Translate this page into:
Molecular based associations of bacterial pathogens with citrus: Implications for toxicology and disease management
⁎Corresponding authors. iramzaheer00@gmail.com (Iram Zaheer), shaziairam@fjwu.edu.pk (Shazia Iram)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
The losses associated with the bacterial diseases of citrus leaves and fruits have been reported throughout all of Pakistan's citrus-growing regions and a lot of work was done on the morphological characterization of pre and post-harvest pathogens. However, this issue has not been addressed in Khanpur, which is a major citrus-producing city in Pakistan and can help in the economic growth of the country. The citrus fruits in the area under study exhibit disease symptoms during pre and post-harvesting, but the actual disease agents have not yet been identified. The retrospective object of the current research is to study the pathogenic variation along with the investigation of bacterial disease incidence i.e. citrus canker, bacterial spots, & HLB in citrus fruit. During a survey conducted in November 2019 and 2020, bacterial isolates from symptomatic citrus fruits were obtained from randomly chosen orchards in Khanpur.The disease assessment was estimated by determining prevalence, severity, and disease incidence (DI). The bacterial isolates were identified morphologically and further through 16S rRNA gene amplification. The identified species were Pseudomonas syringae, Pseudomonas viridiflava, Serratia marcescens, Xanthomonas axonopodis pv citri, and Candidatus liberibacter. In the pathogenicity test, the Xanthomonas strains responsible for the canker disease caused the most severe disease symptoms. To our knowledge, this is the first report on pathogens of citrus fruits in Khanpur. Given the significant contribution of citrus fruits to household income and foreign exchange in Pakistan, this study illustrates the molecular-based association of bacterial pathogens of citrus and the toxic behavior of pathogens which helps to provide recommendations on genetic testing and to develop management programs for disease management. The molecular identification of citrus pathogens will empower the citrus growers, researchers, and regulatory agencies to make informed decisions that will protect citrus crops, reduce economic losses, and promote sustainable agriculture. It is also likely to play an increasingly vital role in the future of citrus cultivation..
Keywords
Citrus canker
Diagnosis
Aggressiveness
Pathogenicity
16S rRNA marker
Phylogeny
Yield loss
Fruit quality
1 Introduction
The Citrus fruit belongs to a well-known plant family named Rutaceae, which is grown all over the world in tropical and subtropical climates. Citrus is said to have originated in the southern Himalayan region, the entire northeastern part of India, and neighboring China, which leads the world in both production and area among tree fruits (Wu et al., 2018). Due to its great nutritional value as a source of sugar, organic acids, amino acids, vitamin C, organic sugar, and minerals including calcium and magnesium, it plays an essential role in human health. It is used throughout the world in both fresh and processed forms.
Citrus stands out among all of the major fruits produced worldwide. According to statistics from the Food and Agriculture Organization of the United Nations for 2021, citrus fruits were the second most produced fruit globally, with 161.8 million tons grown on more than 10.2 million hectares. It is possible to single out China, Brazil, and India as the top three producers. The greater production of tangerines and other citrus fruits is mostly responsible for China's high productivity. Brazil is the biggest exporter of orange juice and the biggest producer of oranges. India is also the world's top producer of limes and lemons. In addition, it is important to recognize that Mexico, Spain, the United States of America, Turkey, Egypt, Nigeria, and Iran are among the top citrus producers in 2021 (Gonzatto and Santos, 2023).
Citrus fruits make up a major portion of Pakistan's GDP, which is mostly driven by the agricultural sector. 98 % of Pakistan's citrus fruit production comes from Punjab, and Kinnow is the kind that is grown here in the greatest quantity. According to the Pakistan Bureau of Statistics, Pakistan produced approximately 2.5 million metric tons of citrus fruit in 2020–21, ranking it as the 10th greatest citrus grower globally. Pakistan is the world's top producer of Kinnow. Citrus fruit production in Pakistan increased by an average of 3.95 % per year, from 495,000 tons in 1972 to 2.33 million tons in 2021 (Pakistan Bureau of Statistics, 2021).
The citrus crop is produced in large quantities and sold as fresh as well as processed fruit byproducts. All fruit portions can be eaten; this is a characteristic of citrus fruits only. The inner mesocarp rind and the outside exocarp are used to extract flavorings and pectin, respectively. The peel is used to extract essential oils for usage in the pharmaceutical and cosmetic industries. Citrus pulp pellets that are made from the pulp and peel after the juice and oils have been extracted are used as animal feed. Products made from seeds include seed oils, dried seed-pressed cakes, and seed meals (Bampidis and Robinson, 2006; Choi, 2006; Bermejo and Cano, 2012).
The accomplishment of long-term disease and pest management measures is essential for citrus output. Citrus is among the few fruit crops that are susceptible to a variety of deadly diseases that are constantly evolving and that can drastically reduce or eliminate citrus output. Citrus hosts a variety of viruses and pests, like many tropical and subtropical crops (Narciso et al., 2012). Citrus fruit production for the processed market might not require as strict pest control as citrus fruit production for the fresh fruit market. But maintaining profitable citrus plantations devoted to processing or fresh-fruit markets as well as developing a certification system needs a thorough grasp of the pathogen's biology (Berk, 2016).
Citrus fruits are an essential crop in Pakistan, and several diseases can affect citrus trees in the country. These diseases can have a significant impact on citrus fruit quality and overall production. Some common citrus bacterial diseases reported in Pakistan, include citrus canker caused by the pathogen Xanthomonas axonopodis) (Tahir and Aslam, 2019), citrus tristeza virus (CTV) (Atta et al., 2017), citrus greening also known as the Huanglongbing, (HLB) caused by the Candidatus liberibacter (Javed, 2016; Khan et al., 2019).
