Translate this page into:
MiR-15a regulates bicuspid aortic valve calcification via TGF-β signaling pathway via inhibiting Smad7
⁎Corresponding authors. yongfengshao30@hotmail.com (Yongfeng Shao), junjie.du@njmu.edu.cn (Junjie Du)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Background
Bicuspid aortic valve is a heritable heart valve disease with characteristic early onset of valve calcification and stenosis with rapid progression compared with tricuspid aortic valve. Strong evidence indicates that many miRNAs and their target genes are involved in the development of BAV calcification. This study was designed to investigate miR-15a and Smad7 expressions in BAV and their mechanism of regulating calcification.
Methods
Immunohistochemistry and western blotting were used to explore the expression levels of Smad7 and Runx2 in human aortic valve tissue. The expression of miR-15a, Smad7 and Runx2 were detected by RT-PCR. The relationship between miR-15a and Smad7 was assessed by dual-luciferase assay. MiR-15a was overexpressed in cultured porcine valve interstitial cells (PVICs) and the variations of Smad7 and Runx2 mRNA were evaluated by RT-PCR, as well as the changes of Smad7 and Runx2 protein. Alizarin reds staining was used to verify the calcification inhibition effect of miR-15a in cultured PVICs under calcification induction.
Results
Compared with calcified TAV, remarkable higher expression of Smad7 and Runx2 were found in calcified BAV, significantly lower expressions of miR-15a and higher expressions of Smad7 and Runx2 mRNA, along with negative correlation between expressions of miR-15a and mRNA level of Smad7. MiR-15a-overexpressed PVICs shown upregulation of miR-15a, downregulation of Smad7 and Runx2 expression. PVICs under calcification induction shown significantly reduced calcification in miR-15a-overexpressed group.
Conclusions
The lower miR-15a expression in BAV results in the high expression of Smad7, thereby reducing antagonism of Smad2/3–Smad4 complex to runx2 and promoting the progress of BAV calcification.
Keywords
Bicuspid aortic valve
Calcification
miR-15a
Smad7
Runx2
- AS
-
aortic valve stenosis
- BAV
-
bicuspid aortic valve
- BMPs
-
bone morphogenetic proteinsm
- HDACs
-
class II histone deacetylases
- Runx2
-
Runt-related transcription factor
- SMADs
-
suppressors of mothers against decapentaplegic
- TAV
-
tricuspid aortic valve
Abbreviations
1 Introduction
Bicuspid aortic valve (BAV) is a congenital heart valve deformity and its prevalence ranges from 0.5% to 2%. Compared with tricuspid aortic valve (TAV), BAV patients are more susceptible to suffer aortic valve stenosis (AS) due to valve calcification, infective endocarditis and aortic dilatation (Padang et al., 2013; Cripe et al., 2004; Shah et al., 2018). Fifty percent of patients under age of 30 with BAV suffered from ascending aortic dilatation, which rised to 88% in people elder than 80 (Grewal et al., 2014). As a result, 20–50% of BAV patients have to suffer artificial valve replacement surgery (Lindman et al., 2013).
One of key osteogenic regulator Runt-related transcription factor 2(Runx2) is a decisive factor in VICs osteoblastic differentiation contributing to valve calcification (Olga et al., 2017). Besides, there are proteins such as bone morphogenetic proteins (BMPs) (Du et al., 2017; Sun et al., 2012; Wang et al., 2015) Osteopontin (OPN), Alkaline Phosphatase (ALP), Matrix Metalloproteinase (MMP), Sry Related HMG box-9 (Sox9), Matrix Gla-Protein (MGP) and Nitric Oxide Synthase (NOS) were also reported to be involved in valve calcification (Song et al., 2015; Yong et al., 2016; Zeng et al., 2017; Li et al., 2017; Chiyoya et al., 2018; Abdellatif, 2015).
Sustained growth studies indicate that many miRNAs and their target genes are involved in the development BAV calcification. MiRNAs are non-coding small molecule RNAs, which are non-protein-coding gene regulators. They are recognized as negative post-transcriptional regulators of their target genes (Condorelli et al., 2014). Studies have shown that TGF-β signaling pathway and suppressors of mothers against decapentaplegic (SMADs) were involved in the progression of aortic valve calcification (Du et al., 2017; Vishal et al., 2016; Yan et al., 2009; Ming et al., 2018; Miller et al., 2011). For Smad7, a well-known negative regulator of TGF-β signaling pathway, which is a newly discovered gene involved in BAV calcification progression via TGF-βsignal pathway that blocks the phosphorylation of SMADs related receptors leading increased expression of Runx2 and promotes progression of valve calcification (Du et al., 2017; Yan et al., 2009). Smad7 expression is also regulated by miR-15a in other tissues or cell lines (Kishore et al., 2011; Skalsky et al., 2012), but currently little is known about miR-15a regulation of aortic valve calcification in BAV.
Our study firstly explored Smad7 and Runx2 differential expression levels in BAV and TAV by immunohistochemical examination and western blotting. Then, significantly lower miR-15a expression was found in BAV than that in TAV. Moreover, We explored the potential regulation mechanism of BAV calcification by miR-15a and its target Smad7 via TGF-β signaling pathway.
2 Materials and methods
2.1 Human aortic valve tissue collection
According to the approval of the ethics committee and patient informed consents, we collected aortic valves diagnosed as BAV or TAV according to the preoperative echocardiography and postoperative anatomy. The fresh isolated valve leaflets were sectioned into small parts, froze in liquid nitrogen for RT-PCR and western blotting, embedded in paraffin after fixed in Paraformaldehyde for immunohistochemistry.
