Translate this page into:
Morphological and molecular phylogenetic analyses of the apicomplexan parasites, Eimeria media and Eimeria stiedai, infecting the domestic rabbits, Oryctolagus cuniculus
⁎Corresponding author. rabdelgaber.c@ksu.edu.sa (Rewaida Abdel-Gaber)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The eimerian infection is one of the most serious infections that can decrease rabbit productivity since it can lead to serious diseases. There is little information about Eimeria media and Eimeria stiedai infections in Saudi Arabia, and molecular data is particularly weak. To establish the prevalence, morphological and molecular characterization of E. media and E. stiedai isolated from spontaneously infected rabbits, the current study was conducted. Ten healthy rabbits, Oryctolagus cuniculus, whose feces were collected and kept at the laboratory were examined for protozoan parasite infection using the floatation method. Purified oocysts were used to extract DNA, which was then used in a polymerase chain reaction (PCR) with primers that amplified a partial sequences of the 18S rDNA gene. Seven rabbits had coinfections with two eimerian species. Sporulated oocysts of E. media had an obvious micropyle and were oval, measuring 24.92–30.03 (28.04) µm in length and 16.33–19.63 (18.31) µm in width. In contrast, E. stiedai were ellipsoid and measured 31.03–36.47 (33.79) µm in length and 18.21–20.93 (19.32) µm in width. The identity of the species of Eimeria parasites detected from the host (rabbits) was verified by the results of the sequences for the 18S rDNA gene. Both organisms (E. media and E. stiedai) grouped with rabbit eimerian parasites with distinct association with the group that contains oocysts residual body. Sequences from E. stiedai revealed insertions on two sites that had never been detected in E. stiedai sequences previously deposited in GenBank. The current parasite species are closely related to the previously described and deposited E. media and E. stiedai and are deeply embedded in the genus Eimeria (family Eimeriidae). This study emphasized the significance of combining taxonomy with morphological and genetic data in the identification of Eimeria species.
Keywords
Eimeria media
Eimeria stiedai
Rabbits
Saudi Arabia
1 Introduction
Coccidiosis is caused by obligate intracellular apicomplexan protozoan parasites of the genus Eimeria (family Eimeriidae), which is one of the most common parasitic disease in domestic animals such as rabbits and chickens (Petrova et al., 2022). Some researchers have previously claimed that domestic rabbits in Saudi Arabia had Eimeria species (Kasim and Al-Shawa, 1987; Toula and Ramadan, 1998; Bashtar et al., 2003; Shazly et al., 2005; Al-Mathal, 2008; Al-Quraishy, 2012; Dkhil et al., 2013; Abdel-Baki and Al-Quraishy, 2013). The most common way for the vulnerable host to become infected is through consuming sporulated oocyst in contaminated food or water (Hamid et al., 2019). Eimeria species go through a complex monoxenous life cycle after endogenous (intra-host) development (merogony and gamogony) and sporogony in the environment (Petrova et al., 2022).
In rabbits (Oryctolagus cuniculus), there are 15 recognized Eimeria species 14 species of which colonize the gastrointestinal tract (E. intestinalis, E. magna, E. piriformis, E. media, E. exigua, E. flavescens, E. coecicola, E. vejdovskyi, E. roobroucki, E. perforans, E. oryctolagi, E. nagpurensis, E. agnotsa, E. irresidua, and E. matsubayashi) while Eimeria stiedai is the 15th species inhabits the biliary ducts of the liver (Shil and Roy, 2021). Most eimerian species that infect rabbits affect production, depending on their pathogenicity, they may induce slower development, reduced feed conversion, and increased mortality rate (El-Shahawi et al., 2012). A normal fecal investigation usually reveals that a rabbit can have concurrent infections from multiple Eimeria species (Jithendran and Bhat, 1996).
According to several studies, morphological features, the time for sporulation, the affected site by infection, and signs of disease are the most frequently used criteria to identify Eimeria species (El-Shahawi et al., 2012). The various degrees of overlap among all these biological characteristics may occasionally make precise identification of Eimeria species problematic. The molecular characteristics of the Eimeria species have recently been determined using some genetic markers (Kvičerová et al., 2008). The 18S rDNA gene, one of the most prevalent loci, has been utilized to investigate the inter- and intra-species variation among Eimeria isolates, as well as their phylogenetic relationship (Ogedengbe et al., 2011). Furthermore, molecular identification approaches for Eimeria species from chickens and rabbits have been established using the internal transcribed spacer 1 (ITS-1) region (Su et al., 2003). Many apicomplexan parasites have the mitochondrial cytochrome oxidase (COI) gene locus, which has been extensively used for genotyping and species identification, including Eimeria species from turkey, ferret-badger and skink (Imai and Barta, 2019).
