Translate this page into:
MicroRNA expression profiling in type 2 diabetes patients treated with liraglutide
*Corresponding author: E-mail address: mohammad.irshad@dasmaninstitute.org (M. Irshad); shhaque@jazanu.edu.sa (S. Haque)
-
Received: ,
Accepted: ,
Abstract
Type 2 diabetes (T2D) is a chronic metabolic condition characterized by impaired blood glucose regulation. Liraglutide, a glucagon-like peptide-1 (GLP-1) receptor agonist, is a commonly used drug for T2D management. Despite scientific advancements, the molecular mechanism underlying liraglutide therapy in T2D remains poorly understood. The study aimed to identify key microRNAs (miRNAs) and uncover the mechanisms of action of liraglutide in T2D by employing an integrated systems biology approach. The miRNA expression dataset, GSE223538, containing data from T2D patients treated with and without liraglutide, was retrieved from NCBI’s Gene Expression Omnibus (GEO) database. The dataset comprised 32 samples (13 control and 19 treated). Raw FASTQ reads were processed by trimming 3’ adapter sequences using the fastx_clipper tool from the FASTX-Toolkit. Reads shorter than 18 nucleotides were discarded, and the remaining reads were consolidated into unique sequences for streamlined mapping and analysis. Five miRNAs – hsa-miR-9-5p, hsa-miR-22-3p, hsa-miR-19b-3p, hsa-miR-132-3p and hsa-miR-93-5p – were found to be significantly linked to genes involved in the PI3K/Akt, MAPK, and FOXO1 signaling pathways. These findings suggest that liraglutide’s therapeutic effects may be mediated through miRNA-regulated mechanisms that modulate PI3K/Akt and other associated signaling pathways. In turn, these pathways regulate the cellular processes that enhance β-cell function, promote insulin secretion, and increase glucose uptake in patients with T2D. The results indicate that these miRNAs provide important insights into the mechanisms through which liraglutide reduces T2D risk, potentially guiding the approach for the development of novel biomarkers, targeted therapies, and precision health strategies. Additionally, the findings lay the groundwork for further experimental substantiation of the key pathways involved in liraglutide therapy.
Keywords
Liraglutide
miRNA
Network medicine
Type 2 diabetes
1. Introduction
Type 2 diabetes (T2D) is a chronic metabolic condition that affects approximately 6.3% of the global population (Khan et al. 2020). T2D develops when β-cells fail to meet the increased demand for insulin required to maintain normal glycemic levels. Persistent chronic hyperglycemia damages nerves and blood vessels, making T2D a major risk factor for both microvascular and macrovascular problems (Al-Ozairi et al. 2024; Mansour et al. 2023). Obesity in individuals with diabetes further exacerbates the burden of diabetes- and obesity-linked health problems (Al Ozairi et al. 2024; Cui et al. 2021). These challenges underscore the demand for effective therapeutic approaches to manage diabetes and its associated complications.
Several glucose-lowering agents are available for T2D management. Recently, the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD) have recommended the use of glucagon-like peptide-1 (GLP-1) receptor agonists for individuals with obesity and T2D (Davies et al. 2022). In the recent past, liraglutide, a GLP-1 receptor agonist with almost 97% structural homology with human GPL-1, has emerged as a prominent option for T2D management (Knudsen and Lau 2019). By mimicking the human GLP-1, liraglutide controls blood glucose levels by improving insulin release, decreasing glycogen secretion, and reducing elevated glucose levels (Knudsen and Lau 2019). Liraglutide improves β-cell function, promotes glucose-dependent insulin secretion, increases satiety, slows gastric emptying, and supports weight loss (Santilli et al. 2017).
Besides having glucose-lowering effects, liraglutide also supports weight management and reduces cardiovascular risk, which are critical concerns in T2D management (Howell et al. 2019). Furthermore, reduction of weight and improvement in glycemic levels have also been observed in liraglutide-treated individuals with type 1 diabetes (Al-Ozairi et al. 2023).
