Translate this page into:
Molecular identification and phylogenetic analysis of cytochrome b gene from Garra tibanica an endogenous species from Saudi Arabia
⁎Corresponding author. afrefaei@ksu.edu.sa (Abdulwahed Fahad Alrefaei)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Garra tibanica is ray-finned freshwater fish belonging to the cyprinid. Previous studies have reported that this specie is a native fish species found in Saudi Arabia. However, this specie was never been assessed using molecular markers to find out its phylogenetic relationship with other species in the area. This study investigates the genetic diversity between of the Garra tibanica collected from Medina province, Saudi Arabia based on phylogenetic analysis using the cytochrome b gene. The phylogenetic tree constructed from the cytochrome b sequence indicates that Garra tibanica is more closely related to Garra sahilia and Garra sharq than Garra rufa, Garra gymnothorax, which may reflect the allopatric speciation of Garra according to their existence in nearby geographic regions. To our knowledge, this report is the first to describe the species of Garra in Saudi Arabia based on phylogenetic analysis using the cytochrome b gene.
Keywords
Garra tibanica
Cytochrome b
Molecular markers
PCR
Phylogenetic analysis
Sequencing
Saudi Arabia
1 Introduction
Freshwater fish have a range of morphological adaptations coming from genetic divergence and/or phenotypic flexibility, which result in adaptive radiation (Kerschbaumer and Sturmbauer, 2011; Jacquemin and Pyron, 2016) and further evolution (Dwivedi, 2020). The family cyprinidae is considered the biggest and most diverse freshwater fish. Their shapes, sizes, and biology, are variable (Dwivedi, 2020; Kirchner et al., 2021).
Nearly 83 % of freshwater fish show a high degree of endemism in the Arabian Peninsula, with about 17 % of taxa assessed to be threatened with extinction while 3 % are near threatened and 20 % are listed as incomplete data (García et al., 2015). The greatest threats are habitat loss and degradation due to the alteration of environmental systems habitat and an increase in cultivation, together with pollution and the current trend of climate change and precipitation decline (Moran, 2009). The continuous rise in temperature due to climate change has a strong impact on physiology, behavior and reproduction of fish populations and trophic interactions (Rijnsdorp et al., 2009; Moran, 2009).
On the Arabian Peninsula, 31 species of freshwater fish have been described (Hamidan and Shobrak, 2019, Freyhof et al., 2021); Among them 15 species are endemic to Arabia, while other six have a broad distribution (Freyhof et al., 2015; Freyhof et al., 2021). Recent study has shown that most of the fresh water fish found in Saudi Arabia belong to the family Cyprinidae (Alotaibi et al., 2020). There are four genera of Cyprinid fishes in Arabian Peninsula which include Garra, Acanthobrama, Cyprinion and Carasobarbus (Playfair, 1870; Krupp, 1983; Alkahem, 1983).
The Garra is descended from labeonine cyprinids which consist of more than 160 species and have a wide distribution from Southeast Asia to West Africa (Fricke et al., 2018; Yang et al., 2012). Garra fish have moderate body size (usually less then 20 cm in length) with adapted mental discs for sucking and scraping algae scrapers (Kottelat, 2020). Behrens-Chapuis et al. (2015) have studied the labeonine cyprinid genera, where members of Crossocheilus, Hemigrammocapoeta, Tylognathus and Typhlogarra were found to be nested within Garra based on mtCOI data. A new species of Garra, Garra jordanica was described by Hamidan et al. (2014) from dead sea and has discussed the relation with G. ghorensis, G. tibanica and G. rufa.
Aljohani (2019) reported that the Arabian Peninsula faces urgent conservation issues with decreasing numbers of flowing springs and increasing levels of water conductivity in surviving springs since 1990 due to overpumping of groundwater. This had a huge impact on fish tolerance and distribution, and now only five native species are surviving in Saudi Arabian and Omani Springs. The ability to tolerate mild to extremely high conductivity. Garra tibanica is the only specie habitated in Saudi Arabian springs.
They are found at three sites in Saudi Arabia (Wadi Damad, Jizan; Wadi al-Bagarah and Ein al-Hamah, Khaibar) and in Yemen (Hamidan and Shobrak, 2019). It was listed as Least Concern by the IUCN in 2015 (The IUCN Red List of Threatened Species 2015). However, due to the reduction of the quality and extent of existing habitats due to extraction of water this specie face big threats.
