they emphasized that TP53 Pro72Arg polymorphism is extensively examined and significantly linked to cancer risk (Tian et al., 2016). In current research, we observed a significant correlation between SNP rs1042522 (Pro72Arg) and CRC in our Saudi cohort. These findings are consistent with numerous previous studies investigating CRC and other cancer types (Dahabreh et al., 2013). Consequently, it is plausible that this SNP may also be associated with the progression of CRC. The mutated amino acid is bigger in size than wild type. The mutation introduces a charge at this position; this can cause repulsion between the mutant residue and neighboring residues. The mutation might harm hydrophobic interactions with other molecules on protein surface (Fig. 2). The wild-type residue is proline. Prolines are widely known as rigid and therefore induce a special backbone conformation which may be required at this position. The mutation can disturb this special conformation (Thomas et al., 1999). The mutation might cause a loss of hydrophobic interactions with other molecules on the surface of the protein. Further, we have plotted LD plots for each individual SNP to sought their association with nearby SNPs. The LD plot demonstrates that, out of the three evaluated polymorphisms, none of the SNPs indicated r2 > 0.80. As presented in Fig. 3, the local LD plot was plotted by using the online SNP Annotation and Proxy Search tool (https://www.broadinstitute.org/mpg/snap/ldplot.php). The highest r2 values for the SNPs studied were 0.749 for rs1042522, 0.749 for rs1642785, and 0.585 for rs9894946 (Fig. 3). The regional association LD plot showed several positions near the rs1042522 SNP with high LD (r2 = 0.749). SNP rs1042522 showed a close association with rs1642785 (r2 = 0.749), highlights their close association within the genetic landscape. This suggests that variations in rs1042522 are highly likely to co-occur with variations in rs1642785. Such insights into LD patterns are invaluable, as they provide a deeper understanding of the genetic architecture and co-inheritance patterns among these SNPs.
5 Conclusion
Current findings illustrated that the GG genotype of SNP rs1042522 was significantly associated with an increased risk of CRC, especially in individuals aged <57 years and males. This SNP may influence CRC risk by increasing the susceptibility of colon cells to DNA damage. We also observed that SNP rs1642785 showed a significant protective effect against CRC in patients <57 years. Our findings suggest that the SNPs rs1042522 and rs1642785 may be useful markers for identifying individuals at an increased risk of CRC. This information could be used to develop targeted strategies for prevention and treatment tailored to this specific group of patients. Further research, including functional investigations and larger cohort studies, is imperative to unravel the intricate molecular mechanisms underlying CRC pathogenesis and to pave the way for more effective interventions.
Authors' contributions
Abdullah M Alhadheq contributed sampling, software analysis, and experiments validation, and data curation. Narasimha Reddy Parine performed the formal analysis. Abdullah M Alhadheq and Saad Alkahtani contributed in study conceptualization. Jilani Purusottapatnam Shaik contributed in methodology. Saad Alkahtani, coordinated the project administration, and evaluated TP53 genotyping. Mohammad Alanazi, writing the original draft and contributed in SNP analysis of genotyping. Nada H. Aljarba, also, contributed in SNP evaluation. Rana Alhadheq performed the statistical analysis. Abdullah M Alhadheq and Saad Alkahtani edited the final version. All authors have approved the final article.
Availability of data and materials
The data generated or analyzed in this article are online publicly available without request.
Acknowledgements
This work was funded by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2023R62), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. Researchers Supporting Project number (RSP2023R26), King Saud University, Riyadh, Saudi Arabia.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Association of single nucleotide polymorphisms in Wnt signaling pathway genes with breast cancer in Saudi patients. PLoS One. 2013;8(3):e59555.
- [Google Scholar]
- The effect of Poly (ADP-ribose) Polymerase-1 Gene 3′ untranslated region polymorphism in colorectal cancer risk among Saudi cohort. Dis. Markers 2016
- [Google Scholar]
- Association of SCN1A gene polymorphism with antiepileptic drug responsiveness in the population of Thrace, Greece. Arch. Med. Sci.. 2017;13(1):138.