2.2 Cell culture, transfection and calcification induction
We isolated and cultured PVICs as previously described by Junjie Du et al. (Du et al., 2017). The PVICs were explant in growth medium [Dulbecco’s modified Eagle’s medium (DMEM),10% fetal bovine serum (FBS),1% penicillin–streptomycin (Invitrogen, USA)] passaged by trypsin digestion, transfected by Lipofectamine 3000 Transfection Kit (Thermo Fisher Scientific, USA) with miRNA mimic, mimic negative control (Ribobio Co, Guangzhou, China). Calcium and phosphate-enriched culture medium (DMEM containing HEPES, 6% FBS, 0.2 mmol/L CaCl2 and 0.75 mmol/L Na5P3O10) was used to induce PIVCs calcification under Different interventions. PIVCs were stained for Alizarin Red S Staining when obvious calcium crystals were found under microscope.
2.3 Reverse transcription-polymerase chain reaction (RT-PCR)
RNAiso Plus (Takara, Japan) was applied to extract miRs or mRNAs in human aortic valve leaflets and cultured PVICs. We use PrimeScript RT reagent Kit with gDNA Eraser (Takara, Japan) to reverse transcribe the extracted miRNAs or mRNAs.
Target genes expression were quantified by using of SYBR Premix Ex TaqTM II (Takara, Japan) on 7900HT Real-Time Polymerase Chain Reaction System (ABI, USA) and normalized to GAPDH (Takara, Japan) for mRNAs and U6 for miRNAs analyses. The sequences are listed in Table 1.
Name
Direction
Sequence
U6
forward
5′-GTGGGGAGAAGAGGACAGGAC-3′
reverse
5′-GTGGTACCCACTTTCGCACA-3′
hsa-Smad7
forward
5′-TTCCTCCGCTGAAACAGGG-3′
reverse
5′-CCTCCCAGTATGCCACCAC-3′
ssc-Smad7
forward
5′-CCAGAACCTATTTCCCATGC-3′
reverse
5′-GGCACTGGGTGAGCAATACT-3′
hsa-Runx2
forward
5′-AGCCACCTACCACAGAGCTA-3′
reverse
5′-GGATGAGGAATGCGCCCTAA-3′
ssc-Runx2
forward
5′-GGACGAGGCAAGAGTTTCAC-3′
reverse
5′-GTGGATTAAAAGGACTTGGTGC-3′
hsa-GAPDH
forward
5′-AGAAGGCTGGGGCTCATTTG-3′
reverse
5′-AGGGGCCATCCACAGTCTTC-3′
ssc-GAPDH
forward
5′-ATGCCATCCACAGGCATC-3′
reverse
5′-GTAGTACTCACTTCGGCACC-3′
2.4 Western blotting analyses
Samples and cells were treated with a sample buffer containing 100 mmol/L Tris-HCl (pH 6.8), 2% SDS, 0.02% bromophenol blue, and 10% glycerol. The protein concentrations were quantified by the Micro BCA™ Protein Assay Kit (Thermo Scientific, Shanghai, China) according to the manufacturer’s instructions. After transferring the proteins to polyvinylidene difluoride membranes and blocking with 5% Bovine Serum Albumin, the membranes were incubated with primary antibodies at 4℃ overnight and then were incubated with peroxidase-linked secondary antibodies specific to the primary antibodies for 2 h at room temperature.
2.5 Dual-luciferase reporter assay
293 T cells were cultured by using growth medium [DMEM, 10% FBS, 1% nonessential amino acids, 1% L-glutamine, and 1%penicillin-streptomycin (Invitrogen, USA)]. The pmirGLO dual-luciferase miR target expression vector (pmirGLO) (Madison WI, USA), Quik Change XL Site-Directed Mutagenesis Kit (Agilent Technologies, USA) and Lipofectamine 3000 (Invitrogen, USA) were used according to the manufacturer's instructions. Human Smad7 3′-UTR including the predicted binding site of miR-15a was amplified by RT-PCR and was inserted into the 3′-UTR downstream of the firefly luciferase gene of pmirGLO vector (pmirGLO-UTR) by using the XbaI and SacI restriction sites. We tested and analyzed the luciferase activity after cellular transfection by Dual-Glo luciferase assay system.
2.6 Alizarin red s staining
PVICs were washed with phosphate-buffered saline before being fixed in 4% paraformaldehyde for 15 min at room temperature. Then the cells were fixed in 95% ethanol for 30 min and washed with double distilled water carefully. Cells were then underwent Alizarin Red S staining process according to the manufacturer’s instructions. The optical density of the samples at 570 nm were detected by a spectrophotometer.
2.7 Histologic examination and immunohistochemistry
Smad7 and Runx2 were detected in the fixed tissue sections. The sections were preprocessed in Vectastain® ABC reagent (Vector, Burlingame, CA, USA), and treated with 3,3′-Diaminobenzidine before being counterstained with hematoxylin. Image-J was employed for signal half-quantitation analyses. All antibodies used in this study were purchased from Abcam (MA, USA).
2.8 Statistical analysis
Statistical analysis was performed by using Prism version 7.0 (GraphPad Software Inc., San Diego, California). The data were presented as the means ± SD. Statistical significance was evaluated by p < 0.05.