In recent years taxonomist characterized organisms into groups based on meristic, morphometric and anatomical characteristics. This method need more molecular attributes used to provide the effectiveness from which phylogeny between taxa analysed and inferred and understand (AlMutair, 2016). Moreover, the results obtained from phylogenetic morphology are not always in agreement with molecular analyses (Zheng et al., 2010).
DNA barcoding is a quick and accurate method in biodiversity research for taxonomic identification, characterization and defining new species. It can determine the genetic and evolutionary relationship from molecular, morphological, and distributional data. DNA barcodes are accurate tools to identify a variety of freshwater fish (Behrens-Chapuis et al., 2015, Bhattacharya et al., 2016). The mtDNA has a constant rate in the substitution of nucleotides over evolutionary time, and this property is useful in determining the divergence period (DeSalle et al., 1987). Markers used in fish biodiversity research include: the cytochrome C oxidase subunit 1 (COI), 16S ribosomal RNA (16S), the 12S ribosomal RNA (12S) and cytochrome b (cyt b) genes (Claver et al., 2021, Yousefi, 2013). Linacre (2012) has mentioned that the loci of choice for taxonomy, phylogenetics, and forensic science are on the mitochondrial genome; being predominantly either the cytochrome b (cyt b) or the cytochrome oxidase I genes (COI). Tobe et al. (2009) recommend use of the cytochrome b gene in the study of inter-species variation as this provides more information from a small fragment.
The aim of this study was to investigate the genetic diversity among the specimens of Garra tibanica collected from Wadi Khadrah, Medina province, Saudi Arabia to assess the genetic differentiation and explore the potential phylogenetic relationships between these species and other Garra species via sequencing of the cytochrome b gene.
2 Materials and methods
2.1 Sample collection
Twelve specimens of Garra tibanica Garra tibanica were collected from Wadi Khadrah (N17 39 410, E42 40 665) between December 2019 and January 2020 in Medina province, Saudi Arabia (Table 1). The morphometric and meristic characters of the fish were examined to identify their characteristics. The collected fish were transported on dry ice to the laboratory at Zoology department, King Saud University, where they were stored at −80 °C for further analysis. “All procedures for the samples collection were carried out in strict accordance with the recommendations by the Research Ethics Sub-Committee (REC) of the College of Sciences at the King Saud University (KSU) in Riyadh, Kingdom of Saudi Arabia (KSA) (Ethics ReferenceNo: KSU-SE-22–14)”.
ID
Sample No.
Species
GenBank
Accession No.
1
1GT
Garra tibanica
OM540814
2
2GT
Garra tibanica
OM540815
3
3GT
Garra tibanica
OM540816
4
4GT
Garra tibanica
OM540817
5
5GT
Garra tibanica
OM540818
6
6GT
Garra tibanica
OM540819
7
7GT
Garra tibanica
OM540820
8
8GT
Garra tibanica
OM540821
9
9GT
Garra tibanica
OM540822
10
10GT
Garra tibanica
OM540823
11
11GT
Garra tibanica
OM540824
12
12GT
Garra tibanica
OM540825
2.2 DNA extraction for polymerase chain reaction
Biopsies were taken from the dorsal fin (avoiding taking the skin with the biopsies), and DNA extracted by DNAzol (Invitrogen, UK). The method is as follows: take 25–50 mg of fish tissue and place on glass slide, homogenize by repeated press with an edge sterile glass slide until yielding a very soft homogenate, then transferred to a 1.5 ml Eppendorf tube containing 500 µl of DNAzol. The mixture was vortexed and centrifuged for 2 min at 10,000 rpm at room temperature. The supernatant was saved and pellet was discarded. 500 ul of Absolut ethanol was added to supernatant and tubes were inverted gently many times until DNA clouds appeared, or small fragments precipitate, and stored at room temperature for 3 min, then centrifuged for 10 min at 10,000 rpm. The supernatant was discarded pellet was left to air dry for 30–40 min. The DNA pellet was dissolved in 50 ul nuclease free double distilled water. The concentration of the DNA was obtained by using NanoDrop 8000 (Thermo Scientific), using software version ND-8000 V2.2.1.
