Translate this page into:
Molecular detection of blaNDM and blaOXA-48 genes in Carbapenem-Resistant Klebsiella pneumoniae isolates from a tertiary care hospital
⁎Corresponding author at: Central Research Laboratory, Meenakshi Academy of Higher Education and Research (Deemed to be University), Chennai, India. n_arunagiri@yahoo.co.in (Narasingam Arunagirinathan)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background
The incidence of carbapenem-resistant Klebsiella pneumoniae (CRKP) infections is increasing globally. In India, around 65% of K. pneumoniae isolates are resistant to carbapenem antibiotics. The blaNDM is the predominant carbapenem-resistant gene in CRKP isolates. However, blaOXA-48 has also been reported to be increasing in recent days.
Methods
K. pneumoniae strains isolated from the clinical specimens of patients attending a tertiary care hospital in Hyderabad from June 2021 to May 2022 were included in this study. Resistance to carbapenems and other antibiotics was screened using Kirby-Bauer’s disc diffusion method. Phenotypic tests such as the Carbapenemase Nordmann Poirel (CarbaNP) test, Modified Carbapenem Inactivation Method (mCIM), and EDTA Carbapenem Inactivation Method (eCIM) were used to detect the carbapenemase producers. The genes responsible for carbapenemase production were detected by the real-time polymerase chain reaction (RT-PCR) method.
Results
Out of 1265 K. pneumoniae strains isolated, 241 (19 %) isolates showed resistance to any one of the carbapenem antibiotics tested. Phenotypic tests such as CarbaNP, mCIM, and eCIM revealed that 94.6 %, 97.1 %, and 95.4 % of CRKP isolates were carbapenemase producers, respectively. About 66.4 % of CRKP isolates harbored blaNDM, 17.4 % harbored blaOXA-48, and 16.2 % harbored both blaNDM and OXA-48 genes. None of the isolates tested positive for blaKPC, blaVIM, and blaIMP genes.
Conclusion
In this study, blaNDM is the most prevalent carbapenemase gene in CRKP isolates. Furthermore, blaOXA-48 and the co-existence of both blaNDM and blaOXA-48 genes were also found. CRKP is emerging as a serious threat among drug-resistant bacterial pathogens which would complicate the treatment of bacterial infections with available antibiotics.
Keywords
Carbapenem-resistant Klebsiella pneumoniae
CarbaNP test
Antibiotics
OXA-48
Real-time-PCR
1 Introduction
Carbapenems are the last resort drugs used to treat multidrug-resistant (MDR) bacterial infections. The incidence of carbapenem resistance is increasing across the globe. Treatment failure and eventually high mortality were reported among individuals with carbapenem-resistant bacterial infections (Falagas et al., 2014; Xu et al., 2017). In the clinical settings, high carbapenem resistance was reported in Klebsiella pneumoniae isolates (Veeraraghavan et al., 2017). In India, around 65 % of K. pneumoniae was reported for carbapenem resistance in 2016 (ICMR Annual Report, 2021). Due to their significant public health threat, the World Health Organization (WHO) declares that Carbapenem-resistant K. pneumoniae (CRKP) is one of the critical groups of drug-resistant pathogens for which new therapeutics need to be developed (World Health Organization, 2017). According to the Ambler classification, K. pneumoniae carbapenemase (blaKPC) was classified as a Class A enzyme which is widespread in Greece, Italy, and the United States. Metallo-β-lactamases (MBLs), New Delhi Metallo-β-lactamase (blaNDM), Verona Integron encoded Metallo-β-lactamase (blaVIM), and Imipenem carbapenemase (blaIMP) are classified as class B enzymes which are frequently reported in India, China, Japan, Russia, and Australia. Oxacillinases (blaOXA-48) are class D enzymes that are endemic in Turkey and also reported in France, Belgium, and North Africa (Nordmann et al., 2011).
In India, blaNDM is the most prevalent carbapenemase gene, and now blaOXA-48 and co-expression of both blaNDM and blaOXA-48 genes are increasingly reported (Tilahun et al., 2021; Veeraraghavan et al., 2017). Timely detection of CRKP infection and epidemiology of carbapenem-resistant genes are vital to optimize antibiotic therapies and develop infection control policies. In this study, we aimed to determine the distribution of carbapenem-resistant K. pneumoniae in a tertiary care hospital in Southern India.