Even though rabbits are a significant animal protein source, there has been very little information on the parasitology of intestinal parasites (Atta et al., 1999; Abu-Akkada et al., 2010; Abdel-Gaber et al., 2019). In this investigation, rabbit-isolated Eimeria media and Eimeria stiedai were described morphologically and characterized molecularly using the partial 18S rDNA sequences.
2 Materials and methods
2.1 Animals collection
Ten domestic rabbits, Oryctolagus cuniculus, were collected from Riyadh’s local markets (Saudi Arabia). Each Rabbit was maintained separately in wire-floored batteries at optimal conditions (temperature and humidity) and fed commercial rabbit feed (without the use of reference anticoccidial drugs). The institution’s policies on the handling and use of animals in research (approval number KSU-SE-22–38) were followed in the care and rearing of the animals.
2.2 Parasitological examination
Using screens just below the cages, fresh fecal samples were taken, instantly excreted and then tested for Eimeria infection. Following the procedure outlined by Abdel-Baki and Al-Quraishy (2013), positive samples were collected and concentrated using the floatation technique. To allow for oocyst sporulation, the oocysts were collected and placed in 2.5% (w/v) potassium dichromate (K2Cr2O7) and incubated for 5–7 days at 25 ± 2 °C. The sporulated oocysts were stored at 4 °C until they were used after being repeatedly washed in phosphate-buffered saline (PBS).
2.3 Species identification
Using the keys previously mentioned by Catchpole and Norton (1979), the oocysts were identified based on their size (length/width), morphological features, and internal structures. A calibrated micrometer was used to measure about 50 sporulated oocysts. All measurements were taken in micrometers (µm) and were shown as a range with mean between brackets.
3 Molecular methods
3.1 DNA extraction
Purified oocysts were subjected to DNA extraction using the method of Zhao et al. (2001) which included lysis buffer and Cetyl–Trimethyl Ammonium Bromide (CTAB) buffer (consisting of 2% w/v CTAB, 1.4 M NaCl, 0.2% β-mercaptoethanol, 20 mM EDTA, 100 mM TRIS), with minor modification by using 1.3% N-Sodium Dodecyl Sulphate (SDS) versus 1.3% N-lauroylsarcosine. The isolated oocysts were then treated with sodium hypochlorite (5–6%), and incubated for 45 min at 65 °C in the lysis buffer. The mixture was then incubated for a further one hour at 65 °C with 350 μl of CTAB buffer. Following the manufacturer’s steps, sporulated oocysts were also used for DNA extraction using the Isolate II fecal DNA extraction kit (Meridian Bioscience, London, UK).
3.2 18S rDNA amplification and sequencing
The partial 18S rDNA gene was amplified by PCR using the primers F1E 5′-TACCCAATGAAAACAGTTT-3′ as a forward primer and a reverse primer, R2B 5′-CAGGAGAAGCCAAGGTAGG-3′ (Orlandi et al., 2003). The Polymerase Chain Reaction (PCR) was set up in 25 µl using Bioline Buffer (Bioline, London, UK), which contained 200 µM concentrations each of dNTPs. The reaction mixture was adjusted to a final concentration of 2 mM MgCl2 and 0.2 µM primer concentrations. The PCR mixture was supplemented with DNA Taq polymerase (Bioline). The PCRs were performed in a Multigene™ thermocycler (Labnet International, Inc., Edison, NJ, USA). As an initial denaturation cycle, the amplification program began with 2 min at 94 °C. The cycling conditions were denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s, and extension at 72 °C for 30 s. Before holding the PCR at 4 °C, a final extension at 72 °C for 5 min was permitted. The PCR products were separated by electrophoresis on a 1.5% agarose gel in 1 × Tris–boric acid–EDTA (TBE), stained with ethidium bromide and examined with a UV trans-illuminator. Macrogen’s sequencing facility (Seoul, South Korea) was used for the DNA sequencing.