Even though Khanpur, Pakistan, the site of our research, is a substantial producer of citrus, which is not only a nutritious fruit but also an economically valuable asset for the country, it is being deteriorated due to pathogen invasions and diseases, these diseases have not yet been researched and reported there. An effort has been made in the current study, to summarize the current state of knowledge regarding the primary bacterial pathogens restricting citrus production, the diseases they cause, their diagnosis through the use of molecular markers, toxicity mechanisms, genetic diversity, and the factors influencing the emergence of the causal agents of these diseases. We also discussed the interactions between pathogens and plants. By investigating the epigenetics of citrus diseases in Pakistan, the findings of this study will assist future researchers in locating transgenic lines resistant to HLB and citrus canker. This will also benefit future researchers who are eager to control diseases in an environmentally friendly manner.
2 Materials and methods
2.1 Study area and sample collection
Citrus orchards situated in the Khanpur, Haripur district Pakistan were chosen for this investigation (Fig. S1). Fig. S2 displays the schematic flowchart of the steps followed during this research. Surveys were conducted in December 2019 and December 2020 between 50 randomly chosen Khanpur orchards, and 15 to 20 samples of leaves and fruits with visible disease symptoms were taken from each of the orchards. To evaluate bacterial leaf and fruit diseases using the criteria developed for disease assessment and subsequent investigation, diseased fruit samples were tagged, brought to the Mycology and Ecotoxicology laboratory, and maintained at laboratory conditions (Akhtar and Alam, 2002; Ahmad et al., 2018).
2.2 Disease assessment
Depending on the field layout and orchard type for the evaluation of diseases, a random sample strategy was adopted. To record orchard and tree-wise data, a Performa was created and placed in various areas. To assess the prevalence and severity of bacterial infections, surveys were conducted in 50 citrus orchards spread across 50 randomly chosen sites in Khanpur. Blocks of each orchard were randomly selected, and samples were taken from those blocks. Five plants from each block were examined for disease infestation. Several citrus varieties, including Succri, Moro blood, Malta, and Musambi, were investigated. From each orchard, approximately 30 plants were carefully examined by visual inspection to detect diseases that makes a total of 1500 samples that were evaluated (Pereira et al., 2011). Trees were in the fruiting stage at the time, which made it easier to gather information on diseased fruits. Walking around the circumference of a few chosen trees allowed for a closer look at their trunks, shoots, leaves, and fruits for symptoms of the disease. In addition, leaves and fruits that had peculiar symptoms fell to the ground were seen. All of this data was entered on a main data collecting sheet. To determine differences in incidence and severity, these recorded data were finally analysed and interpreted. Fig. 1 displays a few of the disease signs that have been identified in various orchards.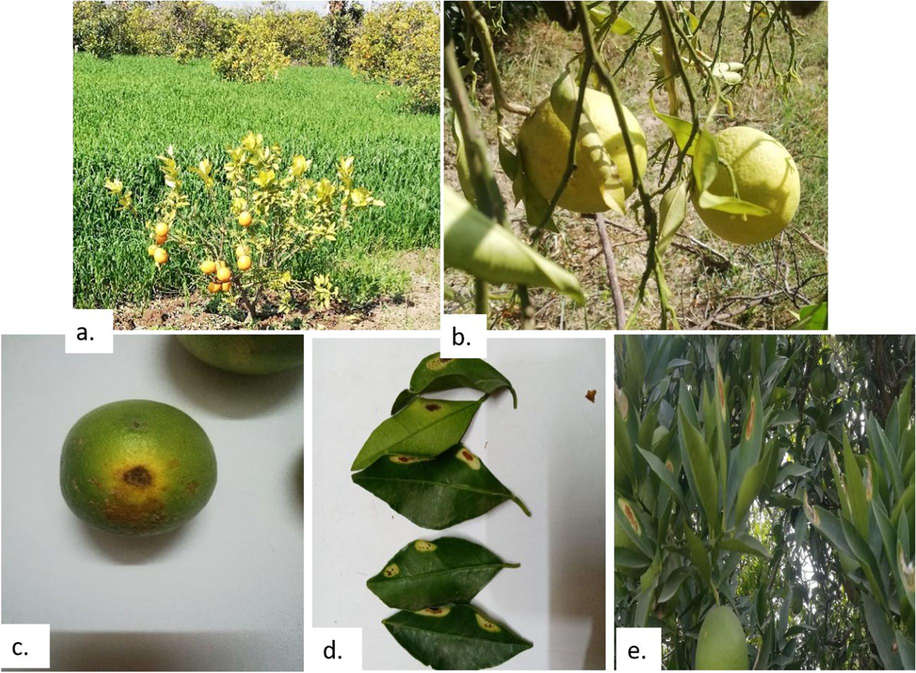
Different citrus bacterial diseases observed in the orchards: a, b) Citrus Greening, c, d) Citrus canker, & e) Bacterial spot.
The following formula was used to determine the disease incidence in each orchard (Fateh et al., 2022).
By Kazmi et al. (2005), the disease severity was assessed using a 0–5 visual rating scale. Where 0 represents healthy plants, 1 represents a 1 to 10 % decline, 2 represents 11 to 20 % decline, 3 represents a 21 to 30 % decline, 4 represents a 31 to 50 % decline, and 5 represents a decline of more than 50 %.
Percent Disease Index was determined by the formula shown below in each orchard (Mc-kinney, 1923; Verma et al., 2022).
2.3 Isolation of bacterial pathogens
The leaf samples were washed first with sterilized distilled water and then sponged with cotton soaked in alcohol under the laminar hood. Then with the aid of a sterile blade and sterilized test tubes, infected leaf tissue was removed. 75 % ethanol was applied to the removed tissues to surface sterilize them. Tissues were crushed, and the suspension was left at room temperature for half an hour in sterilized Phosphate Buffer Saline (PBS) pH 7.4. Fruits with lesions were peeled, and a 2 mm portion of the peel was sliced using a sterile scalpel around the lesion. Application of 75 % ethanol was used to sterilize these tissues. After that, these sliced tissues were cut into small pieces and immersed in PBS buffer. Then 200 µL of both fruits and leaves PBS suspensions were streaked on Yeast Peptone Glucose Agar (YPGA) media plates (Aritua et al., 2006). Different agar plates were streaked with samples from each collection. To prevent contamination, plates were parafilm-wrapped and tagged with the sample ID. After that, the plates were then incubated for 24–48 h at 28–30 °C.