- [Google Scholar]
- Polymorphisms in TP53 and MDM2 combined are associated with high grade endometrial cancer. Gynecol. Oncol.. 2009;113(1):109-114.
- [Google Scholar]
- Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2004;21(2):263-265.
- [Google Scholar]
- Increasing incidence of young-onset colorectal carcinoma A 3-country population analysis. Dis. Colon Rectum. 2020;63(7):903-910.
- [Google Scholar]
- TP53 Arg72Pro polymorphism and colorectal cancer risk: a systematic review and meta-analysis. Cancer Epidemiol. Prevention Biomarkers. 2010;19(7):1840-1847.
- [Google Scholar]
- Genotype misclassification in genetic association studies of the rs1042522 TP53 (Arg72Pro) polymorphism: a systematic review of studies of breast, lung, colorectal, ovarian, and endometrial cancer. Am. J. Epidemiol.. 2013;177(12):1317-1325.
- [Google Scholar]
- Mice deficient for P53 Are developmentally normal but susceptible to spontaneous tumors. Nature. 1992;356(6366):215-221.
- [Google Scholar]
- Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Biophys. Acta. 2021;1876(1):188556
- [Google Scholar]
- Targeting mutant p53 for cancer therapy: direct and indirect strategies. J. Hematol. Oncol.. 2021;14(1):157.
- [Google Scholar]
- Diet, gender, and colorectal neoplasia. J. Clin. Gastroenterol.. 2007;41(8):731-746.
- [Google Scholar]
- MiR-21, miR-34a, miR-125b, miR-181d and miR-648 levels inversely correlate with MGMT and TP53 expression in primary glioblastoma patients. Arch. Med. Sci.. 2017;13(1)
- [Google Scholar]
- DNA damage in cancer development: special implications in viral oncogenesis. Am. J. Cancer Res.. 2021;11(8):3956.
- [Google Scholar]
- TP53 Arg72Pro, mortality after cancer, and all-cause mortality in 105,200 individuals. Sci. Rep.. 2017;7(1):336.
- [Google Scholar]
- The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer. 2009;9(10):749-758.
- [Google Scholar]
- The advantage of women in cancer survival: an analysis of EUROCARE-4 data. Eur. J. Cancer. 2009;45(6):1017-1027. (E. W. Group)
- [Google Scholar]
- Relationship of p53 with other oncogenes, cytokines and systemic lupus erythematosus activity. Tumour Biol.. 2003;24(4):185-188.
- [Google Scholar]
- Genome-wide analysis of sex disparities in the Genetic Architecture of Lung and colorectal cancers. Genes. 2021;12(5):686.
- [Google Scholar]
- FAS/FASL gene polymorphisms in Turkish patients with chronic myeloproliferative disorders. Arch. Med. Sci.. 2017;13(2):426.
- [Google Scholar]
- Cancer of the colon and rectum: potential effects of sex-age interactions on incidence and outcome. Med. Sci. Monit.. 2013;19:203-209.
- [Google Scholar]
- p53 Pro72Arg polymorphism and prostate cancer in men of African descent. Prostate. 2010;70(16):1739-1745.
- [Google Scholar]
- association between TLR-9 polymorphisms and colon cancer susceptibility in saudi arabian female patients. Onco Targets Ther.. 2017;10:1.
- [Google Scholar]
- MGMT promoter methylation as a potential prognostic marker for acute leukemia. Arch. Med. Sci.. 2017;13(6):1433.
- [Google Scholar]
- Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin.. 2021;71(3):209-249.
- [Google Scholar]
- Two polymorphic variants of wild-type p53 differ biochemically and biologically. Mol. Cell Biol.. 1999;19(2):1092-1100.
- [Google Scholar]
- Association between TP53 Arg72Pro polymorphism and leukemia risk: a meta-analysis of 14 case-control studies. Sci. Rep. 2016:6.
- [Google Scholar]
- Increasing incidence of colorectal cancer in young adults in Europe over the last 25 years. Gut. 2019;68(10):1820-1826.
- [Google Scholar]
- Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248(4951):76-79.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2023.102936.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







