Translate this page into:
Molecular phylogenetic investigation of toads based on sequencing of the mitochondrial 16S ribosomal RNA gene
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Amphibians inhabit a wide variety of habitats and environments, such as arboreal, terrestrial and freshwater aquatic ecosystems, and are considered among the vertebrate classes to exhibit one of the most diverse distributions. Their life histories, behaviour and diversity play vital roles in assessing environmental and evolutionary theories. The mitochondrial 16S rRNA gene sequence analysis has been widely used in molecular taxonomy and is frequently used in systematic analyses of families and genera. This study examines taxonomic relationships among 18 specimens of the Arabian toad (Sclerophrys arabica) and the Dhofar toad (Bufo dhufarensis) from Al Madinah Province, Saudi Arabia by estimating phylogenetic relationships using the mitochondrial 16S ribosomal RNA gene (16S rRNA). Results indicate the presence of four new lineages of toads among the samples in this study. The generated phylogenetic tree grouped all samples of Arabian toad in one clade, while the Dhofar toad samples were divided into three subclades. This is the first study to be reported the genetic diversity of Al Madinah Province toads.
Keywords
Toads
16S rRNA
Dhofar toad
Arabian toad
Phylogenetic tree
PCR
1 Introduction
The Arabian Peninsula hosts a unique array of animal and plant species because of the region's dynamic geologic history and changing climate. Peninsular flora and fauna range from limited temperate endemics to disjunct expansions of the Horn of Africa and Eastern Afromontane hotspots (Mittermeier et al., 1999, Myers et al., 2000). The Arabian species is a combination of South Asian, Western Eurasian, and African components, a pattern illustrated even in the comparatively depauperate Arabian bufonids (Portik and Papenfuss, 2015). Portik and Papenfuss (2015) report that South Asian and Western Eurasian lineages are independently derived from continental Africa. Their data suggest that Amietophrynus arabicus and Amietophrynus tihamicus did not colonize the Arabian Peninsula via overwater dispersal or through the southern land bridge from the Horn of Africa. Rather, it is speculated that formation of the Red Sea drove simultaneous divergences in these species; thus, their current distribution in the Arabian Peninsula represents authentic African relics.
Amphibians are regarded as one of the vertebrate classes demonstrating the greatest distributional variety and widest range of ecological environments. Because of their variety of biological features, convergences in form, and diversity of environments they live in, which can require fossorial, terrestrial, or arboreal lifestyles, amphibians are an attractive taxon for researching evolutionary processes (Bossuyt and Milinkovitch, 2000, Wells, 2010). Amphibians have a vast range of reproductive phases, ranging from biphasic life cycles, with larval and adult forms separated by metamorphosis, to systems in which either the larval or adult stage is completely absent, and it can be said that amphibians are the most diversified group of terrestrial vertebrates in terms of reproductive diversity (Wells, 2010).
Saudi Arabia's geographical location between temperate and tropical areas, as well as fluctuations in climatic conditions, make the coastal and riverine oases and wetlands of Saudi Arabia habitat to seven amphibian species of frogs and toads. Four of these are endemic to the Arabian peninsula (Sclerophrys tihamica), Arabian five-fingered frog (Euphlyctis ehrenbergii), Arabian toad (Bufo arabicus) and Dhofar toad (Bufo dhurefencis). The three species also found elsewhere in the world are the Middle East tree frog (Hyla savignyi), marsh frog (Pelophylax ridibundus) and European greek toad (Bufo viradid) (Balletto, 1985). Amphibians in Saudi Arabia have no significant obstacles as a result of their adaptation to a diverse variety of habitats and changed habitat, and the Arabian toad is considered one of the most widely distributed toad species in the Arabian Peninsula (Al-Obaid et al., 2017, Hafez et al., 2017).
Avise (2000) reports that phylogeographic analysis is a strong method for examining the evolutionary history of a species, providing valuable insight into the genesis of current biogeographic and systematic patterns. Molecular phylogenetic tree is used to identify closely related species sequences in order to recognize genes, and non-coding RNAs in newly sequenced genomes in order to explain present and ancient individual genomes, as well as to refine phylogenetic trees (Kellis et al., 2003, Pedersen et al., 2006). The 16S rRNA gene is now globally recognised in the phylogeny of organisms as a suitable, highly conserved and reliable molecular marker that develops slowly and is functionally preserved (Nakahara et al., 2004, EARDLY and Van Berkum, 2005, Pratihar et al., 2016, Hedges et al., 1993, Yamaguchi et al., 2019).
