Phosphoproteomics analysis of hypopharyngeal glands of the newly emerged honey bees (Apis mellifera ligustica)
⁎Corresponding author. apislijk@126.com (Jianke Li)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The most important honey bee queen food royal jelly is produced by the exocrine hypopharyngeal glands (HGs) of the worker honey bees. The HGs exhibits diverse gene and protein that create age-related physiological adaptations. However, limited knowledge is available on how the phosphorylation process is responsible for physiological alterations across the development of HGs in newly emerged bees. This study measured the acinus of HGs and characterized its phosphoproteomics analysis between the newly emerged bees of royal jelly bees (RJBs) and Italian bees (ITBs). Phosphopeptides of HGs were enriched by Ti4+-IMAC reagents, followed by protein identification via Q-Exactive LC-MS/MS. Our findings indicated that the mean acinus size of HGs of newly emerged bees of RJBs was significantly larger (56.18 ± 1.72 µm) than ITBs (45.98 ± 1.62 µm). A total of 1576 phosphopeptides with 1800 phosphosites containing 525 phosphoproteins were identified in RJBs, while 746 phosphopeptides, of which 846 phosphosites correspond to 317 phosphoproteins were identified in ITBs. Most proteins were phosphorylated on 1 residue followed by 2 and 3 residues in newly emerged bees of both bee stocks. In addition, serine phosphorylation was most observed, followed by threonine and tyrosine in both bee stocks. In newly emerged bees of RJBs, the protein metabolic process, glycolytic process, and formation of translation preinitiation were uniquely enriched, while protein translation, peptide metabolic process and elongation were enriched in ITBs. This research shows detailed phosphorylation of HGs and provides helpful information for understanding the biological activities of HGs development in newly emerged bees from both bee stocks.
Keywords
Newly emerged bees
Hypopharyngeal glands
Mass spectrometry
Phosphoproteomics
Phosphopeptides
1 Introduction
The domesticated honey bee (Hymenoptera: Apidae) is an economically eusocial insect and produces various products like honey, royal jelly, and venom, which are used in pharmaceutical drugs and other commercial goods (Grozinger and Flenniken, 2019; Adgaba et al., 2020; Papa et al., 2022). In addition, honey bees are essential pollinators of flowering plant species that cover nearly three-quarters of world food crops (Grozinger and Flenniken, 2019). Many physiological expressions of social insects are linked to exocrine glands, as evidenced by nest-building abilities (Noll et al., 2021).
Moreover, insect glands exhibit diverse functions fluctuating from communication, diet processing, and reproduction to protection, in which hypopharyngeal glands (HGs) are unique characteristics of Hymenopteran insects (Chapman and Chapman, 1998; Cruz-Landim and Costa, 1998; Ahmad et al., 2021). In honey bees, exocrine HGs are responsible for synthesizing the enzyme that metabolizes the nutrient and produces the royal jelly as larval food (Huang et al., 1989; Costa and Cruz-Landim, 1999). The coiled HGs of honey bee workers and queens are situated in an anterior head between the eyes and brain but only well grow in workers (Knecht and Kaatz, 1990). The HGs in newly emerging bees are in a phase of fast development, with tiny, well-structured cell cytoplasm and acinus. HGs are well developed in nurse bees (6–15 days after emergence) with oval acini attached to the axial collecting duct, and secretion of these glands run to the mouth part through the collecting duct (Li et al., 2010). The size of HGs in forager bees (>15 days after emergence) gradually shrink, and the secretion of royal jelly transfer to enzyme synthesis for preparing honey and pollen (Kubo et al., 1996; Ohashi et al., 1997). Comparing HGs among honey bee races is an essential parameter for comprehending the development and physiology of HGs at the molecular level (Liu et al., 2014; Ahmad et al., 2021).
Progression in honey bee genome sequencing and proteomic methods are essential tools for exploring the biology, performance, functioning, neurobiology, and immunopathology in the depths of molecular and biological levels (Hora et al., 2018). The phosphorylation of proteins is the essential post-translational modification (PTMs) that has a significant effect on the biological nature of the protein, which causes changing everything that leads from innate behavior to localization of cell sites and formation of complex structures (Arrington et al., 2017). Post-translation modifications and PTMs sites are essential factors for a better understanding of protein function (Boyer, 2006). The main function of protein phosphorylation involves regulating cell differentiation and development, protein interaction with various molecules, enzyme activity, and maintaining metabolic activity in living cells. One of the most common PTMs is the covalent modification of serine, threonine, and tyrosine in eukaryote with the phosphate group (Hunter, 1995; Raggiaschi et al., 2005; Hernández et al., 2012), which contains almost 90%, 10% and less than 1% of the total protein phosphorylation, respectively. In contrast, arginine and histidine phosphorylation sites are still unclear in eukaryotes (Trentini et al., 2014).