The molecular-level therapeutic effects of liraglutide have been investigated in animal models, but there is limited research available on its effects in human populations. Liraglutide has been reported to inhibit cytokine-induced apoptosis in rat islet-cells, reduce apoptosis triggered by free fatty acids (Bregenholt et al. 2005), and enhance islet function by regulating nine specific microRNAs (miRNAs) involved in pathways, such as PI3k signaling, autophagy, FOXO, and HIF-1 signaling in diabetic rats (Guo et al. 2021). Similarly, liraglutide reduced pancreatic β-cell apoptosis via decreasing miRNA-139-5p expression in diabetic rats (Li et al. 2017). In T2D patients, three specific miRNAs, i.e., miRNA-130a, miRNA-27b, and miRNA-210 have been associated with improved cardiometabolic outcomes following liraglutide treatment (Romaine et al. 2015; Giglio et al. 2020). Whereas, another study reported no substantial difference in expression levels of miRNA between untreated and treated T2D patients (Scherbak et al. 2023).
Microribonucleic acids (miRNAs) are small endogenous non-coding RNAs (∼22 nucleotides in length) that regulate cellular processes through modulation of post-transcriptional gene expression (Singh et al. 2020; Khan et al. 2022). Under physiological conditions, miRNAs play roles in diverse biological processes, including cell development, proliferation, differentiation, apoptosis, and metabolism (Ahmed et al. 2022; Ali et al. 2024; Nizam et al. 2024). To date, nearly 2000 miRNAs have been discovered in humans (Pordzik et al. 2019). Each miRNA can target multiple genes, enabling broad regulatory effects. However, aberrant expression of miRNA due to pathological stress has been reported in chronic diseases like cancer, metabolic diseases, and cardiovascular diseases (Kim and Zhang 2019; Ahmed et al. 2022; Iqbal et al. 2021; Ali et al. 2024). Almost 70 miRNAs were found upregulated, while 100 miRNAs were found downregulated in the blood samples of T2D patients (Kim and Zhang 2019). The ability of miRNA to regulate multiple cellular pathways makes them promising therapeutic targets for addressing cellular dysfunction in disease states (Hanna et al. 2019). The development of miRNA-specific inhibitors represents a promising frontier in drug development (Chakraborty et al. 2017).
Although dysregulated miRNA expression in T2D has been extensively reported, research on the impact of GLP-1 receptor agonists on miRNA expression in T2D patients remains scanty. This study aimed to identify key miRNAs and elucidate mechanisms of action of liraglutide in T2D using an integrated systems biology approach. In this study, we utilized publicly available miRNA databases from the repository and performed an analysis to explore cellular pathways and their roles in diabetes management. Recent advancements in molecular technology provide valuable insights into miRNA’s functions, enhancing our understanding of cellular response to liraglutide.
2. Material and methods
2.1 Retrieval of data and processing
The miRNA expression dataset GSE223538, comprising T2D patients treated with and without liraglutide was retrieved from the Gene Expression Omnibus (GEO) database of NCBI (Scherbak et al. 2023). This study included 32 samples (13 control and 19 treated). Initial processing of raw FASTQ reads involved trimming the 3’ adapter sequence using the fastx_clipper program from the FASTX-Toolkit (Keller et al. 2011). Reads shorter than 18 nucleotides were excluded to minimize non-specific or degraded RNA fragments, which are more likely to arise from technical noise and are less likely to map uniquely to the genome. This threshold is used in small RNA sequencing studies to ensure the reliability of downstream analyses. The batch effects are critical for ensuring robust and reproducible results. To address potential batch effects, the Limma package was applied to normalize the data across different batches (Ritchie et al. 2015). This process was conducted during the data preprocessing stage to mitigate any variability introduced by experimental conditions. The remaining reads were amalgamated into a unique sequence, and their frequencies were recorded for each sample to make the mapping process more efficient.