Some molecular studies have been conducted on toads in Saudi Arabia, but these are few and do not include most regions of Saudi Arabia (Hafez et al., 2017, Al-Qahtani and Amer, 2019). The present study aimed to identify the taxonomic status of the Arabian toad (Sclerophrys arabica) and the Dhofar toad (Bufo dhufarensis). Samples were collected from Wadi Abather in the Al Madinah Province were utilized to compare genetic variation to other taxa, for which nucleotides of the mitochondrial 16S rRNA have previously been sequenced.
2 Materials and methods
Eighteen samples of Arabian and Dhofar toads were collected from Wadi Abather, Yanbu Al-Nakhal, a city in the Al Madinah Province of western Saudi Arabia (24°30′17.0″N 38°37′01.0″E). All individual samples were collected in January 2021 (Table 1) using the toe-clipping method described by Sambrook (1989). Samples were preserved in ethanol before being sent to a laboratory at King Saud University (KSU) and subsequently kept at −20 °C for future examination. DNAzol® (Invitrogen, UK) was used to extract DNA from the tissue samples (tissue was first chopped into small pieces) according to manufacturer instructions. A hand-held glass/Teflon homogenizer was used to homogenize tissue samples. After 40 mg of homogeneous tissue was transferred to a 1.5 mL Eppendorf® tube, 500 µL of DNAzol has been added to each tube and vortexed thoroughly to lyse the cells. Samples were centrifuged for 5 min at 9000g at 4 °C, and the supernatants were collected and transferred to new Eppendorf tubes; 5 mL of absolute ethanol was then added to each tube. Mixtures were mixed for 10 s on a vibrator machine, followed by 8 min of centrifugation at 4 °C at 9,000 g. The liquid was then removed and the DNA pellet washed with 2 mL of 75 % ethanol. DNA pellets were air-dried for 20 min and then resuspended in 1 mL of nuclease-free water (Alrefaei et al., 2022). The extracted DNA was stored at −20 °C. The concentration of DNA in each sample was determined using spectrophotometry based on an absorbance reading at 260 nm. The 16S rRNA was amplified by PCR using the primers 16SL (5′CGCCTGTTTATCAAAACAT-3′) and 16SH (5′CCGGTCTGAAC TCAGATCACG-3′), as published by Palumbi et al. (1991), with the following modifications: to make a 24 µL reaction mix, 10 µL Green Master Mix (2X; Thermo Fisher Scientific, Waltham, MA), 3 µL of 10 pg/µL forward and reverse primer solutions (Macrogen, Seoul, Korea), 7 µL of nuclease-free water, and 1 µL of extracted DNA were added. A negative control was run during each PCR. The following cycling conditions were used for PCR amplification: 94 °C for 4 min, then 40 cycles of heat denaturation at 94 °C for 1 min, followed by primer annealing at 52 °C for 1 min, then a DNA elongation step of 72 °C for 1 min, and then a final extension for 5 min at 72 °C. A 1 % agarose gel was used for each PCR amplification. Amplification was visually confirmed using UV illumination, and all positive PCR products were sent to Macrogen Inc. for sequencing (Seoul, Republic of Korea). The evolutionary relationships between the sequences of species in this study were examined, and a phylogenetic tree was created using the software tool MEGA, Version 11 (Kumar et al., 2018). All samples in this study were subjected to PCR to acquire 16S rRNA in both directions. MEGA software was used to align all 16S rRNA sequence data using the forward and reverse complements of the reverse primer. All sequences acquired in this study were submitted to GenBank, and other samples from various species utilised for phylogeny were downloaded from GenBank. The phylogenetic trees from GenBank and those generated in this study were constructed independently using maximum-likelihood (ML), genetic distance, and the Tamura-Nei models, and were used to analyze the relationships between species using nucleotide sequence analysis (Kumar et al., 2018). The related taxa grouped in the bootstrap test (1000 repetitions) are presented adjacent to the branches using Felsenstein's bootstrap approach. The final data set includes 460 bp of PCR product from all samples. Estimates of evolutionary divergence between sequences were determined using MEGA X (version 10.2.6) as a genetic distance matrix (Kumar et al., 2018). The CLUSTVIS web tool was used to create a genetic distance heatmap (Metsalu and Vilo, 2015).
ID
Sample No.
Species
GenBank Accession No.