3.3 Phylogenetic analysis
The National Center for Biotechnology Information (NCBI) was used to do a BLAST search to find the relevant sequences. The CLUSTAL-X multiple sequence alignment with default parameters was used to directly align the sequences. The alignment that resulted was manually edited with the help of the application BioEdit 7.2.5. The 18S rDNA locus, which is available in the GenBank database, was used to infer phylogenetic relationships between rabbit isolates from related Eimeria species. The collection comprised representative sequences from some related Eimeriidae found in GenBank. Maximum likelihood (ML) and Neighbour Joining (NJ) analyses were carried out to infer the relationships among the 18S rDNA dataset using MEGA X (Kumar et al., 2018). As an outgroup, Toxoplasma gondii (LC 749847.1) was added. 1000 replicates were used to assess the bootstrap support in ML tree constructs.
4 Results
Out of 10 rabbits examined, 7 were infected with mixed Eimeria spp., indicating a prevalence of 70%. Two morphologically distinct eimerian oocysts were detected in the fecal material, which was related to Eimeria media and Eimeria stiedai. E. media was the most predominant species, while infection with E. stiedai was less common. The oocysts of Eimeria species in the present study were illustrated in Figs. 1 and 2. Tables 1 and 2 summarize the morphological and morphometric characteristics of the Eimeria species. + present, - not detected. + present, - not detected.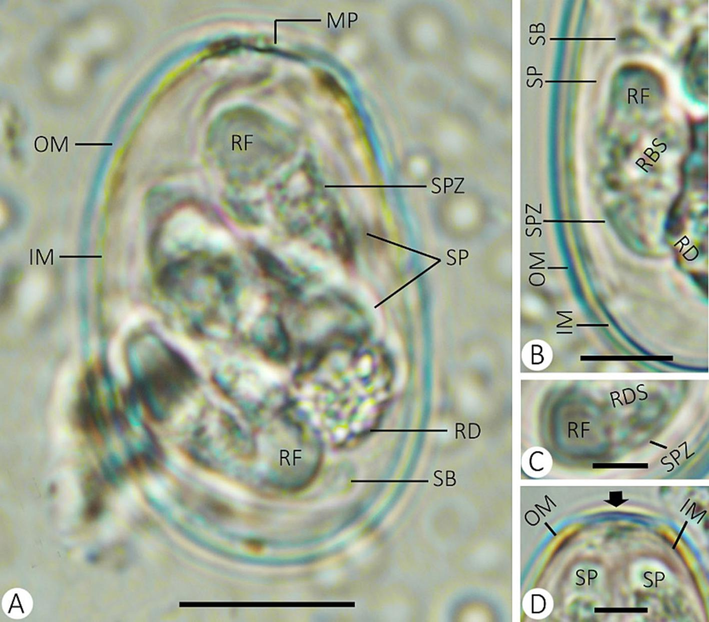
Eimeria media infecting rabbis. (A) sporulated oocyst. (B) sporocyst with sporozoites. (C) refractile body of sporozoite. (D) micropyle of the oocyst. (Note: MP, micropyle; OM, outer membrane; IM, inner membrane; RF, refractile body; RD, residuum; SB, stieda body; SP, sporocyst; RDS, residuum of sporocyst; SPZ, sporozoite) Scale = 10 µm.
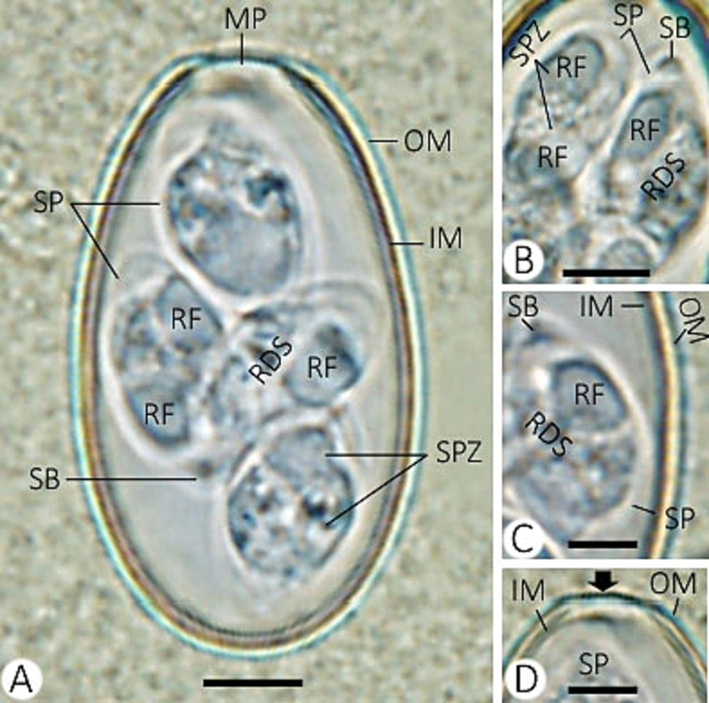
Eimeria stiedai infecting rabbis. (A) sporulated oocyst. (B) sporocysts with sporozoites. (C) refractile and stieda bodies of sporozoite. (D) micropyle of the oocyst. (Note: MP, micropyle; OM, outer membrane; IM, inner membrane; RF, refractile body; SB, stieda body; SP, sporocyst; RDS, residuum of sporocyst; SPZ, sporozoite) Scale = 10 µm.