2.3 PCR amplification of cytochrome b
The cytochrome b gene was amplified using PCR (ProFlex PCR System, Applied biosystem) through utilizing primers L14724 (5′- GACTTGAAAAACCACCGTTG-3′) and H15915 (5′- CTCCGATCTCCGGATTACAAGAC-3′). The reaction mixture consists of 8.5 μl 2 × Taq Plus PCR MasterMix (Solarbio life sciences), 2 μl of forward and reverse primer mix solution, 7.5 μl D.D·H2O and 2 μl of 100 ng/μl DNA. PCR amplification conditions are as follows: 94 °C for three minutes, followed by 35 cycles of 94 °C for one minute and 50 °C for one minute, then 72 °C for one minute, and a final extension at 72 °C for eight minutes. The PCR product was documented under UV light on 1 % agarose gel containing ethidium bromide stain. The products of a PCR were sent to Macrogen, Inc. (Seoul, Republic of Korea) for sequencing.
2.4 Sequence analysis and phylogenetic tree construction
To build a phylogenetic tree in order to analyze the evolutionary relationships among Garra tibanica and related species, using software MEGA X (Kumar et al., 2018). The cytochrome b sequences of Garra tibanica were not known before this study, and these sequences were deposited to public gene data bank with accession numbers OM540814 - OM540825 (Table 1). Fifteen datasets from diverse species used for creating the phylogenetic trees were obtained from GenBank. The phylogenetic trees were built using maximum likelihood (ML) with genetic distance and the Tamura-Nei model (Kumar et al., 2016, Jeanmougin et al., 1998, Tamura and Nei, 1993). We used Felsenstein’s bootstrap method to calculate the associated taxa clustered in the bootstrap test, using 1000 replicates, and the data are written above the nodes. Estimates of Evolutionary Divergence between sequences were calculated as genetic distance matrix with MEGA X (version 10.2.6; (Kumar et al., 2018)). Genetic distance heatmap were ploted using CLUSTVIS web tool (Metsalu and Vilo, 2015).
3 Results
Sequence of the cytochrome b gene was obtained from twelve specimens of Garra tibanica. The amplified cytochrome b gene fragments had a read length 992 base pairs (bp). The ambiguous and gap-containing sites were removed, and the sequence obtained, showed that all samples collected in this study were identical. The divergence levels were compared with the sequences from this study and cytochrome b sequences with related Garra fish species from GenBank. The maximum likelihood estimation of the phylogenetic relationships placed all the samples into five groups (Fig. 1). The clusters, inside groups, were supported by high bootstrap values and revealed that G. tibanica and G. sahilia are related lineage in the same clade with 98.69 % identity and also it is in consistent with traditional morphologically-based inferences. Moreover, genetic distance matrix showed a value of 0,0134 of base substitutions per site between G. tibanica and G. sahilia (Table 2 and Fig. 2).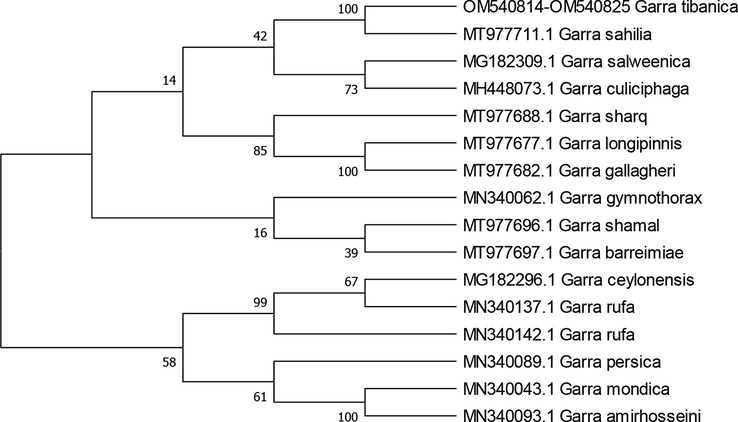

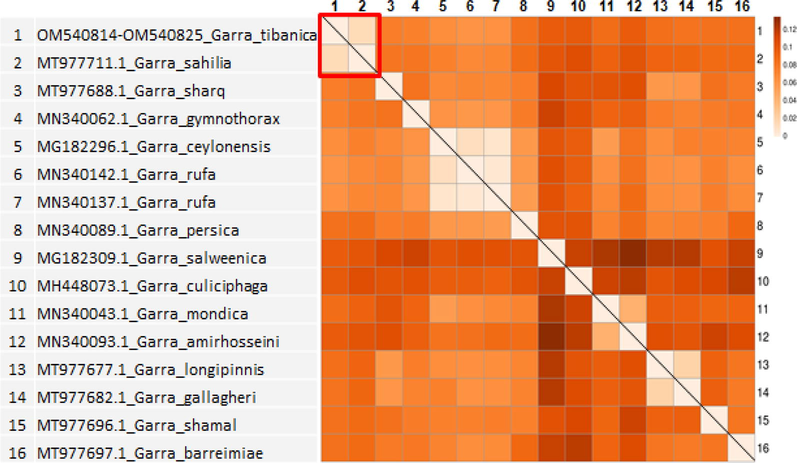
Genetic distance heatmap. The heatmap plot was constructed with genetic distance data (Table 2) using CLUSTVIS web tool (Metsalu and Vilo, 2015). Red square: subgroup with low distance, including G. tibanica and G. sahilia.