1
1SA
Arabian toad Sclerophrys arabica
OL639235
2
2SA
Arabian toad Sclerophrys arabica
OL639236
3
3SA
Arabian toad Sclerophrys arabica
OL639237
4
4SA
Arabian toad Sclerophrys arabica
OL639238
5
5SA
Arabian toad Sclerophrys arabica
OL639239
6
6SA
Arabian toad Sclerophrys arabica
OL639240
7
7SA
Arabian toad Sclerophrys arabica
OL639241
8
8SA
Arabian toad Sclerophrys arabica
OL639242
9
1BD
Dhofar toad Bufo dhufarensis
OL636499
10
2BD
Dhofar toad Bufo dhufarensis
OL636500
11
3BD
Dhofar toad Bufo dhufarensis
OL636501
12
4BD
Dhofar toad Bufo dhufarensis
OL636502
13
5BD
Dhofar toad Bufo dhufarensis
OL636503
14
6BD
Dhofar toad Bufo dhufarensis
OL636504
15
7BD
Dhofar toad Bufo dhufarensis
OL636505
16
8BD
Dhofar toad Bufo dhufarensis
OL636506
17
9BD
Dhofar toad Bufo dhufarensis
OL636507
18
10BD
Dhofar toad Bufo dhufarensis
OL636508
3 Results
Eighteen samples, obtained from eight specimens of the Arabian toad and ten specimens of the Dhofar toad and genetically characterised using 16S rRNA gene amplification, and were deposited in the GenBank database under the following accession numbers: OL639235- OL639242 and OL636499- OL636508 (Table 1). A maximum-likelihood phylogenetic tree was constructed using the 16S rRNA gene fragment with a 460-nucleotide sequence. Results of the ML tree clearly grouped the species sequences in separated clusters with high bootstrap values. Four new lineages were identified among these samples (Fig. 1). This is corroborated by the Genetic Distance Matrix (Table 2 and Fig. 2). The ML tree split the Arabian toad, with only one lineage of all samples that were obtained in this study, whereas sequencing analysis of the Dhofar toad demonstrated three lineages in a clear monophyletic group (Fig. 1).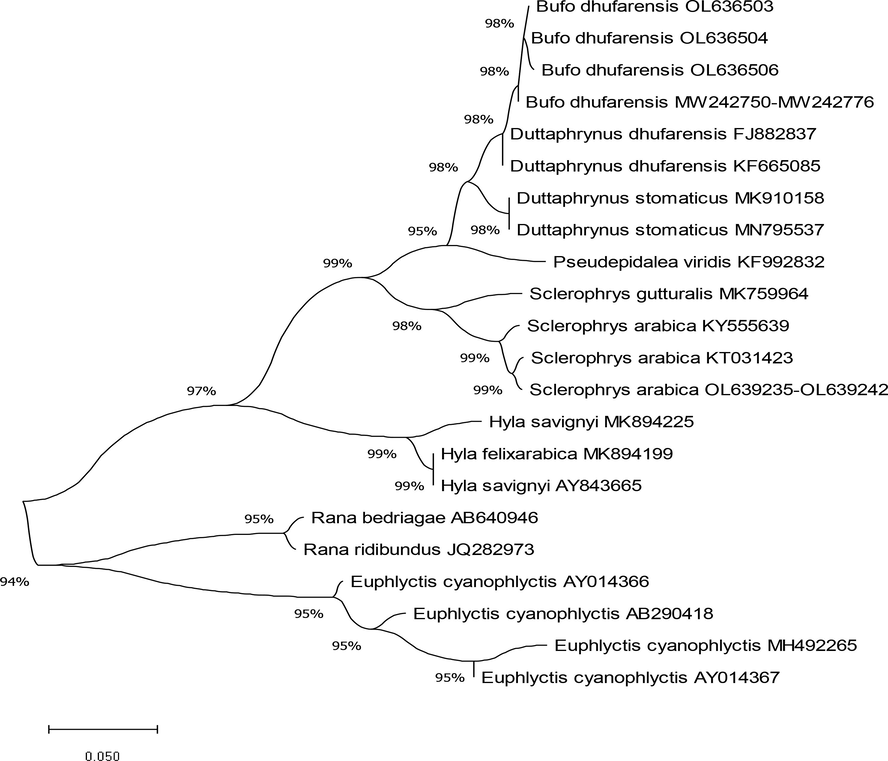
Phylogenetic tree created using the Maximum Likelihood method and Tamura-Nei model (Tamura and Nei, 1993), based on the 16S rRNA gene, indicating the relationships of Arabian toad and Dhofar toad to other toad and frog species. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The NCBI GenBank accession numbers for all sequences are written after each species name. This analysis involved 22 nucleotide sequences. Evolutionary analyses were conducted in MEGA X (Kumar et al., 2018).
