Translate this page into:
Phylogenetic analysis of three endogenous species of fish from Saudi Arabia verified that Cyprinion acinaes hijazi is a sub-species of Cyprinion acinaces acinases
⁎Corresponding author. afrefaei@ksu.edu.sa (Abdulwahed Fahad Alrefaei)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Cyprinion acinaces are ray-finned fish belongs to genus Cyprinion. Previous studies have reported that this species has two subspecies, Cyprinion acinaces acinaces, and, Cyprinion acinaes hijazi, however, the validity of later was always in doubt. In past, this fishes were classified merely based on morphological characteristics, and modern biotechnology related tools were never used, which would had been helpful, to authenticate the subspecies status between closely related species. This is the first study to report the classification of the major endemic fish species of Saudi Arabia based on phylogenetic analysis. The cytochrome b gene sequences of Cyprinion acinaces acinases, and Carasobarbus aponesis were determined for the first time, and were deposited in public Gene data Bank. The phylogenetic tree clearly grouped the species into two major clusters, which are divided into four sub-clusters. The phylogenetic analysis supports the early taxonomic classification and validated that C. acinaes hijazi is a subspecies of C. acinaces acinaces. The phylogenetic tree constructed from the cytochrome b sequence also indicated that B. aponesis found in Saudi Arabia is genetically closely related to C. luteus.
Keywords
Barbus
Cyprinid
Freshwater fish species
Saudi Arabia
Cytochrome b
Sequences
Phylogenetics
- GMYC
General Mixed Yele-Coalescent
- NJ
Neighbor joining
- PTP
Poisson Tree Processes
Abbreviations
1 Introduction
Endemism is an important character of freshwater fishes of the Arabian Peninsula (Krupp, 1983; Hamidan and Shobrak, 2019). A total of 21 species of freshwater fishes have been described from the Arabian Peninsula (Hamidan and Shobrak, 2019), of which 15 are endemic species to Arabia, and six have a wider distribution (Freyhof et al., 2015). Previous research indicates that the species identified from Saudi Arabia belong mainly to the family Cyprinidae. The majority of Cyprinid fishes recorded from the Arabian Peninsula by Playfair (1870), Boulenger (1887), Trewavas (1941), Fowler (1956), Balleto and Spano (1977), Banister et al. (1977), Krupp (1983). Alkahem (1983) belong to the genera Cyprinion, Carasobarbus and Garra. Four species belong to the genus Carasobarbus (Cuvier and Valenciennes, 1842). They are small or medium-sized cyprinids with a short dorsal fin. The lateral line is usually complete and the mouth inferior, with two to four barbels or no barbels at all (Krupp et al., 2008). There are four species belonging to the genus Cyprinion (Borkenhagen et al., 2011). According to Alkahem (1983), the fish are of moderate size, with moderately-sized scales. The last unbranched dorsal ray in large specimens is slightly serrated. The species occurring in South East (SE) Arabia are 70–147 mm in length, and have scales of medium size. In the dorsal fin there are three to four serrated spines, and the mouth is sub-terminal (Krupp, 1983). Specimens of this genus have one pair of barbels, the air bladder is bipartite, and there are seven unbranched anal fin rays. Taxonomic studies carried out on the species in these genera have been restricted mainly to their meristic and morphometric characters. Current taxonomists are using DNA sequencing for the classification of the fish. Studies have been carried out in the Arabian Peninsula, Iran and the Middle East, by Durand et al. (2002). These researchers have studied the phylogeny and biogeography of the Cyprinidae, as inferred from their cytochrome b DNA sequences. A detailed study on the phylogenetic relationships of the family Cyprinidae from Iran, using General Mixed Yele-Coalescent (GMYC) and Poisson Tree Processes (PTPs), plus haplotype network analysis, was conducted by Ghanavi et al. (2016). Borkenhagen et al. (2011) and Borkenhagen (2014) investigated the diversity of the cytochrome b sequences of Carasobarbus species, and based on the mitochondrial cytochrome b gene described a new species, Arabibarbus hadharmi from Yemen. In this study we investigated the degree of genetic diversity of three species (Cyprinion acinaces acinaces, Cyprinion acinaces hijazi, and Carasobarbus apoensis) of freshwater fish collected from the Medina province in Saudi Arabia, and explored the potential relationships between these species by examining the genetic diversity within evolutionary lineages via sequence analysis of the cytochrome b gene.