Sources of E. media species
Shape
Oocyst size
Micropyle
Oocyst residuum
Sporocyst size
Locality
Length
Width
Length
Width
Kheyssin, 1967
–
18.5–33.3
13.3–21.3
–
–
6.6–14.6
5–7
–
Levine and Ivens, 1972
–
19–37
13–22
–
–
17.5
7
–
Pakandl, 1986
Ellipsoid (Type I)
24–33 (29.05)
15–20 (18.18)
+
+
14–17 (15.79)
6–8 (6.67)
Ceské Budĕjovice
Pakandl, 1986
Ellipsoid (Type II)
30–37 (32.88)
17–21 (19.19)
+
+
14–16.5 (15.71)
7–9 (8)
Ceské Budĕjovice
Kasim and Al Shawa, 1987
Ellipsoid-ovoid
25.5–34 (30)
15–22 (18.7)
+
+
9–16 (12.9)
5–8.5 (6.6)
Saudi Arabia
Li et al., 2010
–
21.8–34.6 (29.3)
16.0–21.3 (18.3)
+
+
8.0–17.3 (13.8)
5.3–10.6 (7.6)
Taiwan
El-Shahawy et al., 2012
Ellipsoid-ovoid
19–24 (22.3)
10–15 (12.1)
+
+
7–9 (8.2)
4–6 (4.5)
Egypt
Abdel-Baki and Al Quraishy, 2013
Ellipsoid
19–24 (22)
10–15 (12)
+
+
7–9 (8)
4–6 (4.5)
Saudi Arabia
El-Shahawy and Elgoniemy, 2018
Ellipsoid-ovoid
28.64
16.70
+
+
12.17
7.34
Egypt
El-Sayed et al., 2020
Ellipsoid-ovoid
27.48
17.79
+
+
10.60
6.42
Egypt
Rabie et al., 2022
Ellipsoid
25.34–29.4 (27.44)
16.36–22.11 (18.61)
+
+
11.22–15.88 (13.72)
6.5–8.01 (6.95)
Egypt
Present study
Ellipsoid
24.92–30.03 (28.04)
16.33–19.63 (18.31)
+
+
10.22–15.18 (13.09)
4.34–6.57 (5.47)
Saudi Arabia
Sources of E. stiedai species
Shape
Oocyst size
Micropyle
Oocyst residuum
Sporocyst size
Locality
Length
Width
Length
Width
Aoutil et al., 2005
Ellipsoid
33–40 (37)
19–24 (22)
+
–
18
10
France
Al-Mathal, 2008
Oval
30
10–20
–
–
10
–
Saudi Arabia
El-Shahawy et al., 2012
Ellipsoid
24–29 (26.5)
11–15 (13.1)
+
+
9–11 (9.7)
5–6 (5.6)
Egypt
Abdel-Baki and Al-Quraishy, 2013
Ellipsoid
25–29 (26)
12–15 (13)
+
+
9–11 (10)
5–7 (6)
Saudi Arabia
Ütük et al., 2015
–
34–38 (36.1)
17–21 (19.7)
+
–
12–17 (15.0)
8–10 (8.9)
Turkey
Rabie et al., 2022
Ovoid
27.65–28.44 (28.13)
16.55–17.66 (16.99)
+
–
10.82–11.18 (11.01)
5.67–6.03 (5.89)
Egypt
Present study
Ovoid
31.03–36.47 (33.79)
18.21–20.93 (19.32)
+
–
11.18–15.30 (13.38)
5.68–8.60 (6.94)
Saudi Arabia
4.1 Description of Eimeria media (Fig. 1 and Table 1)
Sporulated oocysts are ellipsoid and measured 24.92–30.03 (28.04) µm in length and 16.33–19.63 (18.31) µm in width, with a noticeable micropyle. The oocyst index (length/width) was 1.53. The wall of the oocyst is double-layered, with a smooth outer layer, somewhat thickened, forming a small ridge at the micropyle, and a membranous inner layer. The oocyst residuum is rounded, and the polar granule is absent. Each oocyst contained four dizoic sporocysts with a pointed end, measuring 10.22–15.18 (13.09) µm in length and 4.34–6.57 (5.47) µm in width. They were ellipsoid and surrounded by a single-layer sporocyst wall. The stieda body and residuum were present. The sporocyst index (length/width) was 2.39. One refractile body is located at the wider end of each sporozoite.