2 Materials &Methods
2.1 Samples and bacterial isolates
This study was carried out in a tertiary care hospital in Hyderabad from June 2021 to May 2022. The ethical approval for this study was obtained (MRNH IEC/TS/75/2022). A total of 1265 K. pneumoniae strains isolated from clinical specimens (wound swabs, urine, blood, pus, sputum, endotracheal secretion, bronchoalveolar lavage fluid, and tracheal aspiration) were included in the study. Other bacterial strains isolated from the study samples were not included in the analysis. Stool samples were excluded from the study as they could contain K. pneumoniae as normal flora from the human intestine.
Bacterial identification was done by standard culture and biochemical tests. Kirby-Bauer’s test was used to determine the antimicrobial susceptibility testing for cefotaxime (CTX, 30 μg), cefoxitin (CX, 10 μg), ceftazidime (CAZ, 30 μg), amikacin (AK, μg), piperacillin/tazobactam (PT, 100/10 μg), gentamicin (GM, 10 μg), ciprofloxacin (CIP, 5 μg), meropenem (MRP, 10 μg), imipenem (IMP, 10 μg), and ertapenem (ETP, 10 μg) (Himedia, India) (Clinical and Laboratory Standards Institute, 2020). K. pneumoniae isolates that exhibit resistance to any one of the carbapenems used in this study were considered as CRKP and they were further screened for phenotypic and genotypic confirmatory tests.
2.2 Phenotypic confirmatory tests
Phenotypic confirmatory tests such as the CarbaNP, Modified Carbapenem inactivation (mCIM), and EDTA Carbapenem inactivation (eCIM) were used to screen carbapenemase-producing K. pneumoniae (Clinical and Laboratory Standards Institute, 2020).
2.3 CarbaNP test
In this test, 100 µL of lysis buffer (20 mM Tris-HCl) was added into two 1.5 mL microcentrifuge tubes (tubes A and B). Loopful colonies from the Muller Hinton Agar (MHA) plate were taken using a 10 µL inoculation loop and emulsified in lysis buffer. The 100 μL of phenol red solution (pH 7.8) containing 0.1 mmol/L ZnSO4 (HiMedia, India) was added in tube A and 12 mg/mL of imipenem/cilastatin was added along with phenol red solution in tube B. Both tubes were incubated at 37 °C for 2 h and color changes were observed every 15 min. If tube B turns yellow or dark yellow, while tube A remains red, the test is considered positive (Lee et al., 2022).
2.4 mCIM and eCIM tests
In these tests, bacterial culture suspension was made in two tubes containing 2 mL of Trypticase soya broth (TSB) (Himedia, India). In one tube, 0.5 mM EDTA was added (eCIM) and in another tube, it was not added. A 10 μg meropenem (MEM) disc was added into both tubes and incubated at 37 °C for 4 h ± 15 min. Then the discs were removed and placed on MHA plates containing lawn culture of E. coli ATCC 25922 (carbapenem susceptible). The plates were incubated at 37 °C for 16 to 20 h (Lee et al., 2022). Results were interpreted as follows:
-
mCIM: The zone of inhibition of 6–15 mm and the presence of colonies were considered positive.
-
An inhibition zone of ≥ 19 mm was considered negative.
-
eCIM: Further increase in inhibition zone of ≥ 5 mm compared to mCIM was considered positive and an increase of < 5 mm was considered negative. Only positive isolates of mCIM were further tested using eCIM.
2.5 Bacterial DNA extraction
The bacterial DNA of CRKP isolates was extracted using the spin column method (Biopro, India). About 5–10 bacterial colonies from the MHA plate were emulsified in 1.0 mL of nutrient broth in a 1.5 mL micro-centrifuge tube and incubated overnight at 37 °C. After incubation, the tube was centrifuged at 8000 rpm for 3 min and the supernatant was discarded. The pellet was suspended in 200 μL of phosphate buffer (pH 7.2) followed by the addition of 200 μL of lysis buffer and incubated for 10 min at 37 °C. Then, 350 μL of a binding buffer was added to precipitate DNA. The whole lysate was transferred to the spin column and centrifuged at 8000 rpm for 1 min and washing was done by adding 600 µL of wash buffer and then centrifuged at 8000 rpm for 30 sec. Finally, the spin column was placed in a new 1.5 mL tube followed by addition of 100 μL of elution buffer and centrifuged at 8000 rpm for 30 sec. Eluted DNA was stored at -20 °C for PCR amplification of target genes.