Our group have been studied phosphorylation of the mandibular gland, royal jelly, honey bee brain, venom gland, and salivary gland, which provided deep knowledge for understanding the biological processes in honey bees (Feng et al., 2013; Li et al., 2013; Han et al., 2014; Bezabih et al., 2017; Li, 2017). Only a few studies have been conducted regarding the phosphorylation of HGs protein across the workers of honey bees (Lu et al., 2013; Qi et al., 2015). However, knowledge is still lacking, so further apply the phosphoproteome approach to HGs to better understand its morphology and physiology at the molecular level and how the protein phosphorylation process controls biological changes, especially in newly emerged bees. Due to recent development in enrichment methodology, Ti4+-IMAC has been used to increase phosphoproteome coverage. Therefore, the primary aim of the research is to measure the acinus of HGs and compare its phosphoproteomics analysis between the newly emerged Royal jelly bees (RJBs) and Italian bees (ITBs). This work demonstrates a comprehensive characterization of phosphorylation of HGs and provides valuable information to understand the biological functions of HGs development in newly emerged bees of both bees’ stock.
2 Materials and methods
2.1 Experimental setting
The workflow explaining and comparing HGs phosphoproteome between the newly emerged RJBs and ITBs is shown in Fig. 1. RJBs and ITBs (Apis mellifera ligustica) colonies were established at “the Institute of Apicultural Research, Chinese Academy of Agricultural Sciences, Beijing, China.” Therefore, ITB queens were imported from Bologna, Italy, while RJB queens were imported from Zhejiang Province, China. Honey bee colonies were managed with the nearly same population of worker bees, broods, honey combs, and availability of an equal amount of chaste berry (Vitex negundo L.) nectar flow to all boxes at the time of June.
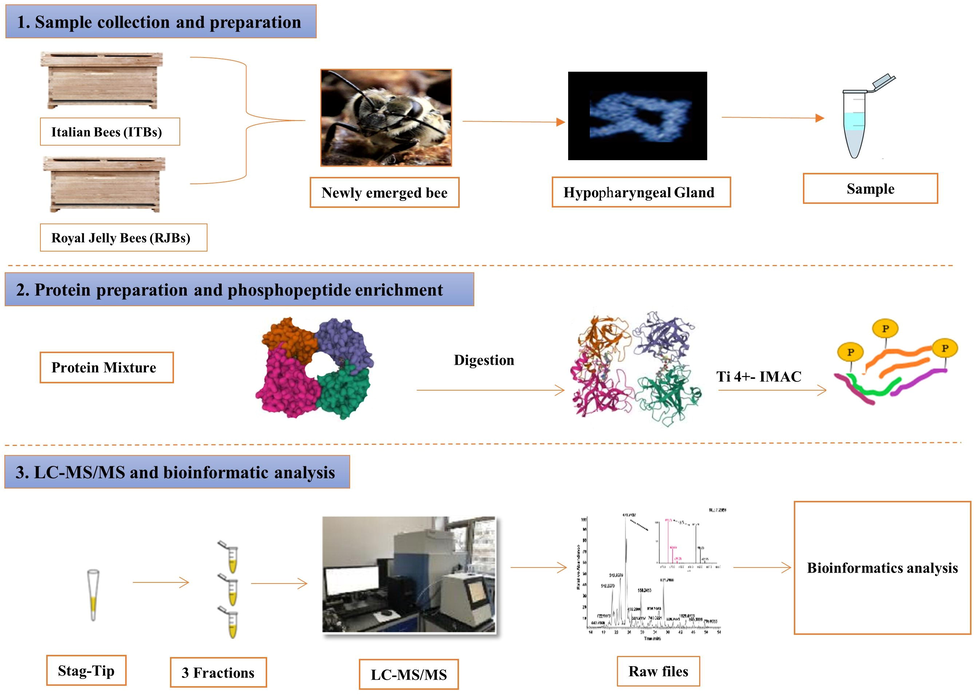
- The overall workflow of the experiments.
2.2 Sampling of hypopharyngeal glands
For the HGs phosphoproteome analysis, five colonies of RJBs, which produced maximum royal jelly, and five ITBs colonies with the lowest royal jelly output were selected for HGs sampling. The newly emerged bees in less than 12 h were taken when it emerged from sealed brood in the incubator. The HGs were dissected of 80 bees of each bee stock in the presence of cold PBS containing protease inhibitors under the operating microscope.
2.3 Measuring of HGs acinus
The average measurement of acini diameter of 40 HGs of each sample type was taken with the help of a scanning electron microscope for statistical analysis with minor modifications in our described method (Li et al., 2010).