2 Materials and methods
2.1 Sample collection
Thirty specimens of the genera Cyprinion and Carasobarbus (eight Cyprinion acinaces acinaces, 11 Cyprinion acinaces hijazi, and 11 Carasobarbus apoensis) were collected from the Wadi Khadrah (N17 39 410, E42 40 665), and Ain Al-Jamma in Khyber (N25 74 754, E03 92 560) in the Medina Region, Saudi Arabia between December 2019 and January 2020. The fish were kept on dry ice and transported to a laboratory at the King Saud University. The specimens were stored at −80 °C for further analysis. The fish were examined for their morphometric and meristic characters, and identified using these characteristics.
2.2 DNA extraction for polymerase chain reaction
Biopsies were taken from the dorsal fin, and was cut into small pieces, and DNA was extracted using DNAzol® (Invitrogen, UK) (https://assets.fishersci.com/TFS-Assets/LSG/manuals/10503.pdf), according to the manufacturer’s instructions. Tissue samples were homogenized using a hand-held glass/Teflon homogenizer. Fifty milligrams of homogenous tissue were transferred to 1 ml Eppendorf® tubes, and then 1 ml of DNAzol was added to each sample and briefly pipetted up and down rapidly at room temperature to lyse the cells. The samples were then centrifuged at 10,000 g for two minutes at 4 °C. To precipitate the DNA, the resulting supernatant was transferred to new Eppendorf tubes, and 1 ml of 100% ethanol was added to each tube, followed by mixing using a vortex machine for 30 s, followed by five minutes of centrifugation at 10,000g at 4 °C. The liquid was then removed, and the DNA pellet washed with 100 µl of 75% ethanol. Samples were centrifuged again for one minute, and the liquid was removed. The DNA pellets were left to air-dry for 30 min before adding 0.5 ml of nuclease-free water to resuspend the DNA. The DNA samples were then stored at −20 °C. The DNA concentration of each sample was measured by spectrophotometry, based on the absorbance at 260 nm. The presence of extracted DNA was confirmed visually on a 1% agarose gel stained with ethidium bromide, under UV light.
2.3 PCR amplification of the cytochrome b gene
PCR was conducted to amplify the cytochrome b gene using the primers H15891 (5′-GTTTGATCCCGTTTCGTGTA-3′) and L15267 (5′AATGACTTGAAGAACCACCGT-3′), as described by Briolay et al. (1998), with the following modifications: reactions were run with 8.5 µl Gree Master Mix (2X; Thermo Fisher Scientific, Waltham, MA), 3 µl of 10 pg/µl of each forward and reverse primer solution (Eurofins Genomics, Germany), 8.5 µl of nuclease-free water, and 1 µl of extracted DNA, to complete a 24 µl reaction mix. Each PCR run included a nuclease-free negative control. PCR amplification was performed using the following cycling conditions: 94 °C for five minutes, followed by 35 cycles of 94 °C for one minute and 50 °C for one minute, then 72 °C for one minute, and a final extension at 72 °C for 10 min. Each PCR amplification was confirmed visually under UV light on a 1% agarose gel. The positive PCR products were submitted for sequencing to Macrogen Inc. (Seoul, Republic of Korea).