2.6 Detection of carbapenem-resistant genes
Extracted DNA was used for the amplification of carbapenem-resistant genes using a Hi-PCR carbapenemase multiplex real-time PCR kit (Himedia, India). The kit has two-tube PCR assays, in tube one, primers and probes for blaKPC, blaNDM, blaVIM, and blaIMP, and in the second tube, primer and probe for blaOXA48 were added. The test was conducted using a 25 μL reaction mixture (Master Mix: 20 μL and DNA template: 5 μL). The PCR thermal conditions used were 95 °C for 10 min for initial denaturation, 95 °C for 05 sec for denaturation, and 60 °C for 1 min for annealing and extension with 45 cycles.
3 Results
Out of 1265 K. pneumoniae isolates, 241 (19 %) isolates were resistant to carbapenem antibiotics. The majority (n = 223, 92.5 %) of the CRKP strains were isolated from individuals admitted to the In-Patient Department (IPD). Table 1 shows the distribution of CRKP strains in various wards. About 76 % of the isolates were obtained from the age group of 41–60 years followed by 24 % from the 21–40 age group (Fig. 1). CRKP strains were majorly isolated from pus samples (31.5 %, n = 82) followed by urine (22.9 %, n = 64) (Table2).
S. No
Ward
No. of CRKP Isolates
Percentage of CRKP Isolates
1
Medical ward
71
29.5
2
Surgical ward
18
7.5
3
Orthopedics ward
13
5.4
4
Pulmonology ward
18
7.5
5
Emergency ward
8
3.3
6
Post-operative ward
7
2.9
7
OBG (Obstetrics & Gynecology) ward
7
2.9
8
OPD (Outpatient Department)
18
7.5
9
SICU (Surgical Intensive Care Unit)
39
16.2
10
MICU (Medical Intensive Care Unit)
29
12.0
11
CCU (Critical Care Unit)
11
4.5
12
ICU (Intensive Care Unit)
2
0.8
Total
241
100.0
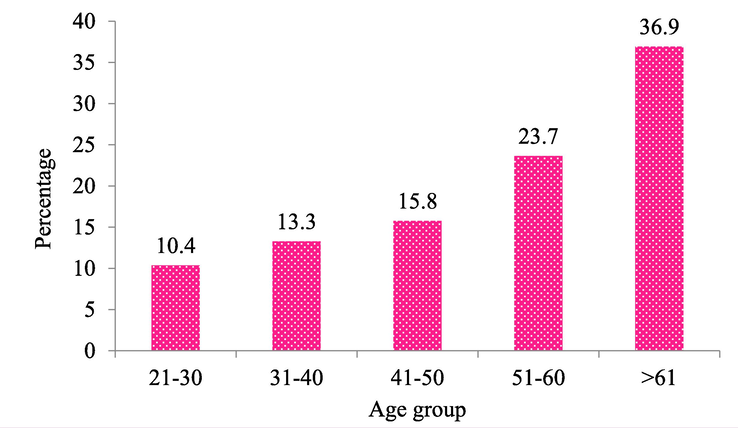
Age-wise Distribution of CRKP Isolates.