2.4 Protein extraction and digestion
The HGs tissues were homogenated in ice by pestle for protein analysis. Then homogenized HGs sample was mixed with a lysis buffer comprising 8 M urea, 2 M of thiourea, 4 percent CHAPS, 20 mM tris base, and 30 mM DTT in ice for 30 min. Then lysate was sonicated (10 × 3 second, 10 intervals) to increase protein solubility, and the sample was centrifuged at (14,000g) for 15 min to eliminate insoluble particles. The supernatant was aspirated gently, and added 4 volumes of TCA to precipitate the protein that improved supernatants for 30 min at −20 °C, after that centrifuging twice to remove the supernatant, tap the tube on clean absorbent towels to dry the protein pellets. The protein pellets were re-dissolved in 80 ∼ 100 μL 5 M urea, vortex the pellets with pipet, and sonication the solution of (10 × 3 sec, 10 intervals). Protein digestion was carried out by the earlier mentioned technique with some minor modifications (Qi et al., 2015; Bezabih et al., 2017). Added 40 mM of (NH4) HCO3, then added DTT to protein solution for 1 h, and at last, alkalize with adding IAA for 1 h in the dark place to make protein denatured and were kept in the reduced state. Using sequencing-grade modified trypsin, the protein sample was digested into peptides at 37 °C for 14 h. At last, 1 μL formic acid was mixed to ceased digestion.
2.5 Phosphopeptide enrichment
The digested HGs samples were mixed in 500 μL of binding solution having 6% of TFA/80% of ACN and treated with 5 mg Ti4+-IMAC magnetic material at 25 °C for 1 h as previously described (Qi et al., 2015). After centrifugation, the supernatant was removed, and phosphopeptide-coated beads were cleaned in 200 L of solution (0.6 percent TFA, 50 percent CAN, and 200 mM NaCl) and (0.1 percent TFA/30 percent ACN), respectively. The enriched phosphopeptide bound was eluted two times with 100 μL (10% NH3) solution stirring for 10 min. At last, three fractions were shared, and phosphopeptides were desalted with solid-phase extraction cartage C18 column. Desalted phosphopeptide sample was dry by a Speed Vac system, dry peptides were re-dissolved in 0.1 of FA, and the final sample was stored for LC–MS/MS analysis at −80 °C.
2.6 Nano LC–MS/MS analysis
From each HGs sample, an 8 μL of phosphopeptide sample containing 0.5 μg of peptides was analyzed by nano HPLC and high-resolution MS/MS. The sample was placed on a trap column, and the flow rate was 5 μL/min prior to analytic separation. Formerly, the peptides consequently were separated into analytical columns. The peptides were eluted within 90 min under the following conditions: from 100 percent buffer A (0.1% FA) to 8% buffer B (0.1% FA, 80% ACN) in five minutes, from 8 to 20 percent buffer B in 55 min, after that from 20 percent to 30 percent buffer B for ten minutes, increase buffer B up to 100 percent for 10 min and additional 100% buffer B for 10 min. After those eluting peptides were ionized by electrospray ionization (ESI) and were analyzed into a Q-Exactive LC-MS (Thermo Fisher Scientific, USA) in 700,000 resolutions for the full scan.
2.7 Database search
The MS/MS spectra of phosphoproteome raw data were retrieved by Xcalibur (Thermo Scientific). All raw data were managed and analyzed by in-house PEAKS software (Bioinformatics Solution Inc. Waterloo, Canada). The database containing protein sequences of A. m with common contaminate integrated with 21,778 total entries. After that, scaffold PTM was used to localize HG phosphorylation sites of HGs by probability via the A scores algorithm (Beausoleil et al., 2006). All MS/ MS of phosphorylation sites were considered to have more than 95% confidence intervals.
2.8 Bioinformatics analysis
Expression levels of protein phosphorylation with three replicates were quantified during three-time points of the HGs of newly emerged bees, and MS raw data were preceded in-house PEAKS. Protein phosphorylation site in HGs of newly emerged bee was identified using Drosophila orthologs uniport identifier as bee protein through uniport database, Drosophila orthologs is a unique identifiers honeybee proteins were identified protein of HGs and was used as functional analysis by Cyto-scape plugin (Zolnierowicz and Bollen, 2000). The significantly developed biological process in the functional Gene Ontology (GO) pathways was determined by right-sided hygrometric tests. Drosophilae genome was chosen as background as a set of GO annotation and then account false discovery rate using Bonferroni test to p-value, and kappa score level is 0.4 in Clue GO. In addition, all descriptive data were analyzed by SPSS (version 26).
