Translate this page into:
Molecular interaction analysis of Sulawesi propolis compounds with SARS-CoV-2 main protease as preliminary study for COVID-19 drug discovery
⁎Corresponding author. sahlan@eng.ui.ac.id (Muhamad Sahlan)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Coronavirus disease 2019 (COVID-19), a respiratory disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is a global health concern, as the World Health Organization declared this outbreak to be a global pandemic in March 2020. The need for an effective treatment is urgent because the development of an effective vaccine may take years given the complexity of the virus and its rapid mutation. One promising treatment target for COVID-19 is SARS-CoV-2 main protease. Thus, this study was aimed to examine whether Sulawesi propolis compounds produced by Tetragonula sapiens inhibit the enzymatic activity of SARS-CoV-2 main protease. In this study, molecular docking was performed to analyze the interaction profiles of propolis compounds with SARS-CoV-2 main protease. The results illustrated that two compounds, namely glyasperin A and broussoflavonol F, are potential drug candidates for COVID-19 based on their binding affinity of −7.8 kcal/mol and their ability to interact with His41 and Cys145 as catalytic sites. Both compounds also displayed favorable interaction profiles with SARS-CoV-2 main protease with binding similarities compared to inhibitor 13b as positive control 63% and 75% respectively.
Keywords
COVID-19
Molecular docking
Potent inhibitor
SARS-CoV-2 main protease
Sulawesi propolis
1 Introduction
Coronavirus disease 2019 (COVID-19), is a respiratory disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is currently considered a global health emergency. Since first being identified in Wuhan, China in December 2019, SARS-CoV-2 has spread widely and infected millions of people globally. On March 11, 2020, the World Health Organization (WHO) declared this outbreak to be a global pandemic. To date, there have been around 1.24 million deaths reported according to WHO as of November 7th, 2020 (World Health Organization, 2020) (Table 1).
No
Compound
Molecular Formula
2-Dimensional Structure
References
1
α-Tocopherol succinate
C32H52O5
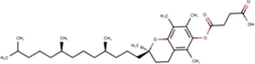
(Sahlan et al., 2019)
2
Xanthoxyletin
C15H14O4
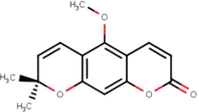
(Sahlan et al., 2019)
3
P-Coumaric acid
C9H10O3
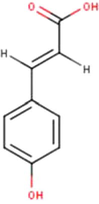
(Mahadewi et al., 2018)
4
Curcumene
C15H22
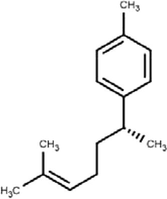
(Mahadewi et al., 2018)
5
Thymol
C10H14O
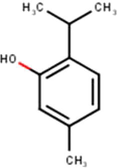
(Mahadewi et al., 2018)
6
Tetralin
C10H12
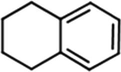
(Mahadewi et al., 2018)
7
Deoxypodophyllotoxin
C22H22O7
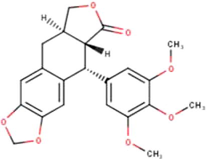
(Sahlan et al., 2019)
8
Sulabiroins A
C22H22O7
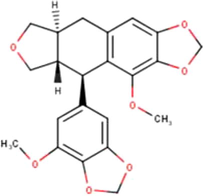
(Miyata et al., 2019, 2020a, 2020b)
9
Sulabiroins B
C23H26O7
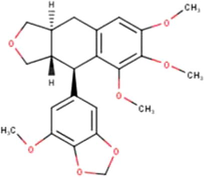
(Miyata et al., 2019, 2020a, 2020b)
10
2′,3′-Dihydro-3′-hydroxypapuanic acid
C25H38O7
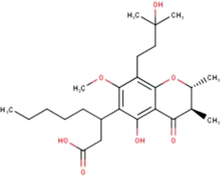
(Miyata et al., 2019, 2020a, 2020b)
11
(–)-Papuanic acid
C25H36O6
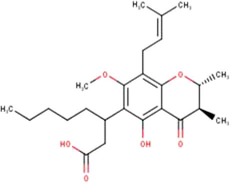
(Miyata et al., 2019, 2020a, 2020b)
12
(–)-Isocalolongic Acid
C24H34O6
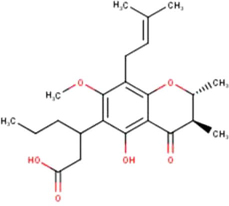
(Miyata et al., 2019, 2020a, 2020b)
13
Isopapuanic acid
C25H36O6
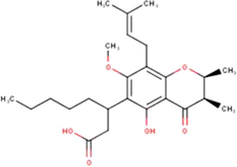
(Miyata et al., 2019, 2020a, 2020b)
14
Isocalopolyanic acid
C24H32O6
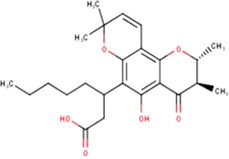
(Miyata et al., 2019, 2020a, 2020b)
15
Glyasperin A
C25H26O7
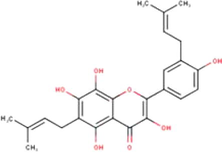
(Miyata et al., 2019, 2020a, 2020b)
16
Broussoflavonol F
C25H26O7
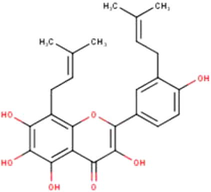
(Miyata et al., 2019, 2020a, 2020b)
17
(2S)-5,7-Dihydroxy-4′-methoxy-8-prenylflavanone
C20H20O5
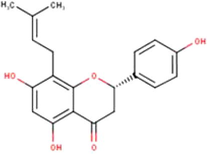
(Miyata et al., 2019, 2020a, 2020b)