S.No
Type of Sample
No. of Isolates
(n = 1265)No. of CRKP Isolates
(n = 241)Percentage of CRKP Strains
1
Pus
260
82
31.5
2
Urine
279
64
22.9
3
Sputum
169
35
20.7
4
Wound swabs
158
19
12.0
5
Endotracheal Secretion
109
12
11.0
6
Bronchoalveolar Lavage fluid
112
12
10.7
7
Blood
153
15
9.8
8
Tracheal aspiration
25
2
8.0
3.1 Antibiotic susceptibility profile
In this study, all carbapenem-resistant isolates were resistant to cefotaxime, cefoxitin, amikacin, and gentamycin. (Fig. 2). The resistance rates for piperacillin-tazobactam, ceftazidime, and ciprofloxacin were 97.9 %, 97.5 %, and 97.1 %, respectively.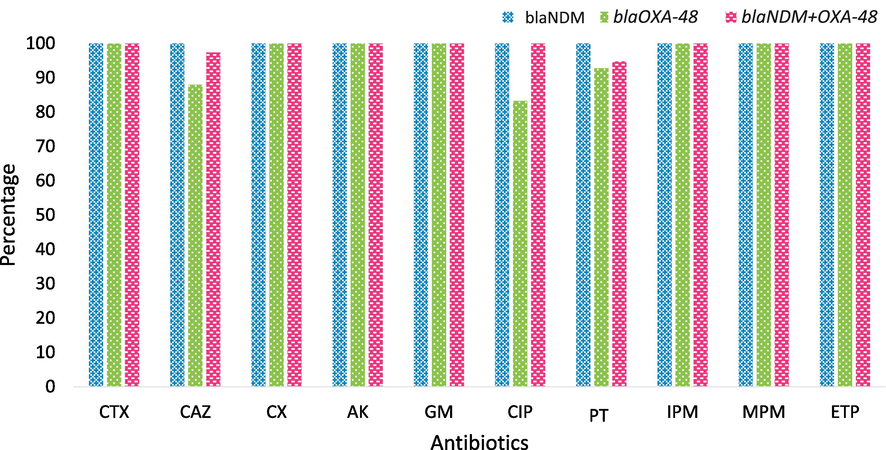
Antibiotics Resistance Profile of CRKP Isolates. CTX – Cefotaxime; CAZ- Ceftazidime; CX- Cefoxitin; AK- Amikacin; GM – Gentamicin; CIP- Ciprofloxacin; PT- Piperacillin/Tazobactam; IPM – Imipenem; MPM- Meropenem; ETP – Ertapenem.
3.2 Phenotypic detection of carbapenemase producers
All the 241 isolates resistant to carbapenem antibiotics were tested for phenotypic screening of carbapenemase production. About 94.6 % (n = 228) of CRKP isolates showed positive for carbapenemase production by CarbaNP test. The mCIM and eCIM tests detected carbapenemase production in 97.1 % (n = 234) and 88.0 % (n = 206) of CRKP isolates, respectively. It was observed that the mCIM test has the highest sensitivity (97.1 %) followed by CarbNP (94.6 %) and eCIM (88.0 %) tests. All three tests showed 100 % specificity for the detection of carbapenemase producers (Fig. 3).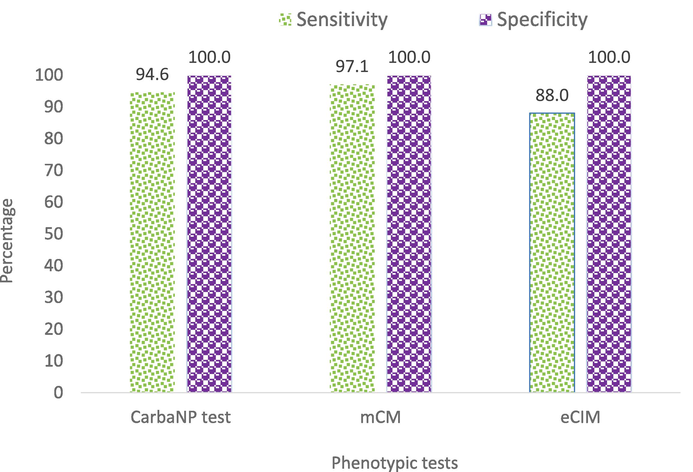
Sensitivity and Specificity of Phenotypic Testes for Detection of Carbapenemases Production.
3.3 Detection of carbapenemase encoding genes
All 241 isolates showed positive for at least one carbapenemase-producing gene tested. The most predominant gene was blaNDM (n = 160, 66.4 %) followed by blaOXA-48 (n = 42, 17.4 %). Thirty-nine (16.2 %) isolates carried both blaNDM and blaOXA-48 genes and none of the isolates carried blaKPC, blaVIM, and blaIMP genes.