3 Results
3.1 Measuring of hypopharyngeal glands acinus size
The results revealed that the acinus size of HGs was more significant in the newly emerged bees of RJBs as compared to ITBs (Fig. 2A). The mean maximum acinus size (56.18 ± 1.72 µm) was observed in HGs of RJBs, followed by ITBs (45.98 ± 1.62 µm) in newly emerged bees (Fig. 2B).
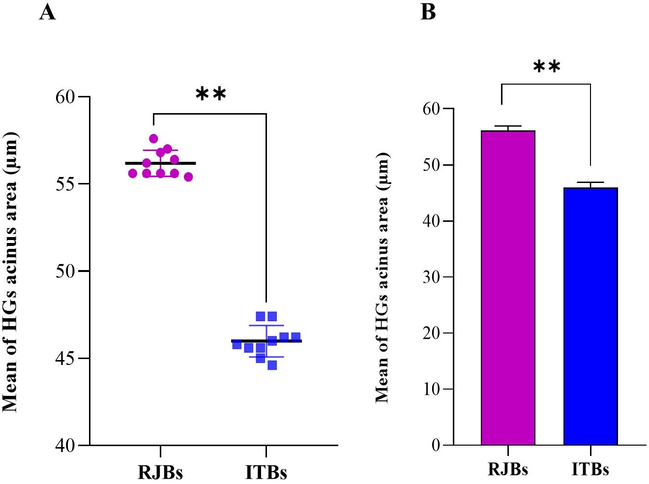
- (A) Mean diameter (µm) of newly emerged bees of hypopharyngeal glands (HGs) acinus of the experimental colonies. (B) The HGs acini mean diameter of newly emerged bees.
3.2 Identification of phosphoproteins and phosphopeptides
The present study used the Ti4+-IMAC phosphopeptides enrichment method, Nano LC–MS/MS analysis, in-house PEAKS search, and an extensive data set of phosphopeptides and phosphorylated protein obtained in the HGs of newly emerged honey bees. In RJBs, a total of 1576 phosphopeptides carrying 1800 phosphorylation sites containing 525 phosphoproteins were identified by IMAC+4 chromatography. In the case of ITBs, 746 phosphopeptides, of which 846 phosphorylation sites corresponding to 317 phosphoproteins were identified (Fig. 3A). Of the total 842 phosphoproteins, 39.67% were unique to RJBs and 14.49% were unique to ITBs, and 45.84% were shared between them (Fig. 3B).
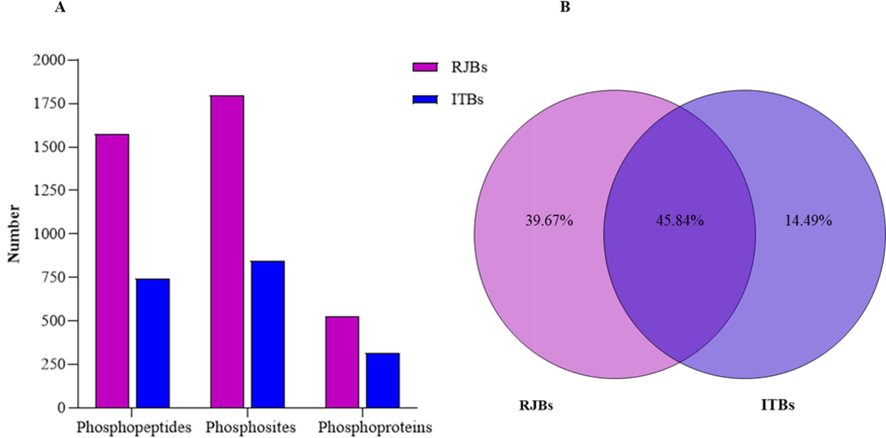
- Overall characteristics of phosphoproteomics of newly emerged bees' hypopharyngeal glands (HGs). (A) The number of phosphopeptides, phosphosites and phosphoproteins were identified between royal jelly bees (RJBs) and Italian bees (ITBs). (B) Percentage of shared and unique phosphoproteins between RJBs and ITBs.
Most HGs phosphopeptides were phosphorylated on a single site, and others were phosphorylated at many sites. In RJBs, 86.93% of phosphopeptides were phosphorylated at 1 residue, 12.56% were phosphorylated on 2, whereas 0.51% were phosphorylated on 3 residues (Fig. 4A). In ITBs, 86.73% of phosphopeptides were phosphorylated on 1 residue, whereas 13.14% were phosphorylated on 2 residues, and only 0.13% were phosphorylated on the single site (Fig. 4B).