2.4 Molecular identification and characterization of bacterial isolates
After the morphological observation of bacterial isolates on agar plates, the gram staining test was carried out for the screening and differentiating of bacterial isolates. To identify the separated strains, epigenetic (molecular) characterization was done utilizing 16S rRNA gene amplification and sequence analysis. Following the protocols of Naseem et al. (2017), from the overnight grown bacterial culture in YPG broth, 1000 μL with an optical density of 0.8 was taken and centrifuged for 10 min at 10,000 rpm to extract the bacterial DNA. Then the pellet formed was suspended in 500 μL of CTAB extraction buffer (NaCl 250 mM, Tris-HCl pH 7.5 200 mM, SDS 0.5 %, EDTA 25 mM, and 2 % PVP), with the supernatant being discarded. After that, the mixture was vortexed and continuously shaken at room temperature for 60 min. Then the lysate was centrifuged for five minutes at 8000 rpm. A fresh eppendorf tube was filled with a 450 μL sample of supernatant and an equivalent volume of isopropanol was added to it and stirred. The mixture was again centrifuged for 10 min at 13,000 rpm. The pellet formed was air dried for some time with the supernatant being discarded and was suspended in 50 μL TE Buffer (EDTA 1 mM, Tris-HCl 10 mM). The extracted DNA sample was run on the agarose gel and purified DNA samples were stored at −20 °C for further processing (Naseem et al., 2017).
2.5 PCR amplification
The PCR amplifications and16S rRNA gene sequencing analysis of 1500 bp fragment was done using the universal primer pair 27F (5-CTTCAACTCAAACGCCGGA-3) and 1492R (5-CATCGGCTGTTCGGGAG-3) (Abu-Obeid et al., 2018). In the Labnet MultiGene OptiMax thermal cycler, reactions were conducted. For each 25 µL reaction mixture, reaction buffer, 25 mM MgCl2, dNTPs, forward reverse primers, and Taq polymerase were included (Thermo scientific). The steps involved are initial denaturation for 5 min at 95 °C, which was followed by 30 cycles of final denaturation at 94 °C for 2 min, then annealing at 55 °C for 2 min, extension for 2 min at 72 °C, and a final extension step at 72 °C for 7 min (Verma et al., 2022). Amplified products were run on 1.2 % agarose gels in 1× TBE buffer for 1 h at a 100 V power source, for identification and visualized in a Gel-Doc Analyzer (InGenius3, Syngene Bio Imaging). A Silica bead gel extraction kit (Thermo Scientific) was used to purify amplified 16S rRNA, and samples were sent to Macrogen Inc. Korea for sequencing analysis.
2.6 Epigenetic and phylogenetic analysis
With the aid of 16S rRNA gene analysis and sequencing, the sequences of isolated bacterial pathogens were obtained. The 16S rRNA gene sequences were constructed and modified using Clustal W Alignment software (Jeon et al., 2014), and BLAST (Basic Local Alignment Search tool) analysis was used to compare the genetic similarity of the strains. Following that, the genomes of all bacterial isolates were deposited in Genbank (DDBJ), and accession numbers were acquired for each. The MEGA-11 software was used to create a neighbor-joining phylogenetic tree and assess the evolutionary relationships between the isolated strains (Tamura et al., 2013).
2.7 Pathogenicity test on citrus leaf
Pathogenicity tests were conducted on fully developed but immature Moro blood and Succri leaves. Leaves were first disinfected in the laminar hood using a 1 % sodium hypochlorite solution. Then leaves were arranged on Petri dishes with the discs of filter paper. A sterilized needle was used to puncture each leaf twice, once on the left side and once on the right side of the principal vein on the abaxial side. The right side punctures of each leaf were then infected with 100 μL of the overnight bacterial culture broth. As a negative control, sterile distilled water was injected into the punctures on the left side. To enhance infection, the petri dishes were labeled and put in a polythene bag and placed in an incubator set at 28 to 30 °C for 3 to 4 weeks. The leaves were observed daily for the development of the symptoms (Naseem et al., 2017)
3 Results
3.1 Isolation of bacterial pathogens
Citrus plants with diseased leaves and fruits yielded fourteen different bacterial isolates that fit the characteristics of species belonging to the four bacterial genera that are Xanthomonas, Pseudomonas, Candidatus, and Serratia. These bacterial isolates produced rounded, creamy yellow to off-white, and mucoid colonies (Fig. 2). Table 1 summarizes the origin, source, and morphological screening results of the isolated bacterial strains that were subjected to further analysis. +++ represents active bacterial growth; ++, represents normal growth rate; + represents mild and delayed growth; - represents negative or no growth.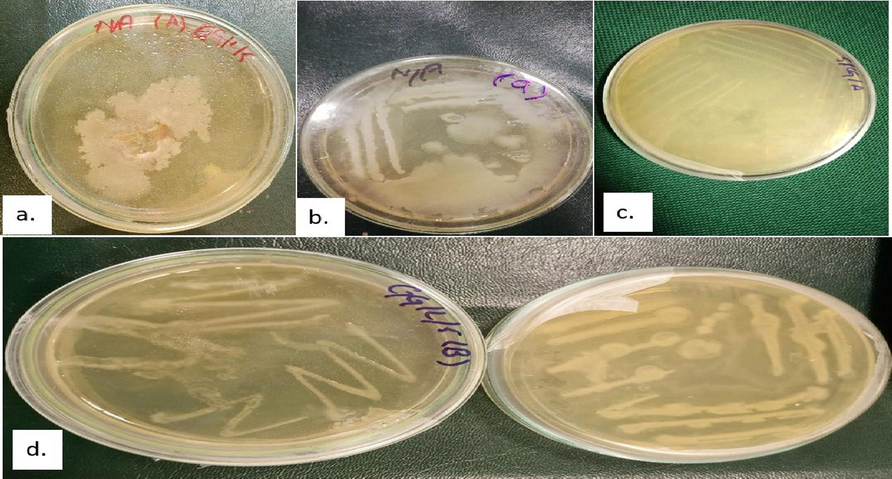
Colonies of Bacterial pathogens isolated from the diseased fruits and leaves samples (a.) Bacterial spot (BS) (b.) Citrus Canker (CCF) (c.) Citrus Canker (CCL) (d.) Citrus Greening (CGF and CGL).