4 Discussion
K. pneumoniae is one of the important bacterial species in the Enterobacteriaceae family and it shows a higher resistance profile to antibiotics, especially to carbapenem antibiotics. In this current study, about 19 % of K. pneumoniae were found to be resistant to carbapenem. In other studies, CRKP was reported as 17.6 % (Sood, 2014) and 27.5 % (Mohanty et al., 2017). We observed that the majority of isolates were obtained from in-patient samples (92.5 %). CRKP strains were isolated in more numbers among the hospitalized patients. In this study, an analysis was not done to differentiate community and hospital-acquired CRKP infections. A previous study from Mumbai, India reported that 19 % of the carbapenem-resistant strains were isolated from out-patient samples (Qureshi et al., 2014). But in this study, it was only about 8 % from out-patient samples. Furthermore, the majority of CRKP isolates were obtained from pus samples (31.5 %) followed by urine (22.9 %). However, in contrast to our study, a study reported that urine samples were the major source of CRKP strains (Nagaraj et al., 2012).
The antibiotic profile showed that all the isolates were resistant to meropenem and ertapenem. Previous studies reported that the Ambler class B (blaNDM) enzymes had the highest carbapenemase activity with a wide range of hydrolytic activity against penicillins, cephalosporins, and carbapenems (Nordmann et al., 2012a). In this current study, the isolates carrying blaNDM were resistant to all tested antibiotics. Changes in the bacterial inoculum size, pH of the culture medium, incubation temperature, and multiple beta-lactamase production could affect the accuracy of the disc diffusion method (Birgy et al., 2012). Therefore, confirmatory testing is essential to detect carbapenemase-producing Enterobacteriaceae.
In this study, CRKP isolates were further tested by CarbaNP test, mCIM, and eCIM to confirm the production of carbapenemase enzymes. CLSI recommends these tests and they are rapid, inexpensive, and have high sensitivity and specificity to detect carbapenem-resistant isolates (Nordmann et al., 2012b; Tamma and Simner, 2018). In this study, using the CarbaNP test, it was found that 94.6 % of K. pneumoniae isolates produced carbapenemases. The average detection time of carbapenemase producers using this method was 24.1 min. In a study, CarbaNP test showed 97.9 % sensitivity and 100 % specificity (Dortet et al., 2014). Furthermore, CarbaNP test identified all blaNDM, blaKPC, blaIMP, and blaVIM-positive isolates as carbapenem producers. However, CarbaNP test failed to detect four blaOXA-48-like gene-positive isolates (Dortet et al., 2014). The low sensitivity was reported for CarbaNP test among blaOXA48 gene-harboring isolates (Armin et al., 2021). But in this study, all OXA-48 gene-positive isolates had shown positivity for carbapenemase production by CarbaNP test. However, negative results were observed for the isolates carrying both blaNDM+OXA48 genes.
The mCIM is a phenotypic test known for its high sensitivity and specificity in detecting carbapenemase producers (Van Der Zwaluw et al., 2015). In the current study using mCIM test, it was identified that 97.1 % of carbapenem-resistant isolates were carbapenem producers. The mCIM test showed 93.3 % sensitivity and 100 % specificity in a study by Yıldız et al. (2017). It is important to monitor the carbapenemase producers since this could complicate the selection of antibiotics for treatment. In India, carbapenem resistance is mainly due to blaNDM and blaOXA-48 enzymes. Other carbapenemase genes such as blaKPC, blaVIM, and blaIMP were rarely seen (Mohanty et al., 2017; Veeraraghavan et al., 2017).
Isolates carrying blaNDM are resistant to many antibiotic classes like β-lactams, fluoroquinolones, and aminoglycosides which were responsible for significant mortality ranging from 18 % to 67 % (Jaggi et al., 2019; Nordmann et al., 2011) due to the treatment failure. In our study, 66.4 % of isolates harbored the blaNDM gene. We have not recorded the patient recovery status for this study. Mohanty et al. in 2017 reported that 65 % of carbapenem-resistant K. pneumoniae harbored the blaNDM gene. The blaOXA-48 has been the second most carbapenemase producer in recent years. This enzyme was first reported in 2004 in Istanbul, Turkey, and from there it was disseminated worldwide (Nordmann et al., 2011). In the current study, 17.4 % of isolates harbored the blaOXA-48 gene, and our results closely correlated with the results of Jaggi et al. (2019) who reported that 15.4 % of CRKP strains harbored the blaOXA-48 gene. In another study, the blaOXA-48 gene was detected in 44.5 % of carbapenem-resistant isolates (Pawar et al., 2020).