3 Results
3.1 The expressions of Smad7 and Runx2 in BAV leaflets were obviously higher than that in TAV leaflets.
Tissue slices of BAV group and TAV group with similar calcification degree evaluated by the degree of calcium deposits were immunohistochemically used for Smad7 and Runx2 expression level detection (Fig. 1A). The results indicated that the Smad7 and Runx2 protein expressions in calcified BAV leaflets were increased than that in calcified TAV leaflets, with statistic difference (Fig. 1B, C). In order to further validate the result, WB was applied to exam Smad7 and Runx2 protein expressions in calcified BAV leaflets and BAV leaflets (Fig. 1D), and the results were statistic analyzed by image lab software. Compared with calcified TAV leaflets, over-expressions of Smad7 and Runx2 were found in calcified BAV leaflets with significant difference (Fig. 1E, F).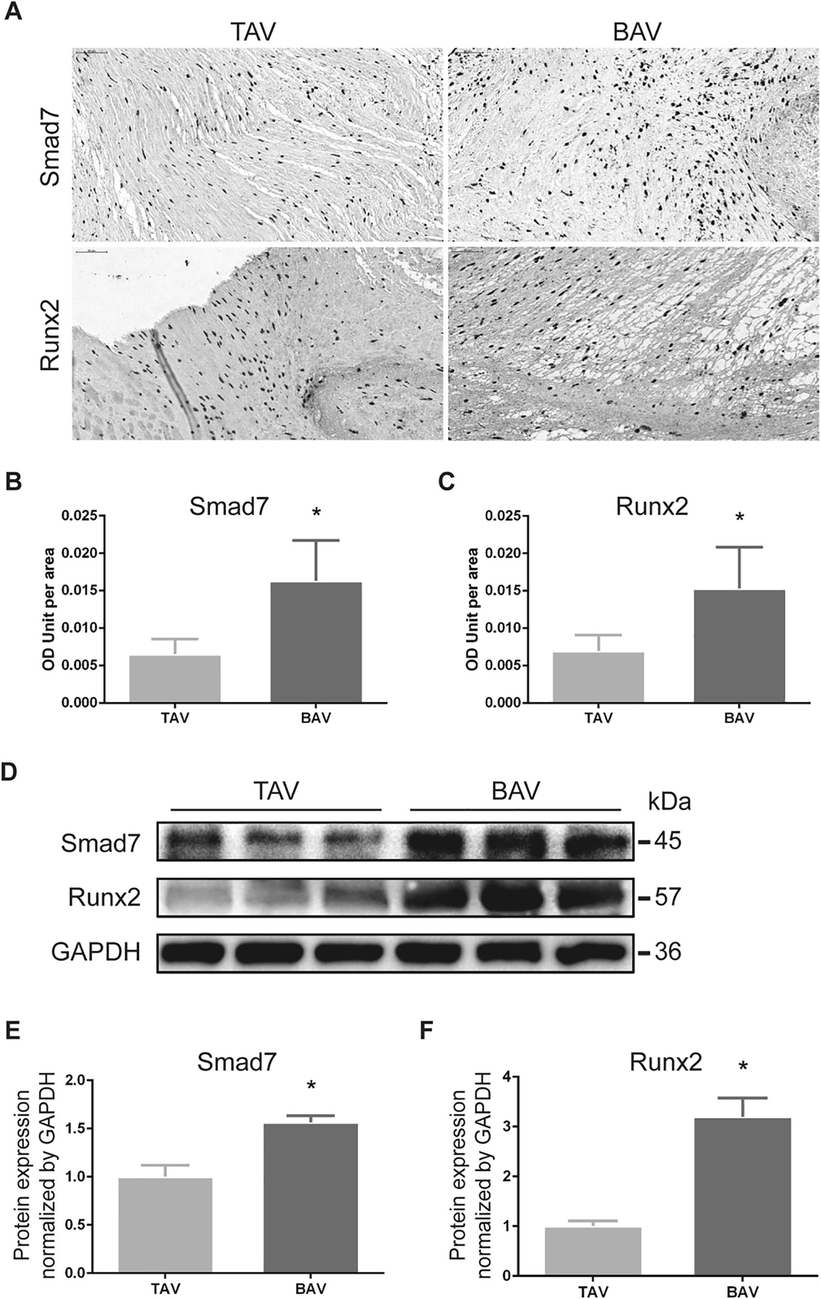
Smad7 and Runx2 were overexpressed in BAV leaflets than that in TAV leaflets by immunohistochemistry and western blotting. (A) Representative figures of tissue immunohistochemistry. (B) and (C) The expressions of Smad7 and Runx2 in calcified BAV leaflets were increased than that in calcified TAV leaflets. (D) Western blotting for Smad7 and Runx2 in BAV and TAV. (E) and (F) Overexpression of Smad7 and Runx2 protein were found in calcified BAV leaflets than that of TAV leaflets. Each protein is presented as a relative level in comparison with GAPDH. The data shown represent the means ± SD. *p < 0.05.