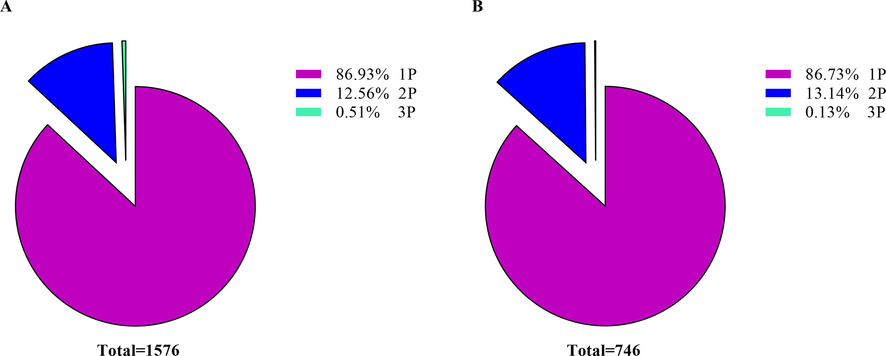
- Disseminated of phosphopeptides depends on the number of phosphosites identified in the hypopharyngeal glands (HGs) of newly emerged bees. (A) The 1P, 2P, and 3P denote a phosphopeptide with one, two, and three phosphosites in royal jelly bees. (B) In Italian bees, the 1P, 2P, and 3P represent a phosphopeptide with one and two phosphosites.
The relative frequencies of serine, threonine, and tyrosine within all identified phosphosites were mentioned (Fig. 5A, B). Out of 1800 phosphosites, serine phosphorylated was most observed in RJBs. That consisted of 1467 (81.5%) in the case of serine phosphorylation, followed by threonine 306 (17%) and tyrosine phosphorylation 27 (1.5%) (see Fig. 5A). Also, in ITBs, serine phosphorylation was most observed. Of the total 846 phosphosites, 700 (84.04%) were identified as serine phosphorylation, while 135 (18.80%) were threonine phosphorylation, and tyrosine phosphorylation was not observed (Fig. 5B).
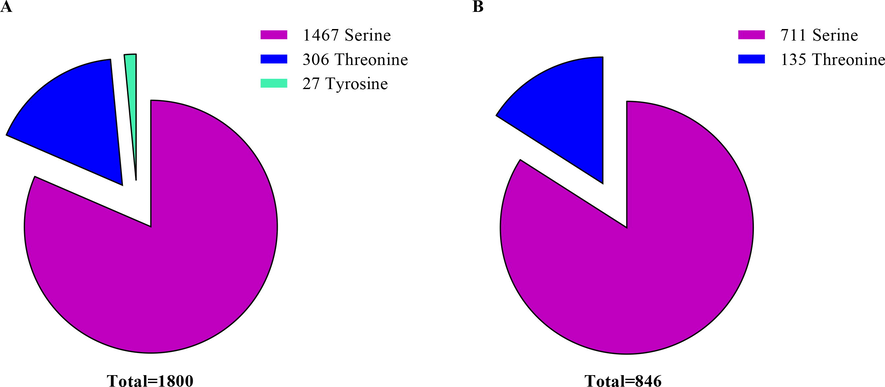
- The relative frequencies of serine, threonine, and tyrosine within all phosphosites in the hypopharyngeal glands (HGs) of newly emerged honey bees.
3.3 Functional enrichment analysis of phosphoproteins
In addition, the identified phosphoproteins were analyzed using Clue Go to identify pointedly enriched KEGG pathways and GO terms. The results demonstrated that in newly emerged bees of RJBs, the most highly represented group of phosphoproteins was related to peptide metabolic process (52.17%), followed by phosphoproteins associated with the formation of translation preinitiation (17.39%), glycolytic process (13.04%), protein metabolic process (13.04%), and translational elongation (4.35%) (see Fig. 6A). Among them, the protein metabolic process, glycolytic process, and formation of translation preinitiation were uniquely enriched (Fig. 6B). In case of newly emerged bees of ITBs, the significantly enriched group of phosphoproteins was related to translation (57.14%), followed by peptide metabolic process (28.57%), and translational elongation (14.29%) (see Fig. 6 C). Among them, protein translation elongation, peptide metabolic process and elongation were uniquely enriched (Fig. 6D).