4.2 Description of Eimeria stiedai (Fig. 2 and Table 2)
Sporulated oocysts are ovoid and measured 31.03–36.47 (33.79) µm in length and 18.21–20.93 (19.32) µm in width, with an observable micropyle. Absence of both oocyst residuum and the polar granule. There are four sporocysts (each with two elongated sporozoites). Each sporocyst is ovoid and measured 11.18–15.30 (13.38) µm in length and 5.68–8.60 (6.94) µm in width. The stieda body is present. Absence of both substieda and parastieda bodies. Granular sporocyst residuum present.
4.3 Molecular analysis
A PCR product of ∼636 bp was successfully amplified and sequenced from the 18S rDNA gene of eimerian isolates. PCR products from two isolates of E. stiedai produced identical sequences for the 18S rDNA whereas one isolate from E. media was amplified and sequenced. Sequences of E. stiedai were given GenBank accession numbers OQ704326 and OQ704327 and sequence from E. media was given GenBank accession number OQ704328. The final alignment of 611 bp of the 18S rDNA was used for the phylogenetic analysis which included 24 sequences with Toxoplasma gondii as an outgroup. Phylogenetic analyses showed two distinct clades for Eimeria species infecting rabbits (Fig. 3). Both E. stiedai and E. media detected in this study clustered with the clade which contained E. intestinalis, E. media, E. perforans, E. coecicola, E. vejdovskyi, and E. stiedai. Another clade grouped other rabbit eimerian parasites including E. exigua and E. piriformis. However, within this clade, E. stiedai formed a subclade that distinguished E. stiedai. There are only two 18S rDNA sequences related to E. stiedai in GenBank (EF694008 and HQ173837) and both were from the Czech Republic. Both sequences were homologous to the recovered sequences. However, the recovered sequences showed significant differences from those other sequences of E. stiedai. Two insertions of 4 and 6 nucleotides at positions 271 and 305 respectively were noted in sequences obtained in the present study. The insertion at position 271 (3 bases) was found in the sequences EF694008 whereas it was not found in sequences HQ173837. The other insertion of 6 bases was missing in sequences HQ173837 and only three bases were missing in sequences EF694008. The sequence of E. media (OQ704328) grouped was homologous to the sequence from E. media (HQ173834) from France with two mutations at positions 174 and 487 of the alignment and both changes transversions T/A and T/G respectively.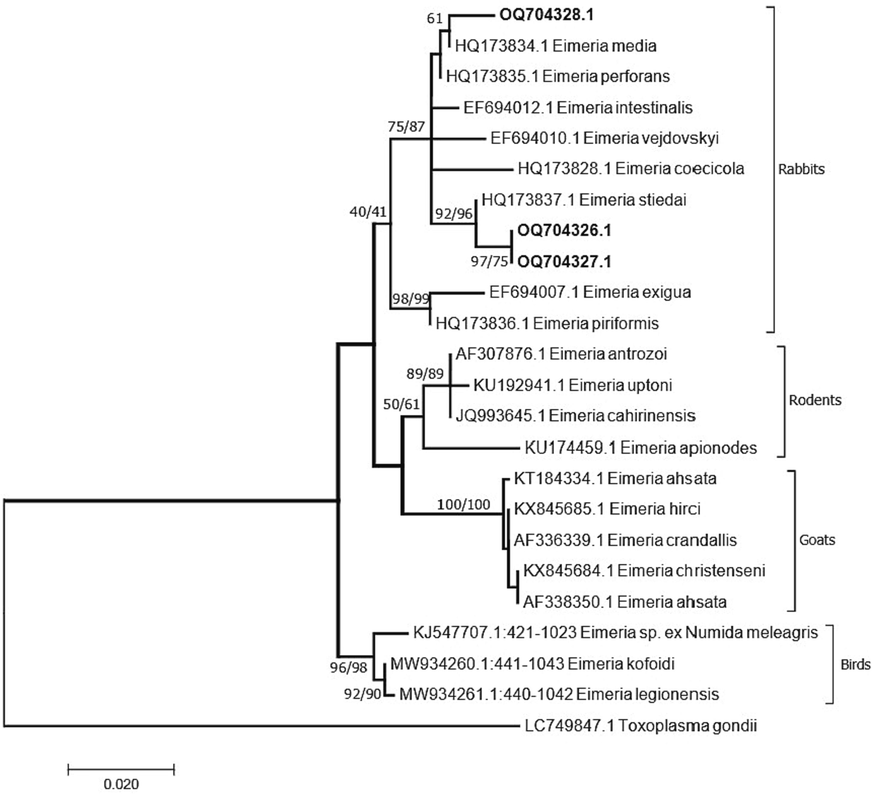
A consensus phylogenetic tree constructed with maximum likelihood (ML) and Neigbour Joining (NJ) methods, showing phylogenetic relationships among Eimeria media and E. stiedai and other eimerian coccidia, with Toxoplasma gondii (Nicolle et Manceaux, 1908), as an outgroup, inferred from the 18S rDNA sequence data generated from the two eimerian species from rabbits (OQ704326-704328, given in bold) and other related taxa from GenBank. Numbers indicated at branch nodes are bootstrap values (ML/NJ). Only bootstraps > 40% are shown.
5 Discussion