3.2 Compared with calcified TAV leaflets, calcified BAV leaflets showed significant lower expressions of miR-15a and higher expressions of Smad7 and Runx2 mRNA
The 65% reduction of miR-15a expression was found in BAV leaflets by RT-PCR (Fig. 2A). The expressions of Smad7 mRNA was significantly increased by 1.85-fold (Fig. 2B), and Runx2 mRNA was significantly increased by 2.62-fold (Fig. 2C). Negative correlation between Smad7 mRNA and miR-15a expressions in BAV and TAV were revealed by Pearson analysis (Fig. 2D).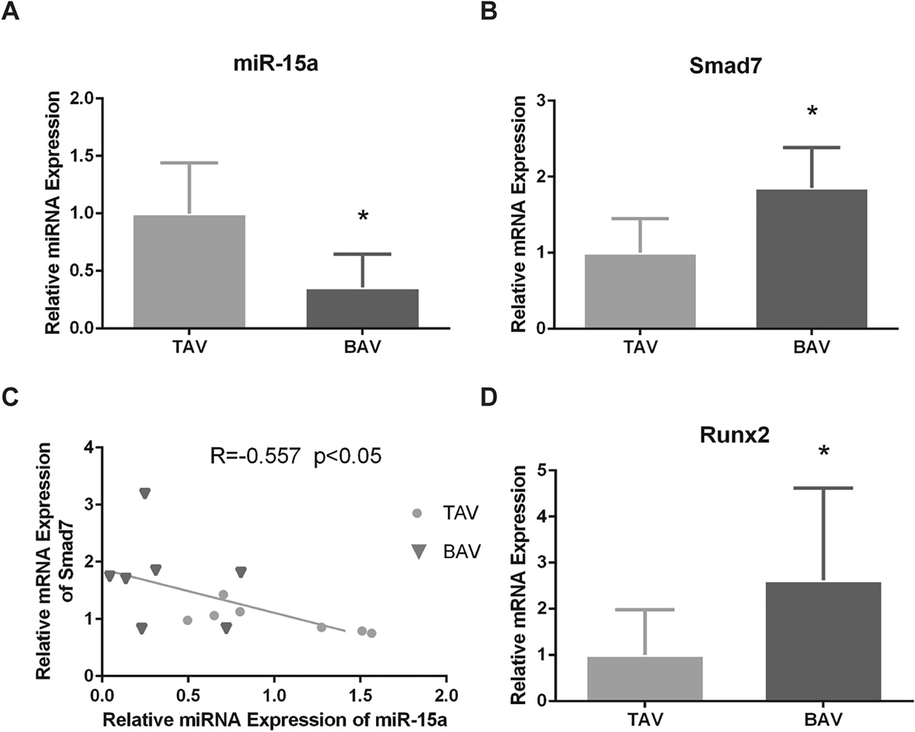
Negative correlation between miR-15a and Smad7 was confirmed in calcified BAV leaflets. (A) The expression of miR-15a in BAV leaflets was significantly reduced than that of TAV leaflets. (B) Higher Smad7 mRNA expression was validated in BAV than in TAV. (C) Pearson’s correlation analysis indicated that Smad7 mRNA level was negatively correlated with miR-15a level (R = − 0.557, P < 0.05). (D) Higher expression levels of Runx2 mRNA were validated in BAV leaflets than in BAV leaflets by RT-PCR. The data are shown as means ± SD. *P < 0.05.
3.3 Dual-luciferase reporter assay validate the interaction between miR-15a and Smad7
By using Target Scan and DIANA, we explored the targeting site of miR-15a on Smad7. We found part of Smad7 mRNA sequence in human was highly consistent with porcine and mouse (Fig. 3A). Furthermore, a putative complementary region of miR-15a in Smad7 mRNA 3′-UTR was found and had been experimental confirmed. Then we conducted Dual-luciferase reporter assay using 293Tcells transfected with wild type or point mutation vectors of 3′-UTR region of Smad7 at the predicted binding site (Fig. 3B). The results indicated that transfection with miR-15a mimic reduced the relative luciferase activity of pGLO-Smad7-3′-UTR-wt (Fig. 3C).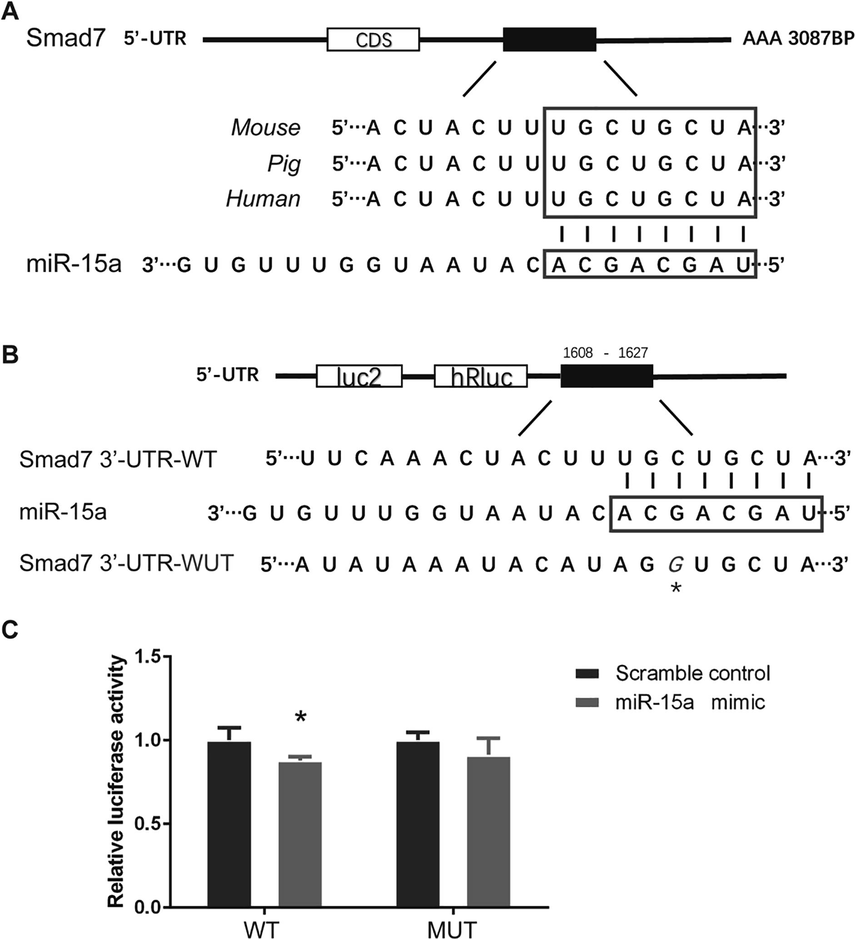
Smad7 is the direct target gene of miR-15a. (A) Predicted binding site of miR-15a on Smad7 in human is highly consistent with that on porcine and mouse. (B) The seed sequence of miR-15a (3′…ACGACGAU…5′) was complementary to the 3′UTR in Smad7 mRNA (5′…UGCUGCUA…3′). In the mutant construct (pmirGLO-UTR-MUT), a cytosine was replaced by a guanine in the 3′-UTR segments of human Smad7 (marked with an asterisk). (C) Transfection with miR-15a mimic reduced the relative luciferase activity of pGLO-Smad7-3′-UTR-wt but not that of pGLO-Smad7-3′-UTR-mut. *P < 0.05.