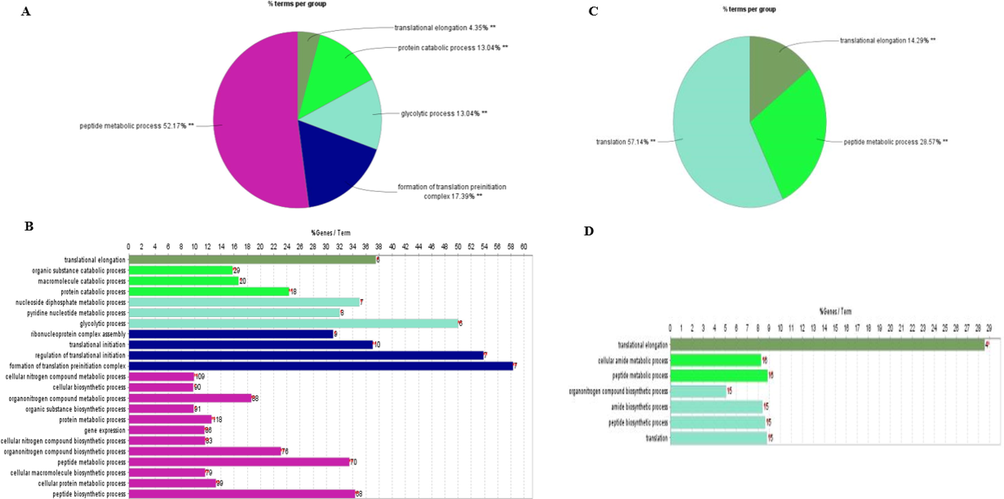
- Enrichment of functional GO term of the phosphoproteins identified in the HG of newly emerged bees. Gene percentage linked to specific phrases is shown on the bars.
4 Discussion
Advances in honey bee genome sequencing and “omics” tools have greatly expanded our understanding of honey bee biology, physiology, and immunology at the molecular and biochemical levels. In addition, proteomic and phosphoproteome have proven to be effective methods for demonstrating the morphology and physiology of the HGs that underpin honey bee biology (Qi et al., 2015; Hu et al., 2019). This study compared the acinus size of HGs and the phosphoproteome analysis of HGs between the newly emerged bees of RJBs and ITBs. Our findings indicated that the acinus size of HGs of newly emerged RJBs was significantly larger than the newly emerged bees of ITBs. Our outcomes were consistent with the previous findings (Li et al., 2010; Qi et al., 2015; Hu et al., 2019).
Remarkably, the acinus area increases progressively from the development of bee workers until 12 days and starts to decrease. It is also documented that the diameter of the acinus of the HGs was twice at nine days old than the length at the first emergence (Lee et al., 2019). Moreover, the development of HGs and their size is negatively affected by various biotic and biotic aspects including honey bee races, food resources, seasonal, and pesticide exposure (Smodiš Škerl and Gregorc, 2015; Omar et al., 2017; Wang et al., 2018; Hu et al., 2019). For instance, the acinus size of HGs fluctuates in different races of honey bee strains across the workers. The acinus size of HGs was smaller in A. mellifera carnica than A. mellifera ligustica in Saudi Arabia (Ali et al., 2019). Smodiš Škerl and Gregorc (2015) observed the larger acini diameter in HGs of A. mellifera carnica nurse bees. Larger glands size was found in nurses of A. cerana as compared to forager and guard bees (Suwannapong et al., 2010). In addition, Zahra and Talal (2008) noticed that supplemental feeding in A. mellifera colonies promoted an increase in the mean acinus size of HGs and its duct length. In newly emerged bees, the cells of HGs are still developing and differentiating in preparation for entering the secretory phase (Qi et al., 2015). The peak size of the gland looks like grapes in which each grape has a distinct lob structure recognized as acinus (plural acini). Therefore, the size of HGs is correlated with bee ages and their age-associated jobs (Li et al., 2010; Corby-Harris and Snyder, 2018; Hu et al., 2019).
Moreover, our results revealed that the abundance level of phosphopeptides, phosphosites, and phosphoproteins was higher in HGs of the newly emerged bees of RJBs in comparison to newly emerged bees of ITBs, even though it is likely not complete. Also, uniquely enriched phosphoproteins were higher in RJBs than ITBs. In this study, many phosphorylation sites in HG proteins were discovered, revealing that the Ti+4-IMAC chromatography technique is a practical approach for characterizing the honey bee phosphoproteome in unprecedented detail. Previously, only a few studies have been on the phosphoproteome analysis of HGs in honey bees. Due to technical limitations, small fractions of phosphopeptides, phosphosites and phoshoryated proteins were recognized in the HGs of RJBs (Lu et al., 2013). However, the advancement of phosphopeptides enrichment techniques increases phosphoproteins' coverage in HGs of honey bees (Qi et al., 2015). A comprehensive analysis of the HGs phosphoproteome demonstrates that a significant number of cellular proteins are phosphorylated in newly emerged bees of HGs. Many phosphopeptides, phosphosites, and phosphorylated proteins were discovered in newly emerged bees of RJBs than the ITBs, indicating the simple signal activity that plays a critical role in cell growth and proliferation. The majority of proteins were phosphorylated on a single residue, followed by double and triple residues in both bee stocks. In addition, most of the phosphorylated events occur on serine and threonine residues. These observations are consistent with the discoveries of Qi et al. (2015).