18
Isorhamnetin
C16H12O7
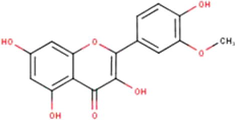
(Miyata et al., 2019, 2020a, 2020b)
19
(1′S)-2-Trans,4-trans-abscisic acid
C15H20O4
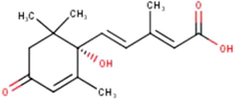
(Miyata et al., 2019, 2020a, 2020b)
20
(1′S)-2-Cis,4-trans-abscisic acid
C15H20O4
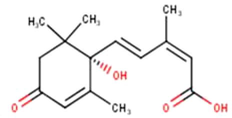
(Miyata et al., 2019, 2020a, 2020b)
SARS-CoV-2 is a single-stranded ribonucleic acid (RNA) enveloped virus from the genus Betacoronavirus, subfamily Orthocoronavirinae, and family Coronaviridae (Zheng, 2020). Two other viruses belong to this genus, namely Middle East respiratory syndrome virus and severe acute respiratory syndrome virus (SARS-CoV). SARS-CoV-2 was identified to have 82% RNA identity with SARS-CoV (Zhang et al., 2020). Both SARS-CoV and SARS-CoV-2 recognize the same receptor in the human body, namely angiotensin-converting enzyme 2. The SARS-CoV-2 genome encodes a trimeric structural spike protein, a homodimeric cysteine proteinase, an RNA polymerase, and several nonstructural proteins (Calligari et al., 2020). The viral pathogenesis causes several symptoms, such as sore throat, running nose, cough, fever, and eventually respiratory failure.
Currently, there is no specific therapy with curative efficacy against the disease (Centers for Disease Control and Prevention, 2020). Several drugs, such as hydroxychloroquine and chloroquine, have been suggested as treatments. However, no studies were adequately powered to prove their efficacy (Pastick et al., 2020). In addition, several reports of serious arrhythmia have been described in patients with COVID-19 who received these compounds (United States Food and Drug Administration, 2020). In addition, efforts are underway to develop vaccines to control the outbreak. However, given the complexity of the virus and the rapid mutation of its single stranded RNA, it may take years to generate effective vaccines (Huanget al., 2020).
Thereby, other compounds should be examined to identify an effective treatment with few or no adverse effects. According to Marcucci, propolis, a resinous bee product, exhibits antiviral activity based on the presence of flavonoids, caffeic acid, and esters of aromatic acids (Marcucci, 1995). The antiviral activities of these compounds occur through the inhibition of viral transmission to other cells, inhibition of viral propagation, and destruction of the outer envelope of the virus (Marcucci, 1995). Various experiments have been conducted to analyze the antiviral activities of the compounds. Research by Yildirim et al. revealed that propolis significantly decreased the number of copies of herpes simplex virus 2 after 48 h of incubation (Yildirim et al., 2016). In addition, Gekker et al. demonstrated that propolis has a destructive effect on the outer envelope of human immunodeficiency virus (Gekker et al., 2005). In terms of antiviral activity in the respiratory tract, another study reported that some propolis compounds have lower IC50 values against human rhinovirus than ribavirin (Kwon et al., 2020).
The potency of propolis as COVID-19 drug has also been observed through some research. Vardhan and Sahoo (2020) have shown that three propolis components, namely limonin, quercetin and kaempferol have inhibitory potential by binding to viral RNA-dependent RNA polymerase (RdRp) with binding energy −9 to −7.1 kcal/mol through molecular docking study (Vardhan and Sahoo, 2020). Molecular docking enables the virtual screening of millions of compounds in a time- and cost-efficient manner (Pinzi and Rastelli, 2019). Hence, this method is widely used as a preliminary study in drug discovery (Meng et al., 2011). Another molecular docking study by Güler et al. (2020) concluded that identified compounds from alcoholic extract of propolis are able to bind with angiotensin-converting enzyme (ACE)-2. Thus, the compounds may be potential to prevent SARS-CoV-2 to bind with ACE-2 (Güler et al., 2020).