3.4 Upregulation of miR-15a expression inhibits Smad7 and Runx2 expression in PVICs
Twenty-four hours after transfected by miR-15a mimic (miR-15a mimic group), significant increased expression of miR-15a was detected in cultured PVICs than transfected by miR-15a mimic negative control (mimic negative control group) or non-transfected (control group) PVICs (Fig. 4A). Meanwhile, significant lower expression of Smad7 mRNA was observed in miR-15a mimic group than that in mimic negative control group and control group (Fig. 4B). Expression of Runx2 mRNA showed the same variation tendency as Smad7 mRNA (Fig. 4C). Thirty-six hours after transfection cellular proteins were extracted and used for WB (Fig. 4D), significant lower protein levels of Smad7 and Runx2 were found in miR-15a mimic group than that in mimic negative control group and control group by WB (Fig. 4E, F). In accordance with Dual-luciferase reporter assay, these results indicated that miR-15a suppressed Runx2 by targeting Smad7.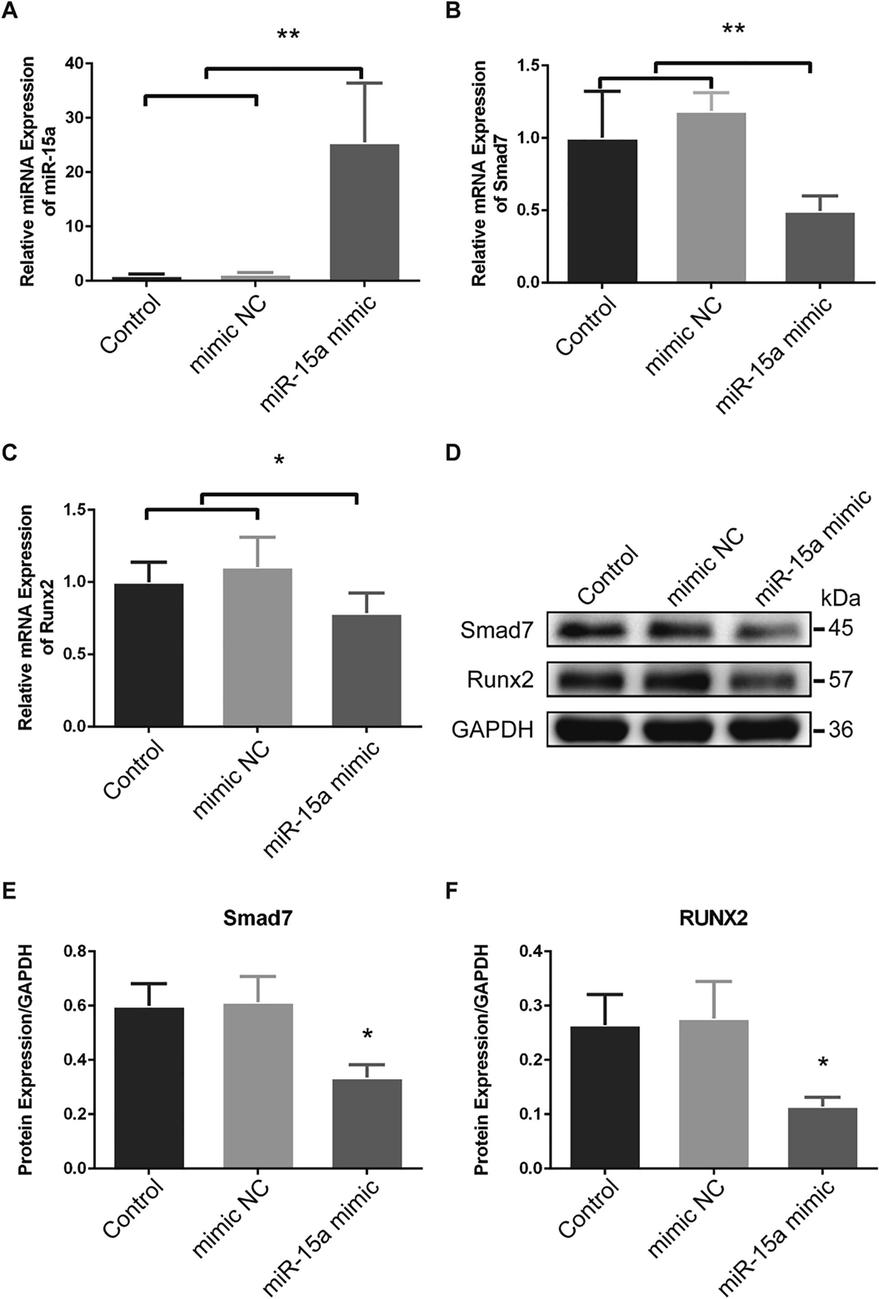
Overexpression of miR-15a inhibits Smad7 and Runx2 expression in PVICs. (A) In vitro cultured PVICs transfected with miR-15a mimic (miR-15a mimic group) showed significant increased miR-15a expression than that in the control group and the mimic negative control group. (B) and (C) Up-regulation of miR-15a decreased both Smad7 and Runx2 mRNA expression. (D) Western blots for Smad7 and Runx2 in cultured PAVICs. (E) and (F) Western blots analyses indicated that the Smad7 and Runx2 protein levels were significant decreased in miR-15a mimic group than that in the control group and the mimic negative control group. The data are shown as means ± SD. *P < 0.05. **P < 0.01.