Strains ID
Origin
Source
Gram’s Staining
Growth on YPG
CCL3
Khanpur, KPK
Leaves
–
+++
CCF3
Khanpur, KPK
Fruit
–
++
CCL2
Khanpur, KPK
Leaves
–
++
CCF2
Khanpur, KPK
Fruit
–
+++
CGL3
Khanpur, KPK
Leaves
–
++
CGF3
Khanpur, KPK
Fruit
–
+
CGL5
Khanpur, KPK
Leaves
–
+++
CGF5
Khanpur, KPK
Fruit
–
++
BSL1
Khanpur, KPK
Leaves
–
++
BSF2
Khanpur, KPK
Fruit
–
+
3.2 Disease incidence
The results showed that the citrus fruit and leaf diseases are prevailing in all the orchards under study. The result of disease incidence revealed that greening disease had a high incidence in orchard number 8 with a percent disease index (PDI) of 31 %, the citrus canker disease had a high incidence in orchard number 3 with a PDI of 18 %, and the bacterial spot had a high incidence in orchard number 5 with a PDI of 44 %. Fig. 3 displays the findings of the disease incidence for all orchards.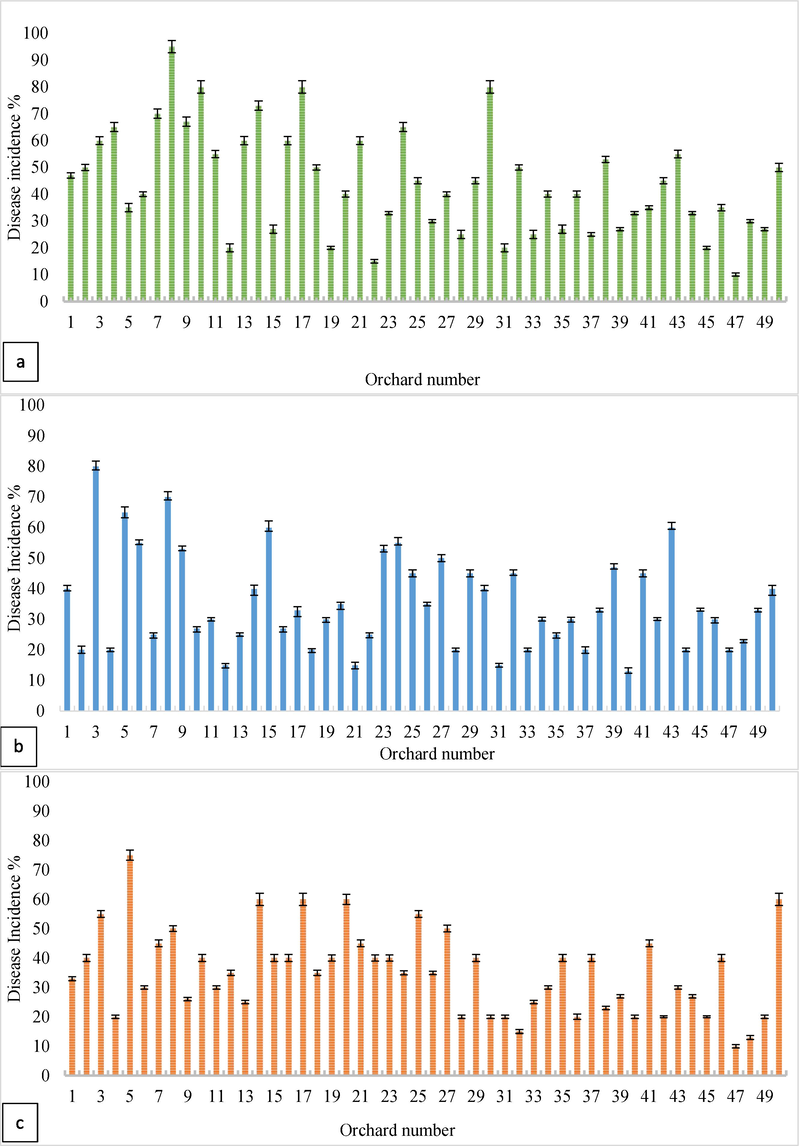
Disease Incidence of (a.) citrus greening, (b.) canker, and (c.) bacterial spot diseases.
3.3 Molecular characterization
The isolated DNA fragments were run with lambda (λ) DNA standards (25 ng/µ) on the gel to check the concentration of isolated DNA samples. Fig. 4a displays the pure DNA bands of the bacterial pathogens isolated from the citrus fruits and leaves diseases.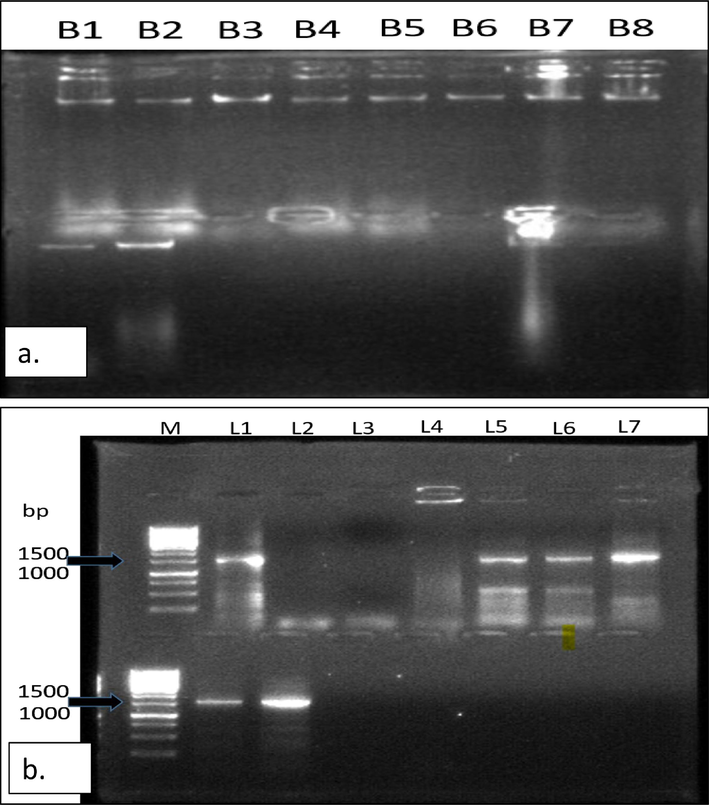
(a.) Pure DNA bands of the isolated bacterial pathogens, (b.)16S rRNA gene amplification of bacterial isolates causing Citrus diseases. Lane 1–7 are CCF2, CCL3, CCF3, CGF5, CGL5 and BSL1 respectively. Lane M represents a 1 kb DNA marker.