Due to the related morbidity and mortality, parasitic infections pose a serious effect on human health as well as a detrimental effect on livestock productivity. The prevalence of parasitic diseases specifically has a detrimental impact on rabbit production and causes a large economic loss (Abdel-Baki and Al-Quraishy, 2013). Therefore, gaining an awareness of intestinal parasite species can help with minimizing the economic rabbit productivity, determining the infection risk, and developing management strategies. According to the available literature, Saudi Arabia has conducted very little research on Eimeria of rabbits (Kasim and Al-Shawa, 1987; Al-Mathal, 2008; Abdel-Baki and Al-Quraishy, 2013). In this study, we attempted to integrate morphological and genetic information on the rabbit-infecting Eimeria species.
In the current study, a significant number of rabbits (70%) had an Eimeria species infection. In earlier studies on rabbits, similar trends were documented (Li et al., 2010; Abdel-Baki and Al-Quraishy, 2013). Herein, mixed species of Eimeria infections were found, which is consistent with reports of El-Shahawy and Elgoniemy (2018) and Rabie et al. (2022) that single Eimeria species infections naturally occur infrequently. This may be the case since some parasite species may be found in food. Eimeria species are distinguished by their oocyst and sporocyst size, morphological features, prepatent duration, and colonization site (El-Sayed et al., 2020). In the feces of the rabbits obtained from Saudi Arabia for the current study, two species of Eimeria, E. media, and E. stiedai, were identified according to these criteria.
The descriptions of the sporulated E. media oocysts are basically in line with what Rabie et al. (2022) reported in their earlier work. These species do, however, differ slightly from earlier descriptions in terms of sporocyst and oocyst size as well as a few other negligible characteristics. Here are a few minor observations: (i) compare to previous studies by El-Shahawi et al. (2012) and Abdel-Baki and Al-Quraishy (2013), the oocysts and sporocysts of E. media in the present study are larger. (ii) the oocyst shape was ellipsoid, except for those identified as ovoidal to ellipsoidal by Kasim and Al-Shawa (1987), El-Shahawi et al. (2012), El-Shahawy and Elgoniemy (2018), and El-Sayed et al. (2020). (iii) The specimens described by Kheyssin (1967) and Levine and Ivens (1972) lacked micropyle and oocyst residuum of E. media.
Our study’s E. stiedai sporulated oocysts are generally consistent with those from Ütük et al. (2015). The following findings can be drawn when contrasting this Eimeria species to other species that have been described: (i) Compared to previous studies by El-Shahawy et al. (2012), Abdel-Baki and Al-Quraishy (2013) and Rabie et al. (2022), the oocysts and sporocysts of E. stiedai in the present study are larger. (ii) All oocysts were ovoid, with the exception of those mentioned as ellipsoid oocysts by Aoutil et al. (2005), El-Shahawy et al. (2012), and Abdel-Baki and Al-Quraishy (2013). (iii) The specimens described by Al-Mathal (2008) lacked the micropyle of E. stiedai. (iv) Other than in El-Shahawy et al. (2012), and Abdel-Baki and Al-Quraishy (2013), the oocyst residuum of E. stiedai was lacking.