3.5 Overexpression of miR-15a relieved calcification process at cellular level
To reveal the function of miR-15a-5p in calcification progress, we induced transfected or non-transfected PVICs in calcification medium. PVICs treated with calcification induction medium had a significant higher calcium deposition (Fig. 5A). PVICs treated with miR-15a mimic relieved the calcification compared with mimic negative control or calcification induced group (Fig. 5B). In conclusion, overexpression of miR-15a alleviated the calcification progress of cultured PVICs.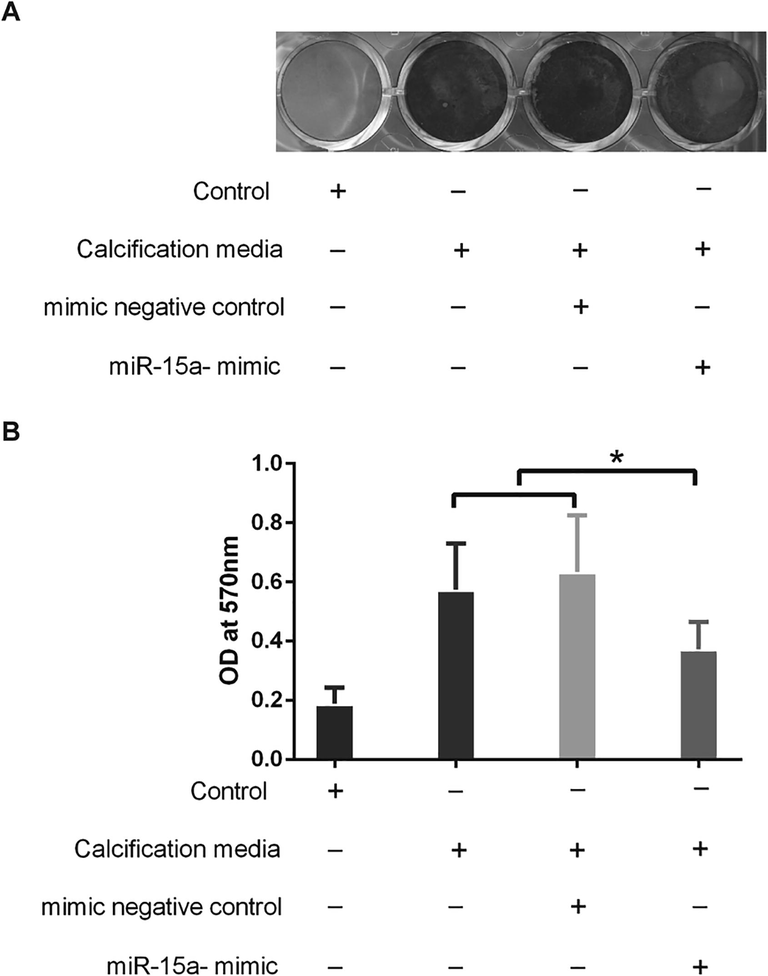
MiR-15a affected calcification process in cultured PVICs. (A) Representative images of Alizarin Red S staining for calcium deposition in cultured PVICs interfered by different methods. (B) Quantification of Alizarin Red S staining suggested that PVICs which cultured in Calcium and phosphate-enriched culture medium treated with miR-15a mimic relieved the calcification compared with that of mimic negative control group or non-transfected group. *P < 0.05.
4 Discussion
BAV is the most common type of aortic valve malformations, accounting for 70% of the aortic valvular dysplasia. BAV Patients are characterized by early onset of calcification, rapid progress and high mortality, and are at high risks of developing valvular regurgitation and aortic dilation, as well as valvular stenosis (AS) which is seen more than 10 years earlier than patients with TAV (Prakash, 2015; Lomas et al., 2010). BAV patients with important AS are more prone to suffer aortic valve replacement surgery with the symptom of thoracodynia and syncope, and are risking complications of the surgery.