2.4 Sequence analysis and phylogenetic tree construction
To investigate the evolutionary relationships among the sequences of the three species in this study we built phylogenetic tree using the software programs MEGA version 7(Kumar et al., 2016) and CLUSTALX version 2.1 (Jeanmougin et al., 1998). The cytochrome b was amplified from all samples by PCR using the primers in both directions. All sequence data were aligned using the forward and reverse complements of the reverse primer, using the MEGA software. Gene fragments were aligned separately and all new sequences obtained in this study were uploaded to GenBank under the accession numbers MT157379, MT157381 and MT157381. Twenty-nine datasets from different species used for constructing the phylogenetic trees were retrieved from GenBank (Fig. 1). The phylogenetic trees of the datasets obtained from GenBank and those obtained in this study were constructed separately, using neighbor joining (NJ) with genetic distance and the Tamura-Nei model (Kumar et al., 2016; Jeanmougin et al., 1998). We used Felsenstein’s bootstrap method to calculate the associated taxa clustered in the bootstrap test, using 1000 replicates, and these data are shown next to the branches.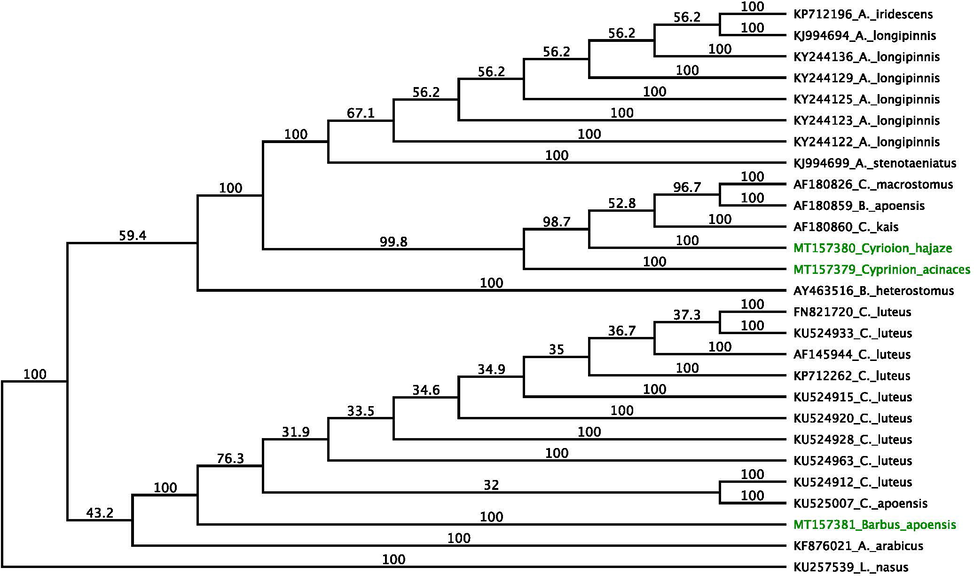
Phylogenetic tree created using the NJ method based on the cytochrome b gene, indicating the relationships of C. acinaces, C. hajaze, and C. apoensis. The NCBI GenBank accession numbers for all sequences are written in front of each species name. C. apoensis was included in the present study as an outgroup. The sequences identified in this study are shown in green.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3 Results
A neighbor-joining phylogenetic tree was constructed from a 455 nucleotide sequence available from all samples (Fig. 1). All clusters were supported by high bootstrap values. Using the reference samples obtained from GenBank, two major groups were identified, composed of C. acinaces and C. hajaze in one clade, and C. apoensis in the other clade.
The relationships of C. acinaces were consistent with traditional morphologically-based inferences, with C. hajaze as the sister lineage of all other species in the same clade. The sequence of C. apoensis clustered with the sequences of Carasobarbus luteus and Carasobarbus apoensis (GenBank: KU525007 and KU524963) with 99.87% identity. This clade was clearly divided from the Cyprinion species, and formed a larger clade as an outgroup of the Carasobarbus species (Fig. 1). All of these sequences are new and are reported here for the first time in Saudi Arabia.
4 Discussion
The taxonomy of some species of the Arabian Peninsula is still not completely resolved, even though, a substantial taxonomic research has been done on the fishes. The fresh water fishes of the Arabian Peninsula mainly belongs to family Cyprinidae, which contains more polyploid species than any other group of fishes (Yang et al., 2015). These fishes need to be re-classified using modern biotechnology tools.
Molecular phylogenetics uses genetic sequence data to understand the relationships between closely related organisms by the comparative study of genes common to both organisms. The use of phylogenetic analysis in taxonomy has become increasingly important, due to the availability of a large amount of sequence data from the public gene data banks (Ziemert and Jensen, 2012).
Comparative genomics helps to identify regions of similarity and differences among genomes. This approach provides a basis for the construction of phylogenetic trees which depict the relationships between clades and taxonomic groups. A phylogenetic tree is a hypothesis pertaining to the evolutionary relationships among groups of organisms. Phylogenetic analysis can be carried out by comparing either whole genomes, or small conserved regions of genes.
The mitochondrial cytochrome genes are relatively highly conserved, and are thus a valuable tool for comparative genomics, and are widely used in phylogenetic studies (Hsieh et al., 2001; Torriani et al., 2009; Irwin et al., 1991).