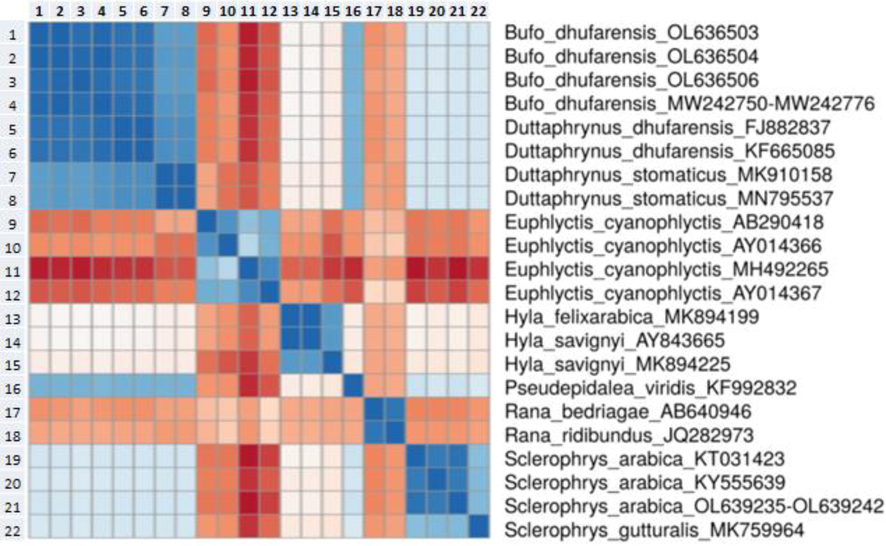
Genetic distance heatmap. The heatmap plot was constructed with genetic distance data (see Table 2) using CLUSTVIS web tool (Metsalu and Vilo, 2015).
Phylogenetic analysis placed all the Arabian toad sequencing in one clade as a sister taxon with high support of the Arabian toad (GenBank: KT031423) (Portik and Papenfuss, 2015), with 99.21 % identity. Inside this clade, however, a subclade which grouped as being sister to the African common toad, Sclerophrys gutturalis, was observed (GenBank: MK759964) (Telford et al., 2019) (Fig. 1). Furthermore, genetic distance matrix showed a value of 0.1205 of base substitutions per site between Arabian toad species (Fig. 2 and Table 2).
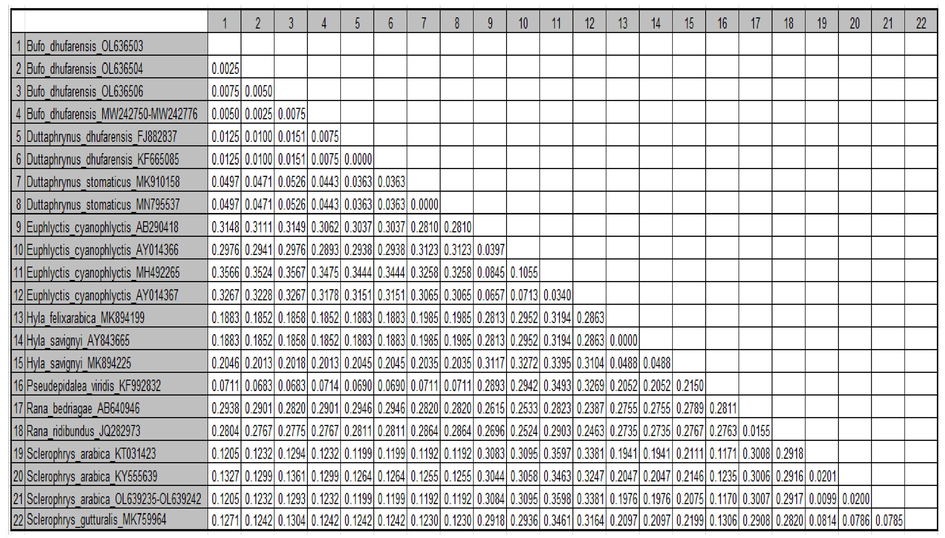
Dhofar toad sequencing was divided into three lineages as monophyletic with strong support. These sequences were demonstrated to be sister taxa of the Riyadh Province species Dhofar toad (GenBank: MW242776), and were also grouped in the same clade as Oman species (GenBank: KF665085 and FJ882837) (Liedtke et al., 2016, Jayawardena et al., 2017), with 99.35 % identity (Fig. 1), and all of these also were supported by the low value of genetic distance in assessing evolutionary divergence (Fig. 2 and Table 2). Notably, all of these sequences are unique in Saudi Arabia toads and are being reported for the first time in this study.