4 Discussion
The maximum-likelihood method was used to identify phylogeny relationship between species of Garra based on cytochrome b gene. The phylogenetic tree constructed in this study which was based on the cytochrome b gene sequences retrieved from GenBank, grouped Garra tibanica with Garra sahilia, Garra salweenica and Garra culiciphaga in one clade, this mean that G. tibanica is very closely related to G. sahilia, while G. rufa, G. gymnothorax and G. ceylonsis were grouped in another clade. This relationship may reflect the allopatric speciation of Garra according to its existence in nearby geographic areas. In order to verify this, further studies should be done using large sample sizes and more molecular tools (DNA barcodes).
The state of all freshwater fish in the Arabian Peninsula, especially Saudi Arabia, could have been changed over the years due to habitat loss, degradation due to altered environmental systems and increase in cultivation (Aljohani, 2019, Freyhof et al., 2015). Moreover the pollution and the current of climatic change and decline in precipitation could also have affected the papulation status of the fish in this region, hence phylogenetic studies using latest molecular biology techniques is an utmost important job to re assess the status of fresh water fish in Saudi Arabia (Moran, 2009; Yang et al., 2012; Macusi et al., 2015; Alotaibi et al., 2020).
Hamidan and Shobrak (2019) recently evaluated the status of fresh water fish species of the area, and collected fish samples between April and May 2013 from 22 sampling sites and reported eight native species and one introduced Cichlid species. Aljohani (2019) after studying 15 springs in Saudi Arabia concluded that Garra tibanica is a native fish species and suggested to reassess the Red List status of the species in the region.
Based on the analysis of cytochrome c oxidase subunit 1 (CO1) gene sequence, (Hamidan et al., 2014) reported that G. tibanica is not related to G. ghorensis genetically, however, it contradicts Krupp (1982) hypothesis based on similar morphology.
5 Conclusions
In this study we investigated, for the first time, the relationships between species of Garra endemic, that were collected from Al Madinah province, in Saudi Arabia using cytochrome b gene. The results of phylogenetic tree constructed that Garra tibanica is more closely related to Garra sahilia and Garra sharq More studies are needed using molecular biology tools to classify the distribution of all species of freshwater fish of genetically in order to determine the degree of similarity and relationship between the existing species. Based on these results further plans should be developed to protect endemic biodiversity, especially endangered and extinct threatened species.
Acknowledgements
“We extend our appreciation to the Deanship of Scientific Research at King Saud University for funding this work through the research group no. RG-1441-307”
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Springs of the Arabian Peninsula: Historical Trends, Current Status and Human Impact in Saudi Arabia. Oman and Jordan, University of South Florida; 2019.
- The freshwater fishes of Saudi Arabia; systematics and conservation. USA: Colorado State Univ; 1983. Ph. D. thesis,
- Almutair, L., 2016. Phylogenetic tree reconstruction inspired by bees algorithm(phylobee), Riyadh, King Saud university.
- Phylogenetic analysis of three endogenous species of fish from Saudi Arabia verified that Cyprinion acinaes hijazi is a sub-species of Cyprinion acinaces acinases. J. King Saud Univ.-Sci.. 2020;32:3014-3017.
- [Google Scholar]
- Adding nuclear rhodopsin data where mitochondrial COI indicates discrepancies–can this marker help to explain conflicts in cyprinids. DNA Barcodes. 2015;3:187-199.
- [Google Scholar]
- DNA barcoding to fishes: current status and future directions. Mitochondrial DNA Part A. 2016;27:2744-2752.