Oocyst morphology can be used to distinguish between Eimeria species, however, it is time-consuming and labor-intensive. At the species level, it is extremely difficult to identify, especially in mixed infections where there aren’t enough oocysts of a particular species. The proper identification of Eimeria species has recently benefited from molecular methods (Ütük et al., 2015; Al Quraishy et al., 2022).
Molecularly, based on the 18S rDNA analysis, sequences from both oocysts detected in this study clustered with the clade of eimerian parasites infecting rabbits. However, two distinct clades were found within eimerian species of rabbits. The first one included E. intestinalis, E. perforans, E. media, E. coecicola, E. vejdovskyi, and E. stiedai whereas the other one included E. exigua and E. piriformis. The first group of eimerian parasites included those which possess oocyst residual body whereas the other group included those without oocyst residual body. This finding coincided with the results outlined by Kvičerová et al. (2008). Within those included in the clade with oocysts residual body, E. stiedai formed a distinct subclade that distinguished it from all other species in the group. It is likely this because E. stiedai inhabits extraintestinal locations where it occurs in the bile ducts, furthermore, it has low host specificity, unlike other eimerian parasites from rabbits which have been reported from the hare (Scholtyseck et al., 1979). Molecular characterization confirmed the identity of the oocysts detected in the present study. However, there was wide variation in the lengths of the size of E. media oocysts ranging from 18 to 37 µm in length and 10–22 µm in width. Molecular data have confirmed the identity of the oocysts detected herein, therefore, it is very important to have the molecular data alongside morphological data to identify eimerian parasites adequately.
Ethical approval
This research was approved by the Research Ethics Committee (REC) at King Saud University (approval number KSU-SU-22–38).
Acknowledgement
This study was supported by the Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2023R437), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia; as well as Researchers Supporting Project (RSP2023R94), King Saud University, Riyadh, Saudi Arabia.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Prevalence of coccidia (Eimeria spp.) infection in domestic rabbits, Oryctolagus cuniculus, in Riyadh, Saudi Arabia. Pakistan J. Zool.. 2013;45(5):1329
- [Google Scholar]
- Prevalence, morphological and molecular phylogenetic analyses of the rabbit pinworm, Passalurus ambiguus Rudolphi 1819, in the domestic rabbits Oryctolagus cuniculus. Acta Parasitol.. 2019;64:316-330.
- [Google Scholar]
- Garlic and hepatic coccidiosis: prophylaxis or treatment? Tropical Animal Health Production. 2010;42(7):1337-1343.
- [Google Scholar]
- Morphological and molecular approaches for identification of murine Eimeria papillata infection. J. King Saud Univ. – Sci.. 2022;34
- [Google Scholar]
- Hepatic coccidiosis of the domestic rabbit Oryctolagus cuniculus domesticus L. in Saudi Arabia. World J. Zool.. 2008;3(1):30-35.
- [Google Scholar]
- Exogenous and endogenous stages of Eimeria perforans naturally infected domestic rabbit (Oryctolagus cuniculus) in Saudi Arabia: light microscopic study. Saudi J. Biol. Sci.. 2012;19:31-34.
- [Google Scholar]
- Eimeria (Coccidia: Eimeridea) of hares in France: description of New Taxa. Parasite. 2005;12:131-144.
- [Google Scholar]
- Tissue residues of some sulphonamides in normal and Eimeria stiedai infected rabbits. Dtsch. Tierarztl. Wochenschr.. 1999;106:295-298.
- [Google Scholar]
- Coccidiosis in rabbits from Saudi Arabia. 1- Exogenous stages of different Eimeria spp. J. Egypt. Ger. Soc. Zool.. 2003;42D:1-10.
- [Google Scholar]
- The species of Eimeria in rabbits for meat production in Britain. Parasitology. 1979;79(2):249-257.
- [Google Scholar]
- Eimeria coecicola: spleen response of Oryctolagus cuniculus. Exp. Parasitol.. 2013;133(2):137-143.
- [Google Scholar]
- Prevalence and morphological identification of Eimeria spp. in domestic rabbit (Oryctolagus cuniculus) in Sharkia province, Egypt. Egyptian Vet. Med. Soc. Parasitol. J.. 2020;16:114-130.