Based on these findings, propolis compounds may have potency for treating COVID-19. Further research is needed to evaluate the ability of propolis compounds to inhibit SARS-CoV-2 replication. Other protein in SARS-CoV-2 that is also considered a potential therapeutic target is main protease (Anand et al., 2003). The enzyme cleaves polyproteins translated from viral RNA into 12 smaller proteins that participate in viral replication (Chen et al., 2020). Therefore, viral replication can be blocked by inhibiting this enzyme (Zhang et al., 2020).
Previously, Zhang et al. developed peptidomimetic α-ketoamides as potential broad-spectrum inhibitors of main protease in Betacoronaviruses and Alphacoronaviruses (Zhang et al., 2020). One such compound, tert-butyl(1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)-amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydro-pyridin-3-yl) carbamate, also known as 13b, inhibited SARS-CoV-2 replication in human Calu3 lung cells. The crystal structure of SARS-CoV-2 main protease in complex with inhibitor 13b is available.
Propolis compounds are diverse according to the region (Alday et al., 2016). Beforehand, Sulawesi propolis compounds from North Luwu have been identified by several research (Mahadewi et al., 2018; Miyata et al., 2019, 2020a, 2020b; Sahlan et al., 2019). Sulawesi propolis previously was known to be produced by Tetragonula aff. biroi. However, recent study rectified that it is actually produced by Tetragonula sapiens (Sayusti et al., 2020). Some research has also proved the health benefits of Sulawesi propolis. Sulawesi propolis exhibits antifungal activity to Candida albicans, C. tropicalis, C. krusei, C. parapsilosis, C. glabrata and Cryptococcus neoformans (Sahlan et al., 2020). Other study showed that Sulawesi propolis is potential to be developed as a non-steroid anti-inflammatory drug and antioxidant agent (Christina et al., 2018; Sahlan et al., 2019). These facts encourage us to do more investigations regarding the health benefits of Sulawesi propolis. The components of antioxidant and flavonoid in Sulawesi propolis may perform antiviral activity. Up to date, there has been no research that aims to evaluate the potency of Sulawesi propolis compounds to treat COVID-19.
In this research, molecular docking was performed to analyze the molecular interaction between SARS-CoV-2 main protease (PDB ID: 6Y2F) and Sulawesi propolis compounds. Molecular docking aims to predict the conformation of the ligand within the receptor and assess the binding affinity (Guedes et al., 2014), which is represented by the docking score (kcal/mol). The docking score and binding characteristics of inhibitor 13b were used to judge whether the propolis compounds are potential inhibitors of SARS-CoV-2 main protease.
Furthermore, this research also performed molecular docking between SARS-CoV-2 main protease and 14b, a modified version of inhibitor 13b. 14b does not feature a Boc group, which predicted to protect the compound as it crosses the cellular membrane (Zhang et al., 2020). Although 14b was mostly inactive in human Calu3 lung cells, this compound may have lower affinity for plasma proteins than inhibitor 13b (Zhang et al., 2020).
2 Materials and methods
2.1 Hardware and software
Molecular docking was performed using an Asus laptop with an Intel® Core™ i7-8550U @1.80 GHz processor, 8 GB of RAM, the Windows 10 Home Single Language 64-bit operating system, and an Intel® UHD Graphics 620 graphics processing unit. The software used in the study included MarvinSketch (ChemAxon, Budapest, Hungary), Autodock Tools 1.5.6 (The Scripps Research Institute, USA), Autodock Vina (The Scripps Research Institute), LigPlot+ (EMBL-EBI, UK) and Visual Molecular Dynamics (University of Illinois, Urbana-Champaign).
2.2 Lipinski’s rule of five (RO5) selection
First, 20 identified Sulawesi propolis compounds from North Luwu, South Sulawesi, Indonesia were selected based on Lipinski’s RO5. The rules aim to assess the solubility and permeability of drug candidates. The RO5 stated that poor absorption or permeability are more likely to occur when a compound has molecular weight greater than 500 g/mol, Log P greater than 5, H-bond donors more than 5 and H-bond acceptors more than 10 (Lipinski et al., 1997). Sulawesi propolis compounds that violates more than 2 rules were thrown away. Lipinski’s RO5 was assessed by using MarvinSketch (ChemAxon, 2018).
2.3 Protein preparation
The SARS-CoV-2 main protease in complex with the α-ketoamide 13b (PDB ID: 6Y2F) was obtained from the Protein Data Bank (http://www.rcsb.org) in the.pdb format (Zhang et al., 2020). The Protein Data Bank is an open access digital data resource that provides 3D structure data for large biological molecules (Berman et al., 2013). The protein and inhibitor 13b file was then separated into two different.pdb files using Visual Molecular Dynamics (Humphrey et al., 1996). Then, the separated protein files were loaded into Autodock Tools 1.5.6 for further preparation consisting of the addition of polar hydrogens, addition of kollman charges and conversion of the file to the .pdbqt format (Forli et al., 2016) for compatibility for docking using Autodock Vina.