The amplified products of 16S rRNA gene having1500 bp amplicon, was obtained using the primer pair 27F and 1492R, that were the custom sequences and were subjected to BLAST analysis to find the sequence homology. The specific amplification was obtained for the bacterial isolates, including CCF2, CCF3, CCL3, CGF5, CGL5, and BSL1 as shown in Fig. 4b.
With the assistance of BLAST analysis, the nucleotide identities of the 16S rRNA gene sequences of the bacterial strains identified in this study have been compared with one another and with other strains published in the GenBank database to explain the phylogenetic evolutionary relationships between the strains (Fig. 5). After 16S rRNA partial sequence analysis a total of 14 species belonging to the 4 bacterial genera were identified as the major bacterial fruit pathogens of citrus plants in the Khanpur orchards. The species are Pseudomonas syringae, Pseudomonas viridiflava, Serratia marcescens, Xanthomonas axonopodis pv citri, and Candidatus liberibacter. These species along with the strains they are showing similarities are summarized in Table 2.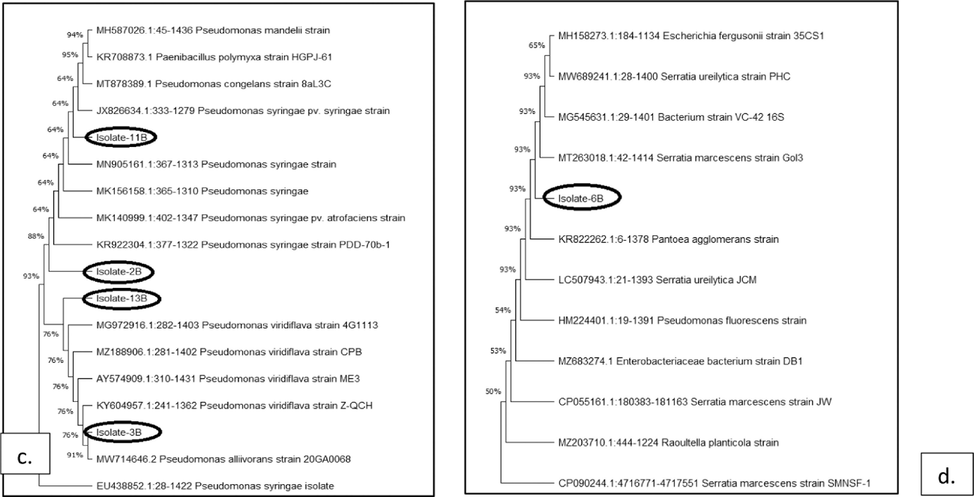
Neighbor-joining tree showing evolutionary links between bacterial strains that cause citrus diseases using Bootstrap methodology to construct the phylogenetic trees (a.) Xanthomonas axonopodis (b.) Candidatus Liberibacter (c.) Pseudomonas syringae and Pseudomonas viridiflava (d.) Serratia marcescens.
Isolate Code
Identity
Source
Host
Location
Similar species
Accession
NumbersSimilarity (%)
Isolate-CB-1
Candidatus liberibacter
Leaves
Citrus sinensis
KPK, Pakistan
Candidatus Liberibacter asiaticus OP108281.1
OQ131081
99
Isolate-CB-2
Pseudomonas syringae
Leaves
Citrus sinensis
KPK, Pakistan
Pseudomonas syringae EU438852.1
OQ131082
99
Isolate-CB-3
Pseudomonas viridiflava
Leaves
Citrus sinensis
KPK, Pakistan
Pseudomonas viridiflava KY604957.1
OQ131083
100
Isolate-CB-4
Xanthomonas axonopodis
Fruit
Citrus sinensis
KPK, Pakistan
Xanthomonas axonopodis pv citri MK818495.1
OQ131084
99
Isolate-CB-5
Xanthomonas citri
Fruit
Citrus sinensis
KPK, Pakistan
Xanthomonas axonopodis pv citri MK818495.1
OQ131085
100
Isolate-CB-6
Serratia
marcescens
Fruit
Citrus sinensis
KPK, Pakistan
Serratia marcescens MT263018.1
OQ131086
100
Isolate-CB-7
Serratia
marcescens
Fruit
Citrus sinensis
KPK, Pakistan
Serratia marcescens CP055161.1
OQ131087
100
Isolate-CB-8
Xanthomonas axonopodis
Leaves
Citrus sinensis
KPK, Pakistan
Xanthomonas axonopodis pv citri MK818495.1
OQ131088
99
Isolate-CB-9
Xanthomonas citri
Fruit
C. sinensis
KPK, Pakistan
Xanthomonas axonopodis pv citri MK818495.1
OQ131089
100
Isolate-CB-10
Candidatus liberibacter
Fruit
Citrus sinensis
KPK, Pakistan
Candidatus liberibacter OP108281.1
OQ131090
98
Isolate-CB-11
Pseudomonas syringae
Fruit
Citrus sinensis
KPK, Pakistan
Pseudomonas syringae MN905161.1
OQ131091
99
Isolate-CB-12
Candidatus liberibacter
Fruit
Citrus sinensis
KPK, Pakistan
Candidatus liberibacter AB480072.1
OQ131092
100
Isolate-CB-13
Pseudomonas viridiflava
Leaves
Citrus sinensis
KPK, Pakistan
Pseudomonas viridiflava MG972916.1
OQ131093
100
Isolate-CB-14
Candidatus liberibacter
Leaves
Citrus sinensis
KPK, Pakistan
Candidatus liberibacter KF712516.1
OQ131094
100
3.4 Pathogenicity test on citrus leaf
The symptoms started to show five days after infection, on the pricking sites in detached leaf pathogenicity tests. Lesions grew larger and changed color from tan to brown after two weeks. After 3 weeks of infection, elevated and spongy lesions—typical canker symptoms—became apparent. There were canker symptoms for CCL3, CCF3, CCL2, and CCF2. Both CCF3 and CCF2 experienced the development of a light brownish hue at puncture sites, which could not be characterized as a canker-like lesion. The right side of the leaf's abaxial surface, where bacterial broth cultures were infected, had canker signs, whereas the left side showed no symptoms. Results of the bacterial cultures CCF3 and CCF5 tests for citrus leaf pathogenicity are displayed in Fig. 6.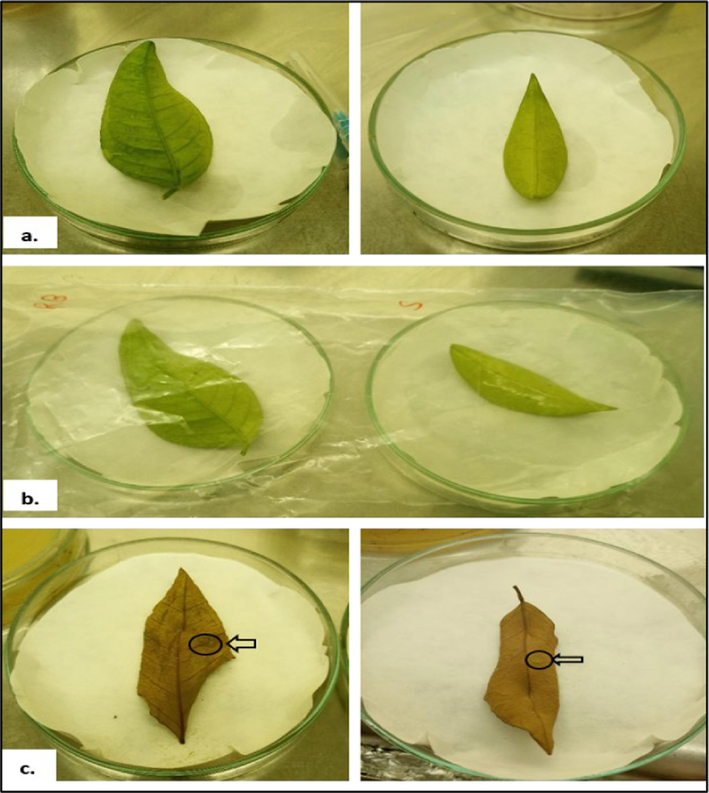