- [Google Scholar]
- Assessing accuracy and completeness of GenBank for eDNA metabarcoding: towards a reliable marine fish reference database. ARPHA Conference Abstracts 2021 Pensoft Publishers, e64671
- [Google Scholar]
- Tempo and mode of sequence evolution in mitochondrial DNA of HawaiianDrosophila. J. Mol. Evol.. 1987;26:157-164.
- [Google Scholar]
- Differentiating three Indian shads by applying shape analysis from digital images. J. Fish Biol.. 2020;96:1298-1308.
- [Google Scholar]
- The status and distribution of freshwater fishes of the Arabian Peninsula. The status and distribution of freshwater biodiversity in the Arabian peninsula. 2015;16
- [Google Scholar]
- 2021. Catalog of Fishes: Genera, Species, References; 2018.
- The status and distribution of freshwater biodiversity in the Arabian peninsula. Switzerland, Cambridge, UK and Arlington, USA: IUCN Gland; 2015.
- Garra jordanica, a new species from the Dead Sea basin with remarks on the relationship of G. ghorensis, G. tibanica and G. rufa (Teleostei: Cyprinidae) Ichthyological Exploration Freshwaters. 2014;25:223-236.
- [Google Scholar]
- An Update on Freshwater Fishes of Saudi Arabia. Jordan. Journal of Biological Sciences. 2019;12
- [Google Scholar]
- A century of morphological variation in Cyprinidae fishes. BMC Ecol.. 2016;16:1-18.
- [Google Scholar]
- Kerschbaumer, M., Sturmbauer, C., 2011. The utility of geometric morphometrics to elucidate pathways of cichlid fish evolution. International Journal of Evolutionary Biology, 2011.
- Hidden diversity—Delimitation of cryptic species and phylogeography of the cyprinid Garra species complex in Northern Oman. J. Zool. Syst. Evol. Res.. 2021;59:411-427.
- [Google Scholar]
- Ceratogarra, a genus name for Garra cambodgiensis and G. fasciacauda and comments on the oral and gular soft anatomy in labeonine fishes (Teleostei: Cyprinidae) Raffles Bull. Zoolog.. 2020;5:156-178.
- [Google Scholar]
- Garra tibanica ghorensis subsp. nov. (Pisces: Cyprinidae), an African element in the cyprinid fauna of the Levant. Hydrobiologia. 1982;88:319-324.
- [Google Scholar]
- Krupp, F., 1983. Fishes of Saudi Arabia: Freshwater Fishes of Saudi Arabia and Adjacen Regions of the Arabian Peninsula, Verlag nicht ermittelbar.
- MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol.. 2016;33:1870-1874.
- [Google Scholar]
- MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol.. 2018;35:1547.
- [Google Scholar]
- Linacre, A. 2012. Capillary electrophoresis of mtDNA cytochrome b gene sequences for animal species identification. DNA Electrophoresis Protocols for Forensic Genetics. Springer.
- The potential impacts of climate change on freshwater fish, fish culture and fishing communities. Journal of Nature Studies. 2015;14:14-31.
- [Google Scholar]
- ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res.. 2015;43:W566-W570.
- [Google Scholar]
- Moran, R. J. 2009. The impacts of climate change on freshwater fish, macroinvertebrates, and their interactions. University of Liverpool.
- PLAYFAIR, R. Note on a freshwater fish from the neighbourhood of Aden. Proc. Zool. Soc. Lond, 1870. 85-86.
- Resolving the effect of climate change on fish populations. ICES J. Mar. Sci.. 2009;66:1570-1583.
- [Google Scholar]
- Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol.. 1993;10:512-526.
- [Google Scholar]
- Cytochrome b or cytochrome c oxidase subunit I for mammalian species identification—an answer to the debate. Forensic Sci. Int.: Genet. Suppl. Series. 2009;2:306-307.
- [Google Scholar]
- Molecular phylogeny of the cyprinid tribe Labeonini (Teleostei: Cypriniformes) Mol. Phylogenet. Evol.. 2012;65:362-379.
- [Google Scholar]
- Yousefi, P. 2013. A Study of Cytochrome C Oxidase Subunit 1 Polymorphism in Malay Population. Universiti Teknologi Malaysia.
- Phylogenetic relationships of the Chinese Labeoninae (Teleostei, Cypriniformes) derived from two nuclear and three mitochondrial genes. Zoolog. Scr.. 2010;39:559-571.
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2022.102390.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