According to a recent study, the Arabian Peninsula is home to twenty-one species of fish, of which fifteen species are endemic to Arabia, and six species are broadly scattered (Hamidan and Shobrak, 2019). The taxonomy of freshwater fish of Arabia was updated in 1983, and two new subspecies, Cyprinion acinaes hijazi and Garra shailia gharbia were added (Krupp, 1983), but the status of the later is always in doubt as, these fish species were classified solely on morphological criteria, and no attempt has previously been made to verify the classification using phylogenetic tools.
One of the main outputs of this study is reporting the new sequence of the mitochondrial cytochrome b genes of three fresh water fish species in Cyprinion family. The cytochrome b gene of Cyprinion acinaces acinases, Cyprinion acinaes hijazi, and Carasobarbus aponesis has not previously been sequenced. The sequence data have been deposited in the NCBI public Gen Bank accession numbers MT157379 (C. acinaces acinases), MT157381 (C. acinaes hijazi), and MT157381 (B. aponesis). The cytochrome b sequence of these species would not only help with the classification of these species, but will provide a valuable tool for comparative genomic analysis in future studies. Marked divergence in the sequence data was found in 14 SNPs between C. acinaces and C. hajaze, and it was clear from the phylogenetic tree that these taxa formed two distinct groups, closely related at the genus level.
The phylogenetic tree which was constructed in this study, grouped these species into two major clusters, which are divided into four sub-clusters. The first cluster comprised C. acinases, C. acinaes hijazi, and C. macrostomum, and this cluster was supported by high bootstrap values (99). The phylogenetic analysis supports the hypothesis that C. acinases, C. acinaes hijazi, and C. macrostomum are closely genetically related species, and that there are no significant differences in the sequences of their cytochrome b genes. These results clearly indicate that although C. acinaes hijazi has some morphological differences from C. acinases acinases, they are genetically closely related, and hence further confirms that the C. acinaes hijazi is indeed a sub-species of C. acinases acinases.
The second cluster contained all of the A. longipinnis genes in one cluster, indicating that these results are robust.
The third cluster comprised a large number of cytochrome b sequences taken from GenBank, which group C. luteus together in one cluster, which was also supported by a high bootstrap value. The phylogenetic tree constructed from the cytochrome b sequences in this study clustered C. apoensis and C. luteus together. Eleven samples of C. apoensis were used in this analysis, and there was no variation among these samples, an observation which indicates that experimental error was not the cause of this result. The question of whether C. apoensis and C. luteus are genetically close should be further investigated by analyzing a larger number of samples with a wider geological distribution.
The existence of hybrids in fresh water fishes were reported which was result of interbreeding after analyzing the Cytochrome b gene sequence between 29 species of European cyprinids (Briolay et al., 1998). In another study the sequence of the cytochrome b gene also provide a valuable taxonomic tool to verify previous taxonomic classification taxonomic status of North Mediterranean species of the genus Barbus (Tsigenopoulos and Berrebi, 2000). Close phylogenetic relationship between Greek and Iberian endemic species was reported based on the complete mitochondrial cytochrome b gene (Zardoya et al., 1999). The analysis of the complete mitochondrial DNA sequence revealed that two Sinocyclocheilus species fall within the Cyprinion-Onychostoma lineage, including Cyprinus, Carassius, and Procypris, rather than among the Barbinae, as previously suggested on morphological grounds (Wu et al., 2010).
5 Conclusions
In this study we investigated, for the first time, the relationships between three species of fish endemic to the Middle East, using a DNA-based phylogenetic approach. We found that the DNA results largely support the existing taxonomy based on morphometric characteristics, although there are some discrepancies which require further investigation. This work will be valuable for researchers interested in the evolutionary history of these endemic fish, and to fishery managers and policy makers.
Acknowledgements
“We extend our appreciation to the Deanship of Scientific Research at King Saud University for funding this work through the research group no. RG-1441-307, and we thank the Researchers Support Services Unit (RSSU) at King Saud University for their technical support.”
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- The freshwater fishes of Saudi Arabia; Systematics and conservation. USA: Colorado State University; 1983. [Ph. D. Thesis]
- Cyprinidi del genera Garra Hamilton 1822, rarecolti Nelo Yemen. Ann. Mus. Civ. Ster. Nat.“Giacomo Doria”. 1977;81:245-287.
- [Google Scholar]
- A new genus and species of cyprinid fish (Actinopterygii, Cyprinidae) from the Arabian Peninsula, and its phylogenetic and zoogeographic affinities. Environ. Biol. Fishes. 2014;97:1179-1195.