Pathogenicity test of canker-causing bacterial strains (CCF3, CCF5) on citrus leaves (a.) 1-week post-inoculation (wpi); (b.)2 wpi; (c.) 3 wpi.
4 Discussion
The climate in Pakistan is favorable for the production of all fruits with commercial value. The largest of them all in terms of output and export is citrus. However, the climate in areas that produce citrus is often warm, which favors the growth of disease-causing organisms and phyto-pathogens. Numerous bacterial diseases have serious economic effects. Citrus canker and HLB, two of the deadliest bacterial plant diseases, are a major problem for Pakistan's citrus industry. According to Gottwald et al. (2002), burning and uprooting infected groves can control the illness in some regions of the world, such as America and Brazil but in emerging nations like Pakistan, where the diseases has spread to the majority of the region's citrus orchards, such eradication is not practical.
One of the other main strategies for controlling these diseases is the use of chemicals and antibiotics, bacterial outbreaks are challenging to contain due to the evolution of resistance against the target pathogen and the lack of effective bactericides also their use in not environment friendly (Balogh et al., 2010). Applications of aminoglycosides have been observed to give plants systemic acquired resistance against bacterial infections, however they have no effect on the culprit responsible for citrus canker (Jones et al., 2012). Others have thought about employing biological remedies like phages to combat phyto-pathogens. The chemical and biochemical tests used to identify isolated bacterial strains showed that they shared characteristics with the plant pathogenic bacteria’s linked to citrus canker, and bacterial spot diseases, including being gram negative, catalase positive, and oxidase negative (Vernière et al., 1998; Naseem et al., 2017).
For molecular characterization, the chosen bacterial strains with disease potential were accessed. The results of this study showed that the best period to sample for the pathogenic bacteria that cause citrus canker is just before the summer solstice. Bacteria grow rapidly on YPG agar plates in 24 to 36 h, while the incubation time period for the toxicity test lasts upto 3 to 4 weeks. Mild chlorosis is seen on all of the leaves during the first two to three days after infection. However, only the strains that carried the virulence genes caused lesions on the pricking sides to enlarge after 1 week of infection. Numerous distinct bacterial species cause a wide range of diseases in citrus.
This is the first report on the bacterial strains that are responsible for the citrus canker caused by Xanthomonas axonopodis, bacterial spot caused by Pseudomonas syringae and Pseudomonas viridiflava, and HLB diseases caused by Candidatus Liberibacter in Khanpur, Pakistan. However, in earlier investigations as reported by Preecha et al. (2018); Ahmad (2020), on the prevalence of citrus canker diseases on Pakistan citrus fruit harvests, only the phenotypic and morphological disease indicators on plants have been evaluated. An effort was made in this study to pinpoint the species that are responsible for the diseases and consequences they were having on the overall citrus crop production.
The isolated bacterial strains by partial sequence16S rRNA gene analysis found the closest relatives from the Xanthomonadaceae, Rhizobiaceae and Pseudomonadaceae families based on clustering. Following a BLAST similarity search, bacterial strains were chosen for relatedness and similarity based on their origin and plant toxicity for the purpose of building phylogenetic trees. On the basis of 16S ribosomal RNA sequencing a neighbor joining phylogenetic tree was constructed for the bacterial strains CCF3, CCL5, and CCF5 that were closely related to the strain Xanthomonas axonopodis pv citri MK818495.1 whereas the bacterial starins CGF5 and CGL5 were closely related to the strain Candidatus Liberibacter asiaticus OP108281.1. The strain BSL1 is closely related to strain Pseudomonas syringae KR922304.1 and the strain BSL2 is closely related to Pseudomonas viridiflava MG972916.1 strain.