In addition, the results demonstrated that in newly emerged bees of RJBs, peptide metabolic process, the formation of translation preinitiation, glycolytic process, protein metabolic process, and translational elongation were significantly enriched. Whereas, in newly emerged bees of ITBs, peptide metabolic process, translation, and translational elongation were significantly enriched. In newly emerged bees, functional classes like pyruvate metabolism, muscle cell growth, and pole plasma mRNA localization are functionally complementary (Qi et al., 2015). Previously studies also reported that synthesis of the ribonucleoprotein complex, fatty acid breakdown, and the highly abundant hexamerins (Hex 110) protein regulate young HGs gland growth and tissue formation in newly emerged bees (Boisvert et al., 2010; Hu et al., 2019). Moreover, further research is needed to use the different phosphopeptide enrichment techniques to compare the phosphopeptide abundances and unveil the physiology of HGs in honey bee workers.
5 Conclusions
In this study, we compared the phosphoproteomics analysis of HGs of newly emerged bees by a phosphorylated-specific enrichment method. Our findings revealed that the acinus diameter of HGs of newly emerged bees of RJBs was significantly more than ITBs. Similarly, a large number of phosphopeptides, phosphosites, and phosphorylated proteins were discovered in newly emerged bees of RJBs than the ITBs. In newly emerged bees of RJBs, the protein metabolic process, glycolytic process, and formation of translation preinitiation were uniquely enriched, while protein translation, peptide metabolic process and elongation were uniquely enriched in ITBs, which increased our knowledge of critical biological processes in HGs of honey bees. In order to further investigate the phosphoproteomics assessment of HGs at various stages of the honey bee life cycle, new phosphopeptides enrichment strategies will be developed to explore the morphology and physiology of HGs in honey bees.
Funding
Financial support is provided by “the Agricultural Science and Technology Innovation Program (CAAS-ASTIP-2015-IAR), the earmarked fund for Modern Agro-Industry Technology Research System (CARS-44) in China, and the National Project for Upgrading Overall Bee-Product Quality of the Beekeeping Industry of China.”. The authors also appreciate the support of the Research Center for Advanced Materials Science (RCAMS) at King Khalid University Abha, Saudi Arabia through a project number RCAMS/KKU/001-21.
Acknowledgements
The first author was financed by “the Chinese Government Scholarship (CGS)-for Ph.D. Program at GS-CAAS”.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Physico-chemical, antioxidant and anti-microbial properties of some Ethiopian mono-floral honeys. Saudi J. Biol. Sci.. 2020;27(9):2366-2372.
- [Google Scholar]
- Ahmad, S., Khan, S.A., Khan, K.A., Li, J., 2021. Novel insight into the development and function of hypopharyngeal glands in honey bees. Front. Physiol., 1853.
- Effect of season and behavioral activity on the hypopharyngeal glands of three honey bee Apis mellifera L. races under stressful climatic conditions of central Saudi Arabia. J. Hymenoptera Res.. 2019;68:85.
- [Google Scholar]
- Recent advances in phosphoproteomics and application to neurological diseases. Analyst. 2017;142(23):4373-4387.
- [Google Scholar]
- A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol.. 2006;24(10):1285-1292.
- [Google Scholar]
- Phosphoproteome analysis reveals phosphorylation underpinnings in the brains of nurse and forager honeybees (Apis mellifera) Sci. Rep.. 2017;7(1):1-16.
- [Google Scholar]
- A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol. Cell. Proteomics. 2010;9(3):457-470.
- [Google Scholar]
- Boyer, R., 2006. Posttranslational modification of proteins: Expanding nature's inventory. Christopher T. Walsh, Roberts & Company Publishers, Greenwood Village, CO, 2005, 576 pp., ISBN 0‐9747077‐3‐2, $98.00,. Wiley Online Library).
- The Insects: Structure and Function. Cambridge University Press; 1998.
- Measuring hypopharyngeal gland acinus size in honey bee (Apis mellifera) workers. JoVE (Journal of Visualized Experiments). 2018;139:e58261.
- [Google Scholar]
- Occurrence and morphometry of the hypopharyngeal glands inScaptotrigona postica Lat. (Hymenoptera, Apidae, Meliponinae) J. Biosci.. 1999;24(1):97-102.
- [Google Scholar]
- Structure and function of the hypopharyngeal glands of Hymenoptera: a comparative approach. J. Comp. Biol.. 1998;3(2):151-163.
- [Google Scholar]
- Novel aspects of understanding molecular working mechanisms of salivary glands of worker honeybees (Apis mellifera) investigated by proteomics and phosphoproteomics. J. Proteomics. 2013;87:1-15.
- [Google Scholar]
- Bee viruses: Ecology, pathogenicity, and impacts. Annu. Rev. Entomol.. 2019;64(1):205-226.