- [CrossRef] [Google Scholar]
- The molecular systematics of the Carasobarbus species from Iran and adjacent areas, with comments on Carasobarbus albus (Heckel, 1843) Environ. Biol. Fishes. 2011;91:327-335.
- [CrossRef] [Google Scholar]
- An account of the fishes obtained by Surgeon-Major ASG Jayakar at Muscat, east coast of Arabia. In: Proc. Zool. Soc.Lond.. Wiley Online Library; 1887. p. :653-667.
- [Google Scholar]
- Molecular phylogeny of cyprinidae inferred fromcytochrome bDNA Sequences. Mol. Phylogenet. Evol.. 1998;9:100-108.
- [CrossRef] [Google Scholar]
- Histoire naturelle des poissons. Paris: P. Bertrand; 1842. p. :465-487.
- Phylogeny and biogeography of the family Cyprinidae in the Middle East inferred from cytochrome b DNA-evolutionary significance of this region. Mol. Phylogenet. Evol.. 2002;22:91-100.
- [CrossRef] [Google Scholar]
- Fishes from Cyprus, Iran, Iraq, Israel and Oman. Bull. Res. Counc. Isr.. 1956;5:260-292.
- [Google Scholar]
- The status and distribution of freshwater fishes of the Arabian Peninsula. In: The Status Distrib. Freshw. Biodivers. Arab, Peninsula. 2015. p. :16.
- [Google Scholar]
- Phylogenetic relationships of freshwater fishes of the genus Capoeta (Actinopterygii, Cyprinidae) in Iran. Ecol. Evol.. 2016;6:8205-8222.
- [CrossRef] [Google Scholar]
- Cytochrome b gene for species identification of the conservation animals. Forensic Sci. Int.. 2001;122:7-18.
- [CrossRef] [Google Scholar]
- Evolution of the cytochromeb gene of mammals. J. Mol. Evol.. 1991;32:128-144.
- [CrossRef] [Google Scholar]
- Multiple sequence alignment with Clustal X. Trends Biochem. Sci.. 1998;23:403-405.
- [CrossRef] [Google Scholar]
- Fishes of Saudi Arabia: Freshwater Fishes of Saudi Arabia and Adjacen Regions of the Arabian Peninsula. Verlag Nicht Ermittelbar; 1983.
- Krupp, F., Schneider, W., Kühne, H., 2008. Die Fischfauna des Nahr al-n/pubmed/9810230“(98 Umwelt und Subsistenz der assyrischen Stadt Dūr-Katlimmu am unteren malst 8, 41–51
- MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol.Evol.. 2016;33:1870-1874.
- [CrossRef] [Google Scholar]
- Note on a freshwater fish from the neighbourhood of Aden. Proc. Zool. Soc.Lond. 1870:85-86.
- [Google Scholar]
- Sequence conservation in the mitochondrial cytochrome b gene and lack of G143A QoI resistance allele in a global sample of Rhynchosporium secalis. Austral. Plant Pathol.. 2009;38:202-207.
- [CrossRef] [Google Scholar]
- British Museum (Natural History) expedition to southwest Arabia. Freshw. Fishes. 1941;1937–8(3):7-15.
- [Google Scholar]
- Molecular phylogeny of north mediterranean freshwater barbs (genus Barbus: cyprinidae) inferred from cytochrome b sequences: biogeographic and systematic implications. Mol. Phylogenet. Evol.. 2000;14:165-179.
- [CrossRef] [Google Scholar]
- The complete mitochondrial genomes of two species from Sinocyclocheilus (Cypriniformes: Cyprinidae) and a phylogenetic analysis within Cyprininae. Mol. Biol. Rep.. 2010;37:2163-2171.
- [CrossRef] [Google Scholar]
- Phylogeny and polyploidy: resolving the classification of cyprinine fishes (Teleostei: Cypriniformes) Mol. Phylogenet. Evol.. 2015;85:97-116.
- [CrossRef] [Google Scholar]
- Phylogenetic relationships of Greek cyprinidae: molecular evidence for at least two origins of the Greek cyprinid fauna. Mol. Phylogenet. Evol.. 1999;13:122-131.
- [CrossRef] [Google Scholar]
- Phylogenetic approaches to natural product structure prediction. Methods Enzymol. Elsevier. 2012;517:161-182.
- [CrossRef] [Google Scholar]