An isolate carrying multiple carbapenem-resistant genes is a major threat to public health since it could have a broad-spectrum resistance profile (Armin et al., 2021; Van Der Zwaluw et al., 2015). In the current study, co-harboring of blaNDM + OXA48 genes was observed in 16.2 % of CRKP isolates and these isolates had shown resistance to multiple classes of antibiotics which could be the major concern in treating patients with carbapenem-resistant bacterial infections. In another study by Veeraraghavan et al. (2017), it was reported that 28 % of isolates produced both blaNDM and blaOXA-48. They also reported that CRKP infections were treated with colistin and tigecycline, in combination with meropenem. This aspect clearly shows that CRKP infections require a high class of antibiotics for treatment and sometimes a combination of drugs is required. It is a serious alarm in clinical settings that CRKP infections could limit the antibiotics for treatment and complicate the recovery of patients from the infections. Many studies reported hospital-acquired CRKP infections and their treatment failure. Infection Control Policies should be strictly implemented in hospitals to control the spread of infectious agents especially multidrug-resistant pathogens (Nagaraj et al., 2012, Falagas et al., 2014, Qureshi et al., 2014, Aldali et al., 2023). Otherwise, these carbapenem-resistant genes can rapidly spread to other bacterial species in Enterobacteriaceae, which could lead to an increased incidence of carbapenem-resistant bacterial infections (Kamalakar et al., 2023).
5 Conclusion
This study outcomes revealed that blaNDM is the most prevalent carbapenemase gene in CRKP isolates. Furthermore, blaOXA-48 and the co-existence of both blaNDM and blaOXA-48 genes are emerging in CRKP isolates. The infections caused by CRKP are a serious public health concern, especially in patients under treatment in hospital settings. Strict implementation of Infection Control Policies would reduce the carbapenem-resistant bacterial infections spreading to the community. Performing regular surveillance studies in hospitals is very important to identify the circulating genes among carbapenem-resistant bacteria which will help to overcome the therapeutic challenges in treating the patients infected with CRKP.
CRediT authorship contribution statement
Sarva Kamalakar: Conceptualization, Formal analysis, Methodology, Writing – original draft, Investigation. Marimuthu Ragavan Rameshkumar: Conceptualization, Data curation, Formal analysis, Validation, Writing – original draft, Writing – review & editing. Tadi Lakshmi Jyothi: Conceptualization, Data curation, Writing – original draft. Raja Sundaramurthy: Conceptualization, Formal analysis, Writing – original draft, Data curation. Balasubramanian Senthamilselvan: Formal analysis, Writing – original draft. Arunagirinathan Nishanth: Formal analysis, Writing – original draft. Chandrasekaran Krithika: Conceptualization, Data curation, Formal analysis, Resources, Writing – original draft, Writing – review & editing. Hissah Abdulrahman Alodaini: Funding acquisition, Writing – review & editing. Ashraf Atef Hatamleh: Funding acquisition, Writing – review & editing. Narasingam Arunagirinathan: Conceptualization, Methodology, Resources, Supervision, Validation, Writing – review & editing, Project administration.
Acknowledgment
The authors would like to express their sincere thanks to the Researchers Supporting Project number (RSP2024R479), King Saud University, Riyadh, Saudi Arabia.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that influence the work reported in this paper.
References
- Hospital-acquired infections caused by carbapenem-resistant Enterobacteriaceae: an observational study. Microorganisms. 2023;11:1595.
- [CrossRef] [Google Scholar]
- Multicentre study of the main carbapenem resistance mechanisms in important members of the Enterobacteriaceae family in Iran. New Microbes New Infect.. 2021;41:100860
- [CrossRef] [Google Scholar]
- Phenotypic screening of carbapenemases and associated β-lactamases in carbapenem-resistant Enterobacteriaceae. J. Clin. Microbiol.. 2012;50:1295-1302.
- [CrossRef] [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing. In: 28th Informational Supplement. CLSI Document M100–S25. Wayne, PA: CLSI; 2020.