FastQC was used to appraise the quality of the processed reads, ensuring high data quality. Further, the miRDeep2 quantifier (Friedländer et al. 2012) was used to map and quantify reads against the latest human reference from miRBase (Ver. 22) (Kozomara et al. 2019). Raw read counts were normalized, and differential expression analysis was performed by DESeq2 package of R (Love et al. 2014). The criteria applied in DESeq2 for the identification of differentially expressed miRNAs were, adjusted False Discovery Rate (FDR), p-value <0.05, a log2 fold change (FC) >1, and a base mean >5.
2.2 Identification of miRNAs targeting differentially expressed genes
miRNA enrichment network tools, MIENTURNET (Licursi et al. 2019) and miRnet (Chang et al. 2020), were used to predict the interaction between miRNAs and differentially expressed genes. These open-source web tools performed statistical analyses through over-representation of miRNA-target gene interactions using an input list of genes and mature miRNAs, thereby retrieving both computational prediction and experimentally validated data from miRTarBase (Huang et al. 2020) and miRDB (Chen and Wang 2020). A co-expression network of target gene-miRNA interactions was constructed using Cytoscape 3.6.1 (Smoot et al. 2011). Further, functional enrichment analyses for target genes and selected miRNAs were performed using REACTOME, WiKi Pathways, KEGG, and human Disease Ontology. The most statistically enriched Gene Ontology (GO) terms were visualized using ggplot2 (Wickham 2011).
2.3 Identification of driver target genes of key miRNA
CytoHubba v.0.1 plugin of Cytoscape was utilized for identifying potential hub genes among the differentially expressed miRNAs (Chin et al. 2014). To identify the potential driver gene(s) targeted by the key miRNA, an eccentric topological property approach was employed for the constructed miRNA-mRNA regulatory network. Eccentric topological properties are critical for identifying the most central nodes in a network, as these eccentric nodes serve as essential driver connectors nodes with significant roles in signal processing, functional integration, and network dynamics.
2.4 Statistical analysis
Statistical analysis was performed using the Signed Rank Tests and Wilcoxon Rank Sum Test (non-parametric test for comparing two groups), with the significance level maintained at p < 0.05.
3. Results
3.1 Expression profile of miRNA in liraglutide treated T2D patients