- [Google Scholar]
- Coccidiosis of domestic rabbit (Oryctolagus cuniculus) in Egypt: light microscopic study. Parasitol. Res.. 2012;110(1):251-258.
- [Google Scholar]
- Coccidiosis of domestic rabbit (Oryctolagus cuniculus) in Egypt: light microscopic study. Parasitol. Res.. 2012;110:251-258.
- [Google Scholar]
- An epidemiological study on endoparasites of domestic rabbits (Oryctolagus cuniculus) in Egypt with special reference to their health impact. Sains Malaysiana. 2018;47(1):9-18.
- [Google Scholar]
- Intestinal and hepatic coccidiosis among rabbits in Yogyakarta, Indonesia. Vet. World. 2019;12(8):1256-1260.
- [Google Scholar]
- Distribution and abundance of Eimeria species in commercial turkey flocks across Canada. Can. Vet. J.. 2019;60(2):153-159.
- [Google Scholar]
- Subclinical coccidiosis in angora rabbits, a field survey in Himachal Pradesh, India. World Rabbit Sci.. 1996;4(1):29-32.
- [Google Scholar]
- Coccidia in rabbits (Oryctolagus cuniculus) in Saudi Arabia. Int. J. Parasitol.. 1987;17:941-944.
- [Google Scholar]
- Life cycles of coccidia of domestic animals. Leningard: Publ House Nauka; 1967. p. :192.
- Phylogenetic relationships among Eimeria spp. (Apicomplexa, Eimeriidae) infecting rabbits: evolutionary significance of biological and morphological features. Parasitology. 2008;135(4):443-452.
- [Google Scholar]
- Prevalence, infectivity and oocyst sporulation time of rabbit-coccidia in Taiwan. Trop. Biomed.. 2010;27(3):424-429.
- [Google Scholar]
- DNA barcoding identifies Eimeria species and contributes to the phylogenetics of coccidian parasites (Eimeriorina, Apicomplexa, Alveolata) Int. J. Parasitol.. 2011;41:843-850.
- [Google Scholar]
- Targeting single-nucleotide polymorphisms in the 18S rRNA gene to differentiate Cyclospora species from Eimeria species by multiplex PCR. Appl. Environ. Microbiol.. 2003;69(8):4806-4813.
- [Google Scholar]
- Two morphological types of oocyst of rabbit coccidia Eimeria media Kessel, 1929. Folia Parasitol.. 1986;33:297-300.
- [Google Scholar]
- Biochemical and pathomorphological investigations on rabbits with experimentally induced hepatic eimeriosis. Macedonian Vet. Rev.. 2022;45(1):53-59.
- [Google Scholar]
- Occurrence of Eimeria species (Apicomplexa: Eimeriidae) in domestic rabbits (Oryctolagus cuniculus) in Qena Governorate, Upper Egypt. J. Parasit Dis.. 2022;46(3):811-832.
- [Google Scholar]
- Transmission of Eimeria stiedai from the rabbit (Oryctolagus cuniculus) to the hare (Lepus europaeus) Acta Veterinaria Acad. Sci. Hungary. 1979;27:365-373.
- [Google Scholar]
- Light and electron microscopy of Eimeria magna infecting the house rabbit, Oryctolagus cuniculus from Saudi Arabia. I. Asexual developmental cycles. Saudi J. Biol. Sci.. 2005;12:1-10.
- [Google Scholar]
- Coccidiosis infection in rabbits and its control. Indian Farmer. 2021;8(3):278-283.
- [Google Scholar]
- Differential diagnosis of five avian Eimeria species by polymerase chain reaction using primers derived from the internal transcribed spacer 1 (ITS-1) sequence. Vet. Parasitol.. 2003;117(3):221-227.
- [Google Scholar]
- Studies on coccidian species of Genus Eimeria from domestic rabbit (Oryctolagus cuniculus domesticus L.) in Jeddah, Saudi Arabia. J. Egypt Soc. Parasitol.. 1998;28:691-698.
- [Google Scholar]
- Molecular detection of Eimeria stiedae in an Angora rabbit. Etlik Veteriner Mikrobiyoloji Dergisi. 2015;26(2):41-44.
- [Google Scholar]
- A simple method of DNA extraction for Eimeria species. J. Microbiol. Methods. 2001;44:131-137.
- [Google Scholar]