- [Google Scholar]
- In-depth phosphoproteomic analysis of royal jelly derived from western and eastern honeybee species. J. Proteome Res.. 2014;13(12):5928-5943.
- [Google Scholar]
- Proteomics improves the new understanding of honeybee biology. J. Agric. Food Chem.. 2018;66(14):3605-3615.
- [Google Scholar]
- In-depth proteome of the hypopharyngeal glands of honeybee workers reveals highly activated protein and energy metabolism in priming the Secretion of Royal Jelly*[S] Mol. Cell. Proteomics. 2019;18(4):606-621.
- [Google Scholar]
- Nature of brood signal activating the protein synthesis of hypopharyngeal gland in honey bees, Apis mellifera (Apidae: Hymenoptera) Apidologie. 1989;20(6):455-464.
- [Google Scholar]
- Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell. 1995;80(2):225-236.
- [Google Scholar]
- Patterns of larval food production by hypopharyngeal glands in adult worker honey bees. Apidologie. 1990;21(5):457-468.
- [Google Scholar]
- Change in the expression of hypopharyngeal-gland proteins of the worker honeybees (Apis mellifera L.) with age and/or role. J. Biochem.. 1996;119(2):291-295.
- [Google Scholar]
- Age-dependent hypopharyngeal gland development and morphometric characteristics in the cross-bred lineage of honeybees reared for high royal jelly production. J. Asia-Pac. Entomol.. 2019;22(3):699-704.
- [Google Scholar]
- Proteome comparison of hypopharyngeal gland development between Italian and royal jelly producing worker honeybees (Apis mellifera L.) J. Proteome Res.. 2010;9(12):6578-6594.
- [Google Scholar]
- Proteome and phosphoproteome analysis of honeybee (Apis mellifera) venom collected from electrical stimulation and manual extraction of the venom gland. BMC Genomics. 2013;14(1):766.
- [Google Scholar]
- Comparative analysis of phosphoproteome between mandibular glands of high royal jelly producing Bees and Italian Bees. Sci. Agricult. Sin.. 2017;50(23):4656-4670.
- [Google Scholar]
- Transcriptome differences in the hypopharyngeal gland between Western Honeybees (Apis mellifera) and Eastern Honeybees (Apis cerana) BMC Genomics. 2014;15(1):744.
- [Google Scholar]
- Phosphoproteome analysis of hypopharyngeal glands of high royal jelly producing bee (Apis mellifera L.) Sci. Agric. Sin. 2013;46:5050-5057.
- [Google Scholar]
- Would wax glands help us to understand the relationships among corbiculate bees? Insectes Soc.. 2021;68(2):191-197.
- [Google Scholar]
- Change in the mode of gene expression of the hypopharyngeal gland cells with an age-dependent role change of the worker honeybee Apis mellifera L. Eur. J. Biochem.. 1997;249(3):797-802.
- [Google Scholar]
- Influence of different pollen diets on the development of hypopharyngeal glands and size of acid gland sacs in caged honey bees (Apis mellifera) Apidologie. 2017;48(4):425-436.
- [Google Scholar]
- The honey bee Apis mellifera: An insect at the interface between human and ecosystem health. Biology. 2022;11(2):233.
- [Google Scholar]
- Phosphoproteomic analysis of protein phosphorylation networks in the hypopharyngeal gland of honeybee workers (Apis mellifera ligustica) J. Proteome Res.. 2015;14(11):4647-4661.
- [Google Scholar]
- Characteristics of hypopharyngeal glands in honeybees (Apis mellifera carnica) from a nurse colony. Slovenian Veterinary Res.. 2015;52(2)
- [Google Scholar]
- Suwannapong, G., Chaiwongwattanakul, S., Benbow, M.E., 2010. Histochemical comparison of the hypopharyngeal gland in Apis cerana Fabricius, 1793 workers and Apis mellifera Linnaeus, 1758 workers. Psyche 2010.
- Chasing phosphoarginine proteins: development of a selective enrichment method using a phosphatase trap. Mol. Cell. Proteomics. 2014;13(8):1953-1964.
- [Google Scholar]
- Investigating the regulation of hypopharyngeal gland activity in honeybees (Apis mellifera carnica) under overwintering conditions via morphologic analysis combined with iTRAQ-Based comparative proteomics. Ann. Entomol. Soc. Am.. 2018;111(3):127-135.
- [Google Scholar]
- Impact of pollen supplements and vitamins on the development of hypopharyngeal glands and on brood number in honey bees. J. Apicult. Sci.. 2008;52(2)
- [Google Scholar]
- Protein phosphorylation and protein phosphatases De Panne, Belgium, September 19–24, 1999. EMBO J.. 2000;19(4):483-488.
- [Google Scholar]