2.4 Ligands preparation
Meanwhile, the test ligands were selected Sulawesi propolis compounds from North Luwu produced by Tetragonula sapiens and 14b using inhibitor 13b as control. The structure of inhibitor 13b and 14b are shown in Fig. 1. Excluding inhibitor 13b, the 2D structures of the ligands were constructed and converted into 3D structures in the .pdb format using MarvinSketch. Then, the ligands were loaded to Autodock Tools 1.5.6 to add polar hydrogens as well as gasteiger charges, and convert the file to the.pdbqt format.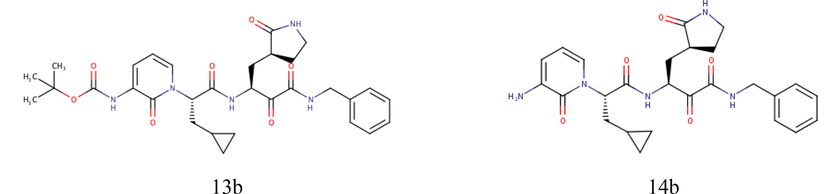
Structure of inhibitor 13b and 14b (Zhang et al., 2020).
2.5 Parameter optimization
Before performing molecular docking between propolis compounds and main protease, the optimal specific search space was determined. The search space is the area within the protein in which the docking simulation will be performed. It may represent the locations of the binding sites. The optimal search space can be predicted via docking between the native ligand of the protein structure, in this case 13b, using the protein that was separated in the protein preparation step. This method is also known as “redocking.” Redocking and the subsequent docking simulation were performed using Autodock Vina. In terms of accuracy, Autodock Vina offers more accurate binding mode predictions than Autodock Tools (Vieira and Sousa, 2019).
Adjustment was performed for three parameters to obtain the optimal specific search space, namely the center of the grid box, number of points in the x, y, and z dimensions, and grid spacing. Based on the redocking simulation, it was found that the binding site had a higher probability to be located at the following coordinates: x = 11.476, y = −1.396, and z = 20.745. The number of points in the x, y, and z dimensions were all set to 25 Å, and the grid spacing was adjusted to 1.0 Å.
After the docking score was obtained, the root mean square deviation (RMSD) of the first rank ligand pose was evaluated. An RMSD of less than 2.0 Å indicates that the redocking procedure is accurate (Ramírez and Caballero, 2018). Thereby, the docking coordinates could be used to dock the propolis compounds onto the protein.
2.6 Molecular docking analysis of propolis compounds onto main protease
To identify which propolis compounds are potential SARS-CoV-2 inhibitors, the compounds were docked into the protein individually using the coordinates and grid parameters obtained from the redocking simulation. The simulations were performed using Autodock Vina with exhaustiveness value set to 64. After the docking score for each compound was obtained, the interactions between the protein and the ligands with the lowest docking scores were analyzed using Ligplot+, which provides 2D visualization of molecular interactions.
3 Results and discussion
This research analyzed the molecular interactions between selected Sulawesi propolis compounds produced by Tetragonula sapiens and SARS-CoV-2 main protease in an effort to identify potential inhibitors. Main protease was selected because of its important role in viral replication. Several main protease structures were available in the Protein Data Bank. Of these, PDB ID 6Y2F was selected because it featured inhibitor 13b bound to the protein. Inhibitor 13b was previously proven to reduce SARS-CoV-2 RNA levels in infected human lung cells with an IC50 of 0.67 ± 0.18 μm (Zhang et al., 2020).
3.1 Lipinski’s RO5 selection result
First of all, 19 Sulawesi propolis compounds from North Luwu were selected based on Lipinski’s RO5 in Table 2. According to the result, (-)-papuanic acid, isopapuanic acid, isocalopolyanic acid and curcumene have Log P of greater than 5.0. In the meantime, a compound, namely α-tocopherol succinate, violates two out of four rules, which are molecular weight and log P parameter. A molecular weight greater than 500 g/mol will lead to poor permeability when a drug molecule penetrates biological membrane through passive diffusion process (Qiu et al., 2016). Meanwhile, Log P represents octanol–water partition coefficient. A drug molecule with high Log P value tends to be more nonpolar and have poorer aqueous permeability (Templeton et al., 2015). Although there are several violations, the number of rules being obeyed is sufficient to indicate that the compounds have good permeability. In addition, there are some tolerable Lipinski’s RO5 parameter value for natural compounds.
No.