4 Discussion
The analysis of 16S ribosomal RNA sequences to establish phylogenetic relationships is a powerful tool used for categorizing various animal species. This study successfully determined the taxonomic status of the Arabian and Dhofar toad in Wadi Abather, Al Madinah Province, in Saudi Arabia by 16S rRNA. The phylogenetic tree and genetic distance heap maps produced confirmed the existence of four new toad lineages among the samples collected from the study area. This experimental approach is significant that it determines the phylogenetic relationship and differences between toad species. With this finding is crucial for providing a better understanding of the genetic variation of toads, as well as for identifying genetic resources of toads, as genetic studies of toads in Saudi Arabia and neighbouring countries are limited.
The 16S rRNA gene has been sequenced and examined extensively and is frequently utilised in systematic investigations of families and genera, as it contains variable regions where conserved areas reflect phylogenetic relationships among species (Fouquet et al., 2007, Smith et al., 2008, Palumbi, 1996, Dufresnes et al., 2019). It has been widely used to determine relationships among amphibian genera, and it has been found that variation within this region is low within species (Harris, 2001). Both Hedges et al. (1993) and Pratihar et al. (2016) found a region of the mitochondrial 16S rRNA gene that is beneficial for revealing elements of amphibian phylogeny and represents an important genetic marker.
Phylogenetic analysis, supported by high bootstrap values, clearly showed that all sequencing obtained from Arabian toads was identical and able to be grouped into one genetic lineage. Moreover, low values in the Genetic Distance Matrix also support the idea of unique genetic lineage for Arabian toads (Table 2 and Fig. 2). This result indicates that the Arabian toad taxa examined in this study were clustered with the Arabian toad (GenBank: KT031423) (Portik and Papenfuss, 2015). The Arabian toad, a common amphibian in Saudi Arabia, showed no genetic variation across this region when using 16S rRNA gene. The resultant trees further indicated that the Dhofar toad was divided into three clades and that ancestors of the clade consisted of the same species from the Riyadh Province (GenBank: MW242776). The genetic variation and relationships among Dhofar toad species are high and show evidence of considerable gene flow, with other species from Oman and the Riyadh Province sharing haplotypes.
Alrefaei et al. (2022), reported that sequence analyses of 27 specimens of Dhofar toad obtained from three different regions in Riyadh Province, Saudi Arabia showed that all the samples collected in this study showed identical sequences depending on the 16S rRNA. However, the results of this study indicate that there are three new lineages of toads among the samples that were collected from study area.
Yang and Rannala (2010) report that phylogenetic analysis employs molecular techniques to identify and analyse the connections among closely related species in systematics and taxonomy. The current study provides a phylogenetic outline of Bufo species found in Saudi Arabia. There are few investigations of genetic variation of these species in Saudi Arabia and thus a paucity of available molecular data. Therefore, additional samples of toad species from around Saudi Arabia are necessary to assess genetic diversity and relationships between individuals of these species.
5 Conclusions
To our knowledge, this study is the first investigated of the relationships of toads that were collected from Wadi Abather, Al Madinah Province, in Saudi Arabia using a DNA-based phylogenetic approach. According to the above-mentioned findings that the mitochondrial 16S rRNA gene support the existing taxonomy based on morphological characteristics, though there are some discrepancies which require further investigation. This work will be valuable for researchers interested in the evolutionary history of toads alike as well as providing an important molecular toolbox for the amphibians ecology community.
Ethical approval
All procedures for the samples collection were carried out in strict accordance with the recommendations by the Research Ethics Sub-Committee (REC) of the College of Sciences at the King Saud University (KSU) in Riyadh, Kingdom of Saudi Arabia (KSA) (Ethics Reference No: KSU-SE-22-20).
Acknowledgments
We extend our appreciation to the Researchers Supporting Project number (RSP-2021/218), King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- An overview of wetlands of Saudi Arabia: Values, threats, and perspectives. Ambio. 2017;46(1):98-108.