Although the mechanisms of BAV calcification is not well elucidated, the osteoblastic phenotype cells transformed from valvular interstitial cells play a key role in the progression (Jenkins et al., 2015). Previous studies, although partially controversial, have shown that several molecular mechanisms and proteins are activated in BAV patients and involved in BAV calcification and stenosis (Magni, 2019).
Although there is no consensus among the researchers about mechanisms of BAV calcification progression as yet, TGF-β signaling pathway and key proteins such as SMADs and Runx2 were found to regulate the induction of cell apoptosis, cell aggregation, and especially promote calcification deposition and calcification nodule formation, which were also involved in other multiple cell processes and functions (Miller et al., 2011; Du et al., 2017; Liu et al., 2018).
In Smad-dependent TGF-β signaling pathway, Smad2/Smad3 suppresses TGF-β-mediated osteoblastic differentiation or cellular calcification by inhibiting Runx2 expression (Furumatsu et al., 2005). Activated Smad3 also represses the function of Runx2 by recruiting class II histone deacetylases4/5 (HDAC4/5) and inhibits the apoptosis of osteoblasts and their differentiation into osteocytes. Without Smad3, TGF-β cannot inhibit osteoblastic differentiation (Li et al., 1998; Alliston et al., 2014). HDAC-4/Smad3 complex can inhibit the overexpression of Runx2 and its induction genes such as MMP-13, thus maintaining the stability of articular cartilage and inhibiting the occurrence of calcification (Chen et al., 2012). Smad2/3 activated by BMP-3 could antagonized osteogenic activity of Smad1/5/8 which recruited Smad4 to transcribed the expression of Runx2, contributing to other osteoblastic gene expression (Franceschi and Xiao, 2003; Afzal et al., 2005).
Smad7, an inhibitory Smad, inhibits TGF-βsignaling by preventing R-Smad nuclei translocation, restraining the phosphorylation of R-Smad, and recruiting E3 ubiquitin ligases Smurf1/2 to promote R-Smads/receptor degradation (Conidi et al., 2013; Heldin et al., 2009). Smad7 interferes Smad2/3–Smad4 complex formation via forming a heteromeric complex, recruits the HECT-type E3 ligase NEDD4-2 to the heteromeric complex and promotes the ubiquitylation and degradation of phosphorylated Smad2/3 (Yan et al., 2016), which could interferes the expression of Runx2. These results and observations suggest that loss of Smad7 would inhibit the osteogenesis process through activating TGFβ/Smad3 signaling.
The negative regulation of Smad7 remains controversial. Smad7 inhibits TGF-β- induced apoptosis of B cells and gastric epithelial cells while promoting TGF-β-mediated human prostatic carcinoma cells apoptosis, which endow Smad7 context dependent peculiarity (Yan et al., 2009). Although Smad7 is known as an anti-fibrotic and anti-inflammatory factor, but its function in calcification has not been fully delineated.
In this study, lower expression of miR-15a and higher expression of smad7 along with over-expressed Runx2 protein were validated in BAV than that in TAV. Transfection with miR-15a mimic lead to lower expression of smad7 and Runx2 both at mRNA and protein level. Furthermore, PVICs transfected with miR-15a mimic showed reduced calcification. Our previous study demonstrated that aberrant expression of miR-195 contribute to BAV calcification via targeting Smad7 (Du et al., 2017). In accordance with our concepts, Li et al illustrated that Smad7 gene knockout mice had reduced osteogenic potential, fewer mineralized nodules, and reduced gene expression of Runx2 (Li et al., 2014).
5 Conclusion
The findings of this study indicated that Smad7 is a target of miR-15a, and is also a promotor of Runx2 by antagonizing Smad2/3–Smad4 complex. The reduced expression of miR-15a implies its contribution to BAV abnormal calcification.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 81974033).
Authors’ contributions
JJD, YFS, Conceived and designed the experiments. JKX, HL, RZ, Performed the experiments. JKX, H L, RZ, JJD, YFS, Analyzed the data. JKX, H L, R Z, JJ D, YFS, Wrote the paper.
Ethics and consent to participate
The present study was granted ethical approval by the First Affiliated Hospital of Nanjing Medical University Ethics Committee. Written informed consent was obtained from the patients.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Plasma levels of matrix metalloproteinase (MMP)-2, MMP-9 and tumor necrosis factor-α in chronic hepatitis C virus patients. Open Microbiol. J.. 2015;9:136-140.
- [Google Scholar]
- Smad function and intranuclear targeting share a Runx2 motif required for osteogenic lineage induction and BMP2 responsive transcription. J. Cell. Physiol.. 2005;204:63-72.
- [Google Scholar]
- TGF-beta-induced Repression of CBFA1 by Smad3 Decreases cbfa1 and Osteocalcin Expression and Inhibits Osteoblast Differentiation. EMBO J.. 2014;20:2254-2272.
- [Google Scholar]
- Chondrocyte-intrinsic Smad3 represses runx2-inducible matrix metalloproteinase 13 expression to maintain articular cartilage and prevent osteoarthritis. Arthritis Rheum.. 2012;64:3278-3289.
- [Google Scholar]
- Matrix Gla protein negatively regulates calcification of human aortic valve interstitial cells isolated from calcified aortic valves. J. Pharmacol. Sci.. 2018;136:257-265.
- [Google Scholar]
- microRNAs in cardiovascular diseases: current knowledge and the road ahead. J. Am. Coll. Cardiol.. 2014;63:2177-2187.