Compounds
Molecular weight (g/mol)
Log P
Number of H-bond acceptor
Number of H-bond donor
Number of violations
1
Sulabiroins A
398.411
2.74
7
0
0
2
Sulabiroins B
414.454
2.55
7
0
0
3
2',3'-Dihydro-3'-hydroxypapuanic acid
450.572
4.33
7
3
0
4
(−)-Papuanic acid
432.557
5.57
6
2
1
5
(−)-Isocalolongic acid
404.503
4.78
6
2
0
6
Isopapuanic acid
432.557
5.57
6
2
1
7
Isocalopolyanic acid
416.514
5.03
6
2
1
8
Glyasperin A
438.476
4.84
7
5
0
9
Broussoflavonol F
438.476
4.84
7
5
0
10
(2s)-5,7-Dihydroxy-4'-methoxy-8-prenylflavanone
340.375
4.19
5
3
0
11
Isorhamnetin
316.265
1.78
7
4
0
12
(1's)-2-Trans,4 trans-abscisic acid
264.321
2.08
4
2
0
13
(1's)-2-Cis,4 trans-abscisic acid
264.321
2.08
4
2
0
14
α-tocopherol succinate
530.790
9.18
4
1
2
15
Xanthoxyletin
258.273
2.01
3
0
0
16
P-coumaric acid
164.160
2.12
3
2
0
17
Curcumene
202.341
5.19
0
0
1
18
Thymol
150.221
3.42
1
1
0
19
Tetralin
132.206
3.27
0
0
0
20
Deoxypodophyllotoxin
398.411
2.63
6
0
0
3.2 Docking score
Selected Sulawesi propolis compounds, inhibitor 13b and 14b then were docked to main protease by using Autodock Vina. Docking simulation by using Autodock Vina produces two results, which are the most stable ligand poses and docking score. The docking score is generated from an empirical calculation that considers the number of hydrophobic interactions and hydrogen bonding (Trott and Olson, 2010). The greater the number of interactions, the greater the tendency for the docking score to be negative. The docking score may become a representative of binding affinity, and it is inversely proportional to the binding stability (Flamandita et al., 2020). The negative docking score indicates that the compound, which in this case acts as a ligand, has the ability to interact with the protein. Meanwhile, the most stable ligand pose will further be used to find out which amino acid residues in the protein interact with the ligand. Thereafter this is referred as interaction profile. By comparing interaction profile between SARS-CoV-2 main protease and propolis compounds with SARS-CoV-2 main protease and inhibitor 13b, the potency of propolis compounds to have the ability to inhibit the protein with the same pathway as inhibitor 13b may be evaluated.
The docking score between main protease and test compounds are available in Table 3. Based on the docking simulation, 13b and 14b bound to main protease with a binding affinity of −8.2 and −7.2 kcal/mol. Meanwhile, among the propolis compounds, the three lowest docking scores were obtained for broussoflavonol F, glyasperin A, and sulabiroins A, with values of −7.8, −7.8, and −7.6 kcal/mol, respectively. Broussoflavonol F and glyasperin A are identified as flavonoids, while sulabiroins A is a derivative of podophyllotoxin compounds (Miyata et al., 2019, 2020a, 2020b). The interaction profiles of these three compounds, inhibitor 13b and 14b with main protease severally then were analyzed. *) Native ligand as a control.
No
Compounds
Docking score (kcal/mol)
1
* Inhibitor 13b
−8.2
2
14b
−7.2
3
Broussoflavonol F
−7.8
4
Glyasperin A
−7.8
5
Sulabiroins A
−7.6
6
Isorhamnetin
−7.5
7
Deoxypodophyllotoxin
−7.3
8
(2S)-5,7-Dihydroxy-4′-methoxy-8-prenylflavanone
−7.1
9
Sulabiroins B
−7.0