The results of this study are in line with findings of Monteiro-Vitorello et al. (2005) who reported in 2005 that the citrus canker is caused by Xanthomonas citri (Xanthomonas axonopodis pv. Citri) belonging to the family Xanthomonadaceae of the Gammaproteobacteria. According to Teixeira et al., [31] three Candidatus species have been reported to cause HLB citrus greening diseases in Brazil that are ‘Ca. Liberibacter asiaticus’ (CaLas),‘Ca. Liberibacter africanus’ (CaLaf), and ‘Ca. Liberibacter americanus’ (CaLam) (Carmo et al., 2005). The isolated species Serratia marcescens is closely related to strain MT263018.1 was identified in the current study is in line with findings of Hasan et al. (2022), who also revealed that the isolates as Serratia marcescens strains are responsible for black rot disease of Citrus sinensis fruits in Bangladesh that reduces the quantity and quality of the citrus and causes enduring losses to the farmers and fruit industry.
The pathogenicity test revealed that the lesion diameter measured after inoculation for all the tested isolated strains produced more significant results than the control which did not show any disease symptoms. However, the effect on the leaves of Kinnow (Citrus nobilis x Citrus deliciosa) was more intense as compared to the other two varieties Sucrri and Moro Blood, which showed an almost similar pattern with minimum lesion diameter after inoculation. The results of the present research have shown that the most aggressive isolates are the CCL3, CCF3, CCL2, and CCF2 for which the high lesion areas were observed and the isolated species from these are Xanthomonas axonopodis and Xanthomonas citri, after the molecular identification by sequence analysis, whereas the other isolates that are CGF5 and CGL5 which are identified as Candidatus liberibacter are moderately aggressive.
Similar results were reported by different researchers in the world who have worked on the pathogenicity tests and virulence of bacterial strains on citrus fruits. A study by Niu et al., (2020), evaluated the pathogenicity of Xanthomonas citri subsp. citri, which is responsible for the citrus bacterial canker disease, on different citrus varieties. The results showed that the aggressiveness of the bacterial strain varied among the citrus varieties, with some showing higher susceptibility to the disease than others.
In another study by Tatineni et al. (2020), citrus huanglongbing disease was reported, in which they evaluated the pathogenicity of Candidatus Liberibacter, the causative agent of the disease, on different citrus fruit varieties. The findings suggested that some citrus varieties were more susceptible to the disease than others, with younger trees showing higher susceptibility.
The bacterial isolates BSL1 and BSL2 which are identified as Pseudomonas syringae and Pseudomonas viridiflava respectively are slightly aggressive. However, a different result was reported by Al-Ali et al. (2021), who investigated the pathogenicity of Pseudomonas syringae, which is responsible for the bacterial blast disease on citrus fruits. Their findings revealed that the bacterial species Pseudomonas syringae were highly aggressive and pathogenic on citrus fruits and caused severe damage to the fruits.
Overall, all the above-discussed study demonstrated that various bacterial isolates can be pathogenic to citrus, causing fruit and tree damage and providing valuable information on the pathogenicity of different strains on citrus plants. The results of the present investigation on the aggressiveness analysis of pathogens can contribute to the development of effective control strategies and resistant varieties against isolated pathogens and the diseases caused by the pathogens on the citrus.
5 Conclusions
The findings of this research revealed that Khanpur, Pakistan's citrus crop is at risk due to bacterial pathogens that are deteriorating this valuable asset of the country. The production of citrus is seriously threatened by the broad-spectrum toxicity of citrus bacterial diseases, the cultivation of vulnerable citrus varieties, and the advent of emerging strains. The disease symptoms observed on the diseased citrus leaves and fruits in this study were no doubt the characteristics of citrus bacterial diseases that are reported elsewhere. The pathogen associated with the diseases were identified as Xanthomonas campestris pv citri, Candidatus Liberibacter, Pseudomonas syringae, Pseudomonas viridiflava, and Serratia marcescens which are responsible for canker, citrus greening, bacterial leaf spots and black rot diseases of Citrus sinensis. In general, knowledge of the centers of origin, the diversity of citrus, the accurate identification of pathogens, and biological and molecular characterizations of their genomes, together with effective sanitation, proper ecological management, certification programs, and the use of healthy planting materials are necessary for the maintenance of productive citrus orchards.
Ethics approval
Not applicable.
Funding
This research received no external funding
CRediT authorship contribution statement
Iram Zaheer: Data curation, Conceptualization. Shazia Iram: Writing – original draft, Supervision, Project administration, Conceptualization. Sibgha Noreen: Writing – original draft, Methodology, Investigation. Seema Mahmood: Visualization, Formal analysis. Amna Bibi: Investigation. Ajaz Ahmad: Validation, Resources, Formal analysis. Prashant Kaushik: Visualization, Validation, Software, Resources, Formal analysis.
Acknowledgement
The authors would like to extend their sincere appreciation to the Researchers Supporting Project Number (RSP2024R350), King Saud University, Riyadh, Saudi Arabia
References
- Detection and Identification of Potato Soft Rot Pectobacterium carotovorum Subspecies carotovorum by PCR Analysis of 16S rDNA in Jordan. Agric. Sci.. 2018;09:546-556.
- [CrossRef] [Google Scholar]
- Ahmad, B., Mehdi, M., Ghafoor, A., & Anwar, H. (2018). Value chain assessment and measuring export determinants of citrus fruits in Pakistan: an analysis of primary data. Pakistan Journal of Agricultural Sciences, 55(3). doi: 10.21162/pakjas/18.6056 Ahmad, K.S., 2018. Exploring the potential of Juglans regia derived activated carbon for the removal of adsorbed fungicide Ethaboxam from soils. Environmental Monitoring A and Assessment. 190, 737. doi: 10.1007/s10661-018-7119-z.