- [Google Scholar]
- Aptamers and their potential to selectively target aspects of EGF, Wnt/β-catenin and TGFβ-Smad family signaling. Int. J. Mol. Sci.. 2013;14:6690-6719.
- [Google Scholar]
- Downregulated MicroRNA-195 in the bicuspid aortic valve promotes calcification of valve interstitial cells via targeting SMAD7. Cell. Physiol. Biochem.. 2017;44:884-896.
- [Google Scholar]
- Regulation of the osteoblast-specific transcription factor, Runx2: Responsiveness to multiple signal transduction pathways. J. Cell. Biochem.. 2003;88:446-454.
- [Google Scholar]
- Smad3 induces chondrogenesis through the activation of SOX9 via CREB-binding protein/p300 recruitment. J. Biol. Chem.. 2005;280:8343-8350.
- [Google Scholar]
- Bicuspid aortic valve: phosphorylation of c-Kit and downstream targets are prognostic for future aortopathy. Eur. J. Cardiothorac. Surg.. 2014;46:831-839.
- [Google Scholar]
- Mechanism of TGF-beta signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr. Opin. Cell Biol.. 2009;21:166-176.
- [Google Scholar]
- Valvular (18)F-fluoride and (18)F-fluorodeoxyglucose uptake predict disease progression and clinical outcome in patients with aortic stenosis. J. Am. Coll. Cardiol.. 2015;66:1200-1201.
- [Google Scholar]
- A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat. Methods. 2011;8:559-564.
- [Google Scholar]
- Klotho suppresses high phosphate-induced osteogenic responses in human aortic valve interstitial cells through inhibition of Sox9. J. Mol. Med. (Berl).. 2017;95:1-13.
- [Google Scholar]
- Smad2 overexpression enhances smad4 gene expression and suppresses CBFA1 gene expression in osteoblastic osteosarcoma ROS17/2.8 cells and primary rat calvaria cells. J. Biol. Chem.. 1998;273:31009-31015.
- [Google Scholar]
- Partial loss of Smad7 function impairs bone remodeling, osteogenesis and enhances osteoclastogenesis in mice. Bone. 2014;67:46-55.
- [Google Scholar]
- Metformin ameliorates TGF-β1-induced osteoblastic differentiation of human aortic valve interstitial cells by inhibiting β-catenin signaling. Biochem. Biophys. Res. Commun.. 2018;500:710-716.
- [Google Scholar]
- Healthcare-associated infective endocarditis: an undesirable effect of healthcare universalization. Clin. Microbiol. Infect.. 2010;16:1683-1690.
- [Google Scholar]
- Bicuspid aortic valve, atherosclerosis and changes of lipid metabolism: are there pathological molecular links? J. Mol. Cell. Cardiol.. 2019;129:231-235.
- [Google Scholar]
- Calcific aortic valve stenosis: methods, models, and mechanisms. Circ. Res.. 2011;108:1392-1412.
- [Google Scholar]
- Mir-29b promotes human aortic valve interstitial cell calcification via inhibiting TGF-β3 through activation of wnt3/β-catenin/Smad3 signaling. J. Cell. Biochem.. 2018;119:5175-5185.
- [Google Scholar]
- NOTCH1 mutations in aortic stenosis: association with osteoprotegerin/RANK/RANKL. Biomed Res. Int. 2017:1-10.
- [Google Scholar]
- The genetic and molecular basis of bicuspid aortic valve associated thoracic aortopathy: a link to phenotype heterogeneity. Ann Cardiothorac. Surg.. 2013;2:83-91.
- [Google Scholar]
- Bicuspid aortic valve aortopathy in adults: Incidence, etiology, and clinical significance. Int. J. Cardiol.. 2015;201:400-407.
- [Google Scholar]
- The viral and cellular MicroRNA targetome in lymphoblastoid cell lines. PLoS Pathog.. 2012;8:e1002484
- [Google Scholar]
- BMP-2 and TGF-β1 mediate biglycan-induced pro-osteogenic reprogramming in aortic valve interstitial cells. J. Mol. Med. (Berl).. 2015;93:403-412.
- [Google Scholar]
- Ex vivo evidence for the contribution of hemodynamic shear stress abnormalities to the early pathogenesis of calcific bicuspid aortic valve disease. PLoS ONE. 2012;7:e48843
- [Google Scholar]
- MicroRNA-590-5p stabilizes Runx2 by targeting Smad7 during osteoblast differentiation. J. Cell. Physiol.. 2016;232:371-380.
- [Google Scholar]
- Notch-Tnf signalling is required for development and homeostasis of arterial valves. Eur. Heart J.. 2015;38:675-686.
- [Google Scholar]
- Smad7 Protein Interacts with Receptor-regulated Smads (R-Smads) to Inhibit Transforming Growth Factor-β(TGF-β)/Smad Signaling. J. Biol. Chem.. 2016;291:382.
- [Google Scholar]
- Regulation of TGF-β signaling by Smad7. Acta Biochim. Biophy. Sin.. 2009;41:263-272.
- [Google Scholar]
- Clinical implication of aortic wall biopsy in aortic valve disease with bicuspid valve pathology. Korean J. Thorac. Cardiovasc. Surg.. 2016;49:443-450.
- [Google Scholar]
- Interleukin-37 suppresses the osteogenic responses of human aortic valve interstitial cells in vitro and alleviates valve lesions in mice. Proc. Natl. Acad. Sci. U.S.A.. 2017;114:1631-1636.
- [Google Scholar]