10
Isocalopolyanic acid
−6.8
11
Isopapuanic acid
−6.8
12
2′,3′-Dihydro-3′-hydroxypapuanic acid
−6.7
13
(−)-Isocalolongic acid
−6.7
14
(−)-Papuanic acid
−6.6
15
Xanthoxyletin
−6.2
16
(1′S)-2-Trans-4-trans-abscisic acid
−6.1
17
(1′S)-2-Cis-4-trans-abscisic acid
−5.9
18
α-Tocopherol succinate
−5.1
19
P-Coumaric acid
−4.9
20
Curcumene
−4.7
21
Thymol
−4.7
22
Tetralin
−4.4
3.3 Interaction profile
The interaction profile analysis was focused on the existence of interaction with His41 and Cys145. These amino acids have an important role as main protease catalytic sites. Thus, propolis compounds must interact with these sites in order to inhibit the activity of the enzyme. The two-dimensional (2D) visualization of the interaction of main protease with 13b, broussoflavonol F, glyasperin A, and sulabiroins A were presented in Fig. 2. The interaction profile between main protease and inhibitor 13b and 14b were firstly analyzed. According to the visualization, the docking simulation successfully captured the interaction of inhibitor 13b with both His41 and Cys145, in line with the results reported by Zhang et al. (2020) as the crystallographer (Zhang et al., 2020). The shortest interatomic distance between main protease with His41 and Cys145 are 3.03 and 3.27 Å respectively. In the meantime, 14b only interacts hydrophobically with His41. Based on this finding, although 14b may have a better in vivo potency due to lower affinity for plasma protein binding, it does not have favorable interaction towards the main protease.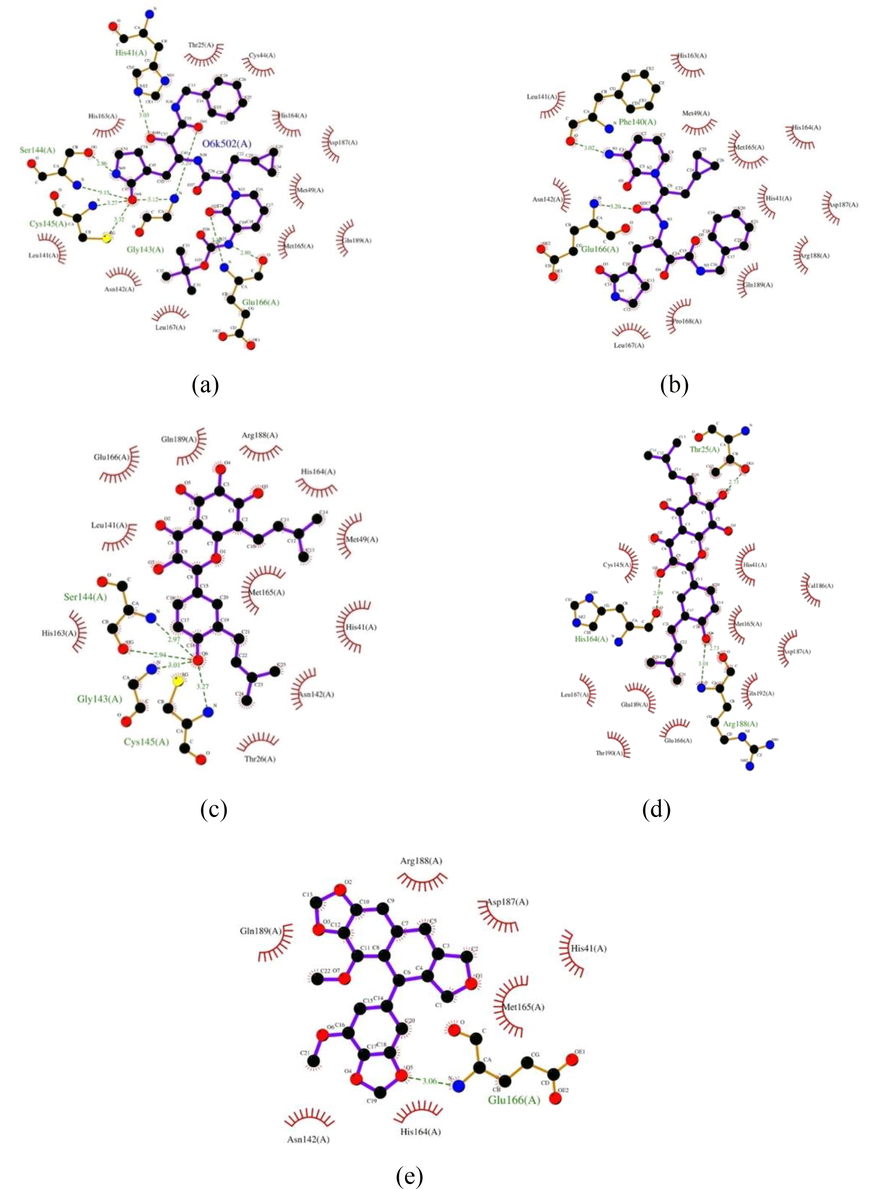
Visualization of the molecular interactions of main protease with various ligands. (a) 13b, (b) 14b, (c) broussoflavonol F, (d) glyasperin A, (e) sulabiroins A. The purple lines denote the ligand structure, whereas the brown lines denote the structure of amino acid residues. The molecular interactions are reflected as dashed lines and arcs. The green dashed lines between atoms represent hydrogen bonds, and the numbers above these lines indicate the length of the bond. Meanwhile, the arcs with spokes radiating toward the ligand atoms represent hydrophobic interactions. The atoms involve in hydrophobic interactions are indicated by the presence of spokes radiating back (Wallace et al., 1995).
The interaction profile between main protease and each propolis compounds were then analyzed. Based on the results, broussoflavonol F forms hydrogen bond with Cys145 and interacts hydrophobically with His41. The interatomic distance between main protease and Cys145 is 3.27 Å. The same value of interatomic distance compared to inhibitor 13b shows that broussoflavonol F may bind to Cys145 as strong as inhibitor 13b. Meanwhile, glyasperin A does not form hydrogen bonds with any catalytic sites. It interacts hydrophobically with both catalytic sites instead. Hydrophobic interaction with the catalytic sites shows that although the binding affinity between the compound and the protein may not as strong as in hydrogen bonds, the compound may still have the potency to inhibit the activity of the enzyme. Lastly, for sulabiroins A, it is shown that this compound only interacts hydrophobically with one catalytic site, which is His41.