- Assessment keys for some important diseases of Mango. Pak. J. Biol. Sci.. 2002;5:246-250.
- [CrossRef] [Google Scholar]
- Pathogenicity of Pseudomonas syringae pv. syringae on citrus fruits. J. Plant Pathol.. 2021;103(3):985-991.
- [Google Scholar]
- Rep-PCR reveals a high genetic homogeneity among Ugandan isolates of Xanthomonas campestris pv. Musacearum Afr. J. Biotechnol.. 2006;6:179-183.
- [CrossRef] [Google Scholar]
- Citrus tristeza virus in Pakistan: A Review. Pakistan J. Phytopathol.. 2017;29(2):273.
- [CrossRef] [Google Scholar]
- Phage therapy for plant disease control. Curr. Pharm. Biotechnol.. 2010;11:48-57.
- [CrossRef] [Google Scholar]
- Citrus by-products as ruminant feeds. A review. Anim. Feed Sci. Technol.. 2006;128:175-217.
- [CrossRef] [Google Scholar]
- Diseases and pests. Citrus Fruit Processing; 2016.
- Analysis of nutritional constituents in twenty citrus cultivars from the Mediterranean area at different stages of ripening. Food Nutr. Sci.. 2012;03:639-650.
- [CrossRef] [Google Scholar]
- Carmo, T., D., Luc Danet, J., Eveillard, S., Cristina Martins, E., Jesus Junior, W., Takao Yamamoto, P., Aparecido Lopes, S., Beozzo Bassanezi, R., Juliano Ayres, A., Saillard, C., Bové, J.M., 2005. Citrus Huanglongbing in São Paulo State. Molecular and Cellular Probes. 19, 173-179. doi: 10.1016/j.mcp.2004.11.002.
- Lipolytic effects of citrus peel oils and their components. J. Agric. Food Chem.. 2006;54:3254-3258.
- [CrossRef] [Google Scholar]
- Occurrence and prevalence of mango decline in the Punjab province of Pakistan. Plant Prot.. 2022;6:11-18.
- [CrossRef] [Google Scholar]
- Gonzatto, M.P. and Santos, J.S. (2023). Introductory chapter: World citrus production and research, IntechOpen. Available at: https://www.intechopen.com/chapters/86388 (Accessed: 24 October 2023).
- Citrus canker: The pathogen and its impact. Plant Health Progress. 2002;3
- [CrossRef] [Google Scholar]
- First report of Serratia marcescens associated with black rot of citrus sinensis fruit, and evaluation of its biological control measures in Bangladesh. F1000Research. 2022;9:1371.
- [CrossRef] [Google Scholar]
- Citrus greening disease (Huanglongbing) in Pakistan: A review. Pakistan J. Phytopathol.. 2016;28(1):1-7.
- [Google Scholar]
- EzEditor: a versatile sequence alignment editor for both rRNA- and protein-coding genes. Int. J. Syst. Evol. Microbiol.. 2014;64:689-691.
- [CrossRef] [Google Scholar]
- Considerations for using bacteriophages for plant disease control. Bacteriophage. 2012;2:208-214.
- [CrossRef] [Google Scholar]
- Incidence and etiology of mango sudden death phenomena in Pakistan. Pakistan J. Phytopythol.. 2005;17:154-158.
- [CrossRef] [Google Scholar]
- Citrus Huanglongbing: A newly emerging epidemic in Pakistan. Phytopathology Research. 2019;1(1):1-12.
- [Google Scholar]
- A new system of grading of plant diseases. J. Agric. Res.. 1923;26:195-218.
- [CrossRef] [Google Scholar]
- Use of carnauba based carrier for copper sprays reduces infection by Xanthomonas citri Subsp. citri and Diaporthe citri in Florida commercial grapefruit groves. Agric. Sci.. 2012;03:962-970.
- [CrossRef] [Google Scholar]
- First report on characterization of citrus disease causing bacteria and related phages isolated in Pakistan. Int. J. Agric. Biol.. 2017;19:857-864.
- [CrossRef] [Google Scholar]
- Differential virulence of Xanthomonas citri subsp. citri on citrus varieties. Plant Dis.. 2020;104(3):779-785.
- [Google Scholar]
- Pakistan Bureau of Statistics. (2021). Agriculture Statistics of Pakistan. Retrieved from http://www.pbs.gov.pk/content/agriculture-statistics, (Accessed: 24 October 2022).
- Pereira, F.M.V., Milori, D.M.B.P., Pereira-Filho, E.R., Venâncio, A.L., Russo, M.d.S.T., Cardinali, M.C.d.B., 2011. Laser-induced fluorescence imaging method to monitor citrus greening disease. Computer and Electronics in Agriculture. 79, 90-93. doi: 10.1016/j.compag.2011.08.002.
- Occurrence of Canker Caused by Xanthomonas axonopodis pv. citri on Pummelo (Citrus maxima (Burm.) Merr.) Cultivar. Tabtimsiam in Nakhon Si Thammarat Province, Thailand and Screening Fungicides, Antibiotics and Antagonistic Bacteria against X. a. pv. citri in Vitro. J. Geosci. Environ. Protect.. 2018;06:1-7.
- [CrossRef] [Google Scholar]
- MEGA 11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol.. 2013;30:2725-2729.
- [CrossRef] [Google Scholar]
- Pathogenicity of Candidatus Liberibacter asiaticus on different citrus varieties. Plant Dis.. 2020;104(1):159-165.
- [Google Scholar]
- Survey on foliar diseases of mango (Mangifera indica L.) in Jabalpur (M.P.) region. Pharma Innov. J.. 2022;11:1901-1904.
- [CrossRef] [Google Scholar]
- Characterization of phenotypically distinct strains of Xanthomonas axonopodis pv. citri from south west Asia. Eur. J. Plant Pathol.. 1998;104:477-487.
- [CrossRef] [Google Scholar]
- Genomics of the origin and evolution of Citrus. Nature. 2018;554:311-316.
- [CrossRef] [Google Scholar]