- [Google Scholar]
- Rapid detection of carbapenemase-producing Enterobacteriaceae from blood cultures. Clin. Microbiol. Infect.. 2014;20:340-344.
- [CrossRef] [Google Scholar]
- Deaths attributable to carbapenem-resistant Enterobacteriaceae infections. Emerg. Infect. Dis.. 2014;20:1170-1175.
- [CrossRef] [Google Scholar]
- Antimicrobial resistance research and surveillance network. New Delhi, India: Indian Council of Medical Research; 2021.
- Carbapenem resistance in Escherichia coli and Klebsiella pneumoniae among Indian and international patients in North India. Acta. Microbiol. Immunol. Hung.. 2019;66:367-376.
- [CrossRef] [Google Scholar]
- Performance of carbapenemase Nordmann-Poirel, Modified carbapenem inactivation, and EDTA carbapenem inactivation methods for detecting carbapenem-resistant Klebsiella pneumoniae isolates. Microb. Drug Resist.. 2023;29:504-509.
- [CrossRef] [Google Scholar]
- Geographic patterns of global isolates of carbapenem-resistant Klebsiella pneumoniae and the activity of ceftazidime /avibactam, meropenem/vaborbactam, and comparators against these isolates: Results from the antimicrobial testing leadership and surveillance (ATLAS) program, 2020. Int. J. Antimicrob. Agents. 2022;60:106679
- [CrossRef] [Google Scholar]
- Identification of carbapenemase-mediated resistance among Enterobacteriaceae bloodstream isolates: a molecular study from India. Indian J. Med. Microbiol.. 2017;35:421-425.
- [CrossRef] [Google Scholar]
- Carbapenem resistance among Escherichia coli and Klebsiella pneumoniae in a tertiary care hospital in south India. Indian J. Med. Microbiol.. 2012;30:93-95.
- [CrossRef] [Google Scholar]
- Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis.. 2011;17:1791-1798.
- [CrossRef] [Google Scholar]
- Identification and screening of carbapenemase-producing Enterobacteriaceae. Clin. Microbiol. Infect.. 2012;18:432-438.
- [CrossRef] [Google Scholar]
- Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis.. 2012;18:1503-1507.
- [CrossRef] [Google Scholar]
- Rising threat of OXA-48 and other carbapenemase encoding genes among carbapenem resistant Enterobacteriaceae in India. J. Pure Appl. Microbiol.. 2020;14:1917-1925.
- [CrossRef] [Google Scholar]
- Epidemiology and clinical outcomes of patients with carbapenem-resistant Klebsiella pneumoniae Bacteriuria. Antimicrob. Agents Chemother.. 2014;58:3100-3104.
- [CrossRef] [Google Scholar]
- Sood, S., 2014. Identification and differentiation of carbapenemases in Klebsiella pneumoniae: A phenotypic test evaluation study from Jaipur, India. J. Clin. Diagn. Res. DOI: 10.7860/JCDR/2014/7027.4614.
- Phenotypic detection of carbapenemase-producing organisms from clinical isolates. J. Clin. Microbiol.. 2018;56:e01140-e01218.
- [CrossRef] [Google Scholar]
- Emerging carbapenem-resistant Enterobacteriaceae infection, its epidemiology and novel treatment options: a review. Infect. Drug Resist.. 2021;14:4363-4374.
- [CrossRef] [Google Scholar]
- The Carbapenem inactivation method (CIM), a simple and low-cost alternative for the CarbaNP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS ONE. 2015;10:e0123690.
- [Google Scholar]
- Carbapenem resistant Klebsiella pneumoniae isolated from bloodstream infection: Indian experience. Pathog. Glob. Health.. 2017;111:240-246.
- [CrossRef] [Google Scholar]
- WHO publishes list of bacteria for which new antibiotics are urgently needed. Geneva: Switzerland; 2017.
- Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann. Clin. Microbiol. Antimicrob.. 2017;16:18.
- [CrossRef] [Google Scholar]
- Performance of CarbaNP and CIM tests in OXA-48 carbapenemase-producing Enterobacteriaceae. Acta. Microbiol. Immunol. Hung.. 2017;64:9-16.
- [CrossRef] [Google Scholar]