For further consideration, binding similarity between interaction profile of main protease with the three propolis compounds and 14b compared to interaction profile of main protease and inhibitor 13b were calculated. Based on the calculation, 14b, broussoflavonol F, glyasperin A and sulabiroins A bound to main protease with the similarity of 71%, 75%, 63% and 44% respectively. The summary of interaction profile between main protease and each test compounds are available in Table 4, while Table 5 summarizes the hydrogen bonds present between main protease and each test compounds. *) Native ligand as a control. *) Native ligand as a control.
No.
Compounds
Hydrogen bonds
Hydrophobic interactions
Number of interactions
Binding similarity
1
* 13b
His41, Gly143, Ser144, Cys145, Glu166
Thr25, Cys44, Met49, Leu141, Asn142, His163, His164, Met165, Leu167, Asp187, Gln189
16
100%
2
14b
Phe140, Glu166
His41, Met49, Leu141, Asn142, His163, His164, Met165, Leu167, Pro168, Asp187, Arg188, Gln189
14
71%
3
Broussoflavonol F
Gly143, Ser144, Cys145
Thr26, His41, Met49, Leu141, Asn142, His163, His164, Met165, Glu166, Arg188, Gln189
14
75%
4
Glyasperin A
Thr25, His164, Arg188
His41, Cys145, Met165, Glu166, Val186, Asp187, Gln189, Thr190, Gln192
13
63%
5
Sulabiroins A
Glu166
His41, Asn142, His164, Met165, Asp187, Arg188, Gln189
8
44%
No.
Compounds
Hydrogen bond distance (Å)
Interacting amino acid
Binding ligand group
Binding amino acid group
1
* 13b
2,80
Glu166
–NH
–O
2,86
Ser144
–NH
–OH
3,03
His41
–O
–NH
3,12
Gly143
–O
–NH2
3,15
Ser144
–O
–NH2
3,20
Gly143
–O
–NH2
3,27
Cys145
–O
–NH2
3,30
Glu166
–O
–NH2
3,32
Cys145
–O
–SH
2
14b
3.02
Phe140
–NH2
–O
3.20
Glu166
–O
–NH2
3
Broussoflavonol F
2.94
Ser144
–OH
–O
2.97
Ser144
–OH
–NH2
3.01
Gly143
–OH
–NH2
3.27
Cys145
–OH
–NH2
4
Glyasperin A
2.73
Thr25
–OH
–OH
2.73
Arg188
–OH
–OH
2.99
His164
–OH
–O
3.01
Arg188
–OH
–NH2
5
Sulabiroins A
3.06
Glu166
–O
–NH2
4 Conclusion
Sulawesi propolis compounds from North Luwu that are produced by Tetragonula sapiens exhibit potential to inhibit SARS-CoV-2 main protease activity. Three compounds, namely broussoflavonol F, glyasperin A, and sulabiroins A, displayed the greatest docking score, with values of −7.8, −7.8, and −7.6 kcal/mol, respectively. The study results suggest that molecular dynamic simulation should be conducted for broussoflavonol F and glyasperin A given that both compounds interact with main protease catalytic sites and have the ability to bind with main protease with binding similarity of 75% and 63% respectively compared to potent inhibitor. Further research should be conducted to verify the potency and safety of broussoflavonol F and glyasperin A to the treatment of COVID-19.
Acknowledgments
The research was financially supported by DRPM Universitas Indonesia through Grant Publikasi Terindeks Internasional (PUTI) Kolaborasi Internasional (2Q2) 2020 No: NKB785/UN2.RST/HKP.05.00/2020 and the authors extend their appreciation to the Researchers supporting project number (RSP-2020/7), King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper..
References
- Advances in pharmacological activities and chemical composition of propolis produced in Americas Beekeep. Bee Conserv. Res. 2016
- [Google Scholar]
- Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science (80-.).. 2003;300:1763-1767.
- [Google Scholar]
- How community has shaped the protein data bank. Structure. 2013;21(9):1485-1491.
- [CrossRef] [Google Scholar]
- Molecular investigation of SARS–CoV-2 proteins and their interactions with antiviral drugs. Viruses. 2020;12:445.
- [Google Scholar]
- Centers for Disease Control and Prevention, 2020. Information for clinicians on investigational therapeutics for patients with COVID-19. 2020-04-13]. <https//www.cdc.gov/coronavirus/2019-ncov/hcp/therapeutic-options.html>.
- ChemAxon. Marvin was used for drawing, displaying and characterizing chemical structures, substructures and reactions, Marvin 18.28, 2018, ChemAxon (http://www.chemaxon.com) (Version 18.28).
- Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CLpro) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Res. 2020;9:129.
- [CrossRef] [Google Scholar]
- Christina, D., Hermansyah, H., Wijanarko, A., Rohmatin, E., Sahlan, M., Pratami, D.K., Mun’Im, A., 2018. Selection of propolis Tetragonula sp. extract solvent with flavonoids and polyphenols concentration and antioxidant activity parameters. In: AIP Conference Proceedings. https://doi.org/10.1063/1.5023967.