- [Google Scholar]
- First molecular identification of Euphlyctis ehrenbergii (Anura: Amphibia) inhabiting southwestern Saudi Arabia. Eur. Zool. J.. 2019;86:173-179.
- [Google Scholar]
- 16S rRNA gene identification and phylogenetic analysis of dhofar toad (Bufo dhufarensis) from riyadh province, saudi arabia. J. King Saud Univ.-Sci.. 2022;34:101972
- [Google Scholar]
- Phylogeography: the history and formation of species. Harvard University Press; 2000.
- Convergent adaptive radiations in Madagascan and Asian ranid frogs reveal covariation between larval and adult traits. Proc. Natl. Acad. Sci.. 2000;97:6585-6590.
- [Google Scholar]
- A river runs through it: tree frog genomics supports the Dead Sea Rift as a rare phylogeographical break. Biol. J. Linn. Soc.. 2019;128:130-137.
- [Google Scholar]
- Use of population genetic structure to define species limits in the Rhizobiaceae. Symbiosis 2005
- [Google Scholar]
- Underestimation of species richness in Neotropical frogs revealed by mtDNA analyses. PLoS ONE. 2007;2:e1109.
- [Google Scholar]
- Phylogeographical identification of some toads from afro-Arabian origin from Egypt and Saudi Arabia, using fragments of mtDNA. Egypt. J. Zool.. 2017;68:275-300.
- [Google Scholar]
- Reevaluation of 16S ribosomal RNA variation in Bufo (Anura: Amphibia) Mol. Phylogenet. Evol.. 2001;19:326-329.
- [Google Scholar]
- Caecilian phylogeny and biogeography inferred from mitochondrial DNA sequences of the 12S rRNA and 16S rRNA genes (Amphibia: Gymnophiona) Herpetol. Monographs 1993:64-76.
- [Google Scholar]
- Species boundaries, biogeography and evolutionarily significant units in dwarf toads: Duttaphrynus scaber and D. atukoralei (Bufonidae: Adenominae) Ceylon J. Sci.. 2017;46
- [Google Scholar]
- Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241.
- [Google Scholar]
- MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol.. 2018;35:1547.
- [Google Scholar]
- No ecological opportunity signal on a continental scale? Diversification and life-history evolution of African true toads (Anura: Bufonidae) Evolution. 2016;70:1717-1733.
- [Google Scholar]
- ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res.. 2015;43:W566-W570.
- [Google Scholar]
- Hotspots: Earth's biologically richest and most endangered terrestrial ecoregions. SA, Agrupación Sierra Madre, SC: CEMEX; 1999.
- Choricystis minor as a new symbiont of simultaneous two-species association with Paramecium bursaria and implications for its phylogeny. Symbiosis 2004
- [Google Scholar]
- The simple fool’s guide to PCR, version 2.0. Honolulu: University of Hawaii; 1991. p. :45.
- Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput. Biol.. 2006;2:e33.
- [Google Scholar]
- Historical biogeography resolves the origins of endemic Arabian toad lineages (Anura: Bufonidae): evidence for ancient vicariance and dispersal events with the Horn of Africa and South Asia. BMC Evol. Biol.. 2015;15:1-19.
- [Google Scholar]
- Phylogenetic relationships linking Duttaphrynus (Amphibia: Anura: Bufonidae) species based on 12S and 16S rDNA sequences. Mitochondrial DNA Part A. 2016;27:2881-2882.
- [Google Scholar]
- Molecular cloning: a laboratory manual. NY: Cold Spring Harbor; 1989.
- Extreme diversity of tropical parasitoid wasps exposed by iterative integration of natural history, DNA barcoding, morphology, and collections. Proc. Natl. Acad. Sci.. 2008;105:12359-12364.
- [Google Scholar]
- Origin of invasive populations of the Guttural Toad (Sclerophrys gutturalis) on reunion and Mauritius Islands and in Constantia, South Africa. Herpetol. Conserv. Biol.. 2019;14:380-392.
- [Google Scholar]
- The ecology and behavior of amphibians. University of Chicago Press; 2010.
- Toads in Qatar: The species present and their probable original source. J. Arid Environ.. 2019;160:91-94.
- [Google Scholar]
- Bayesian species delimitation using multilocus sequence data. Proc. Natl. Acad. Sci.. 2010;107:9264-9269.
- [Google Scholar]