- Molecular docking analysis of podophyllotoxin derivatives in Sulawesi propolis as potent inhibitors of protein kinases. In: AIP Conference Proceedings. AIP Publishing LLC; 2020. p. :20010.
- [Google Scholar]
- Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc.. 2016;11(5):905-919.
- [CrossRef] [Google Scholar]
- Anti-HIV-1 activity of propolis in CD4+ lymphocyte and microglial cell cultures. J. Ethnopharmacol.. 2005;102:158-163.
- [Google Scholar]
- An investigation of ethanolic propolis extracts: their potential inhibitor properties against ACE-II receptors for COVID-19 treatment by Molecular Docking Study. Sci. Prepr. 2020
- [Google Scholar]
- Pharmacological therapeutics targeting RNA-dependent RNA polymerase, proteinase and spike protein: from mechanistic studies to clinical trials for COVID-19. J. Clin. Med.. 2020;9:1131.
- [Google Scholar]
- Antiviral effects and possible mechanisms of action of constituents from Brazilian propolis and related compounds. J. Apic. Res.. 2020;59:413-425.
- [Google Scholar]
- Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev.. 1997;23:3-25.
- [Google Scholar]
- Mahadewi, A.G., Christina, D., Hermansyah, H., Wijanarko, A., Farida, S., Adawiyah, R., Rohmatin, E., Sahlan, M., 2018. Selection of discrimination marker from various propolis for mapping and identify anti Candida albicans activity. In: AIP Conf. Proc. 1933. https://doi.org/10.1063/1.5023939.
- Marcucci, M.C., 1995. Propolis: chemical composition, biological properties and therapeutic activity.
- Molecular docking: a powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug Des.. 2011;7:146-157.
- [Google Scholar]
- Propolis components and biological activities from stingless bees collected on South Sulawesi, Indonesia. HAYATI J. Biosci.. 2020;27:82.
- [Google Scholar]
- Miyata, R., Sahlan, M., Ishikawa, Y., Hashimoto, H., Honda, S., Kumazawa, S., 2020b. Erratum: Propolis Components from Stingless Bees Collected on South Sulawesi, Indonesia, and Their Xanthine Oxidase Inhibitory Activity (Journal of Natural Products (2019) 82: 2 (205− 210). J. Nat. Prod.
- Propolis components from stingless bees collected on South Sulawesi, Indonesia, and their xanthine oxidase inhibitory activity. J. Nat. Prod.. 2019;82:205-210.
- [CrossRef] [Google Scholar]
- Hydroxychloroquine and chloroquine for treatment of SARS-CoV-2 (COVID-19) In: Open Forum Infectious Diseases. US: Oxford University Press; 2020. p. ofaa130.
- [Google Scholar]
- Molecular docking: Shifting paradigms in drug discovery. Int. J. Mol. Sci.. 2019;20:4331.
- [Google Scholar]
- Developing Solid Oral Dosage Forms: Pharmaceutical Theory and Practice. Academic press; 2016.
- Is it reliable to take the molecular docking top scoring position as the best solution without considering available structural data? Molecules. 2018;23:1038.
- [Google Scholar]
- Anti-inflammatory activity of Tetragonula species from Indonesia. Saudi J. Biol. Sci.. 2019;26:1531-1538.
- [Google Scholar]
- Exploration of the antifungal potential of Indonesian Propolis from Tetragonula biroi bee on Candida sp. and Cryptococcus neoformans. Evergr. J.. 2020;7:118-125.
- [Google Scholar]
- Stingless bees (Hymenoptera: Apidae) in South and West Sulawesi, Indonesia: morphology, nest structure, and molecular characteristics. J. Apic. Res. 2020:1-14.
- [Google Scholar]
- Discovering and Developing Molecules with Optimal Drug-like Properties. Springer; 2015.
- AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem.. 2010;31:455-461.
- [Google Scholar]
- United States Food and Drug Administration. 2020. FDA cautions against use of hydroxychloroquine or chloroquine for COVID-19 outside of the hospital setting or a clinical trial due to risk of heart rhythm problems. 2020.
- Vardhan, S., Sahoo, S.K., 2020. Searching inhibitors for three important proteins of COVID-19 through molecular docking studies. arXiv Prepr. arXiv2004.08095.
- Comparing AutoDock and Vina in ligand/decoy discrimination for virtual screening. Appl. Sci.. 2019;9:4538.
- [Google Scholar]
- LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel.. 1995;8:127-134.
- [Google Scholar]
- World Health Organization. 2020. Retrieved from <https://www.covid19.who.int/>.
- Antiviral activity of hatay propolis against replication of herpes simplex virus type 1 and type 2. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res.. 2016;22:422.
- [Google Scholar]
- Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (80-.).. 2020;368:409-412.
- [Google Scholar]
- SARS-CoV-2: an emerging coronavirus that causes a global threat. Int. J. Biol. Sci.. 2020;16(10):1678.
- [Google Scholar]