The miRNA expression profiles of 19 T2D patients treated with liraglutide were compared to those of 13 T2D patients, who did not receive liraglutide treatment. Principal component analysis (PCA) of the miRNA sequencing data showed distinct clustering of individual patient groups with a clear differentiation between T2D patients treated with and without liraglutide (Fig. 1a). Heatmap clustering was performed using the expression patterns of the top 50 miRNAs from both T2D patients treated with and without liraglutide (Fig. 1b). Differential expression analysis identified 33 miRNAs (out of 2886 miRNAs), which were notably diverse between T2D patients treated with and without liraglutide (P < 0.05; Fig. 1c).
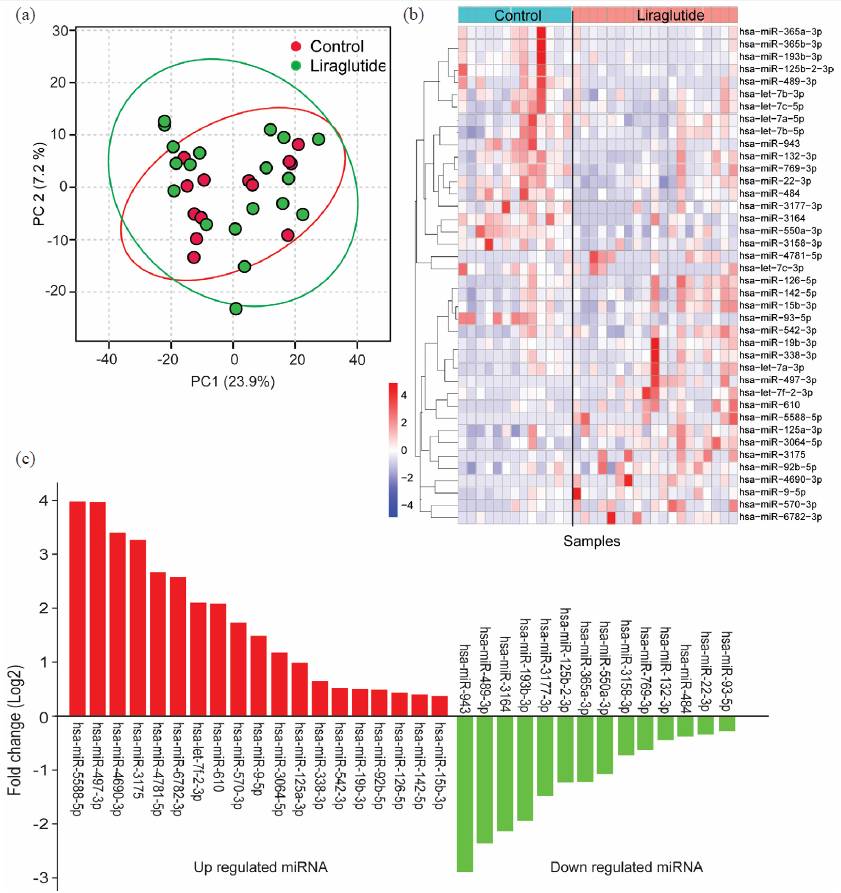
- Expression profile of miRNA. (a) Principal component analysis (PCA) of miRNA expression in people with T2D treated with and without liraglutide, (b) Heatmap of the top 50 differentially expressed miRNAs in people with T2D treated with and without liraglutide, (c) Expression levels of differentially expressed miRNAs in people with T2D treated with and without liraglutide. T2D: Type 2 diabetes. T2D: Type 2 diabetes.
A total of 19 miRNAs, viz., hsa-miR-5588-5p, hsa-miR-497-3p, hsa-miR-4690-3p, hsa-miR-3175, hsa-miR-4781-5p, hsa-miR-6782-3p, hsa-let-7f-2-3p, hsa-miR-610, hsa-miR-570-3p, hsa-miR-9-5p, hsa-miR-3064-5p, hsa-miR-125a-3p, hsa-miR-338-3p, hsa-miR-542-3p, hsa-miR-19b-3p, hsa-miR-92b-5p, hsa-miR-126-5p, hsa-miR-142-5p, and hsa-miR-15b-3p were found significantly overexpressed in T2D patients treated with liraglutide compared to those not received liraglutide treatment. Whereas, 14 miRNAs, hsa-miR-943, hsa-miR-489-3p, hsa-miR-3164, hsa-miR-193b-3p, hsa-miR-3177-3p, hsa-miR-125b-2-3p, hsa-miR-365a-3p, hsa-miR-550a-3p, hsa-miR-3158-3p, hsa-miR-769-3p, hsa-miR-132-3p, hsa-miR-484, hsa-miR-22-3p and hsa-miR-93-5p were found significantly downregulated in T2D patient treated with liraglutide (Fig. 1c).
3.2 Prediction of target genes of miRNAs
miRNAs are key regulators of mRNA expression within cells. They bind to target mRNAs, leading to translation processes. Target prediction was performed separately for differentially expressed miRNAs. A total of 1246 potential target genes for differentially expressed miRNAs in T2D patients treated with liraglutide were predicted using miRNet. The predicted target genes were identified using the miRNA databases. The results confirm the presence of 1246 potential target genes for these differentially expressed miRNAs. Subsequently, a miRNA-gene interaction network consisting of differentially expressed miRNA-gene pairs was constructed using Cytoscape software. The shortest path filter network is illustrated in Fig. 2a. The resulting regulatory network revealed that the miRNA-genes interaction network comprised 164 interacting nodes and 203 edges. Notably, hsa-miR-9-5p targeted 45 genes, hsa-miR-22-3p targeted 28 genes, hsa-miR-19b-3p targeted 23 genes, hsa-miR-132-3p targeted 22 genes and hsa-miR-93-5p targeted 16 genes (Fig. 2a-b).
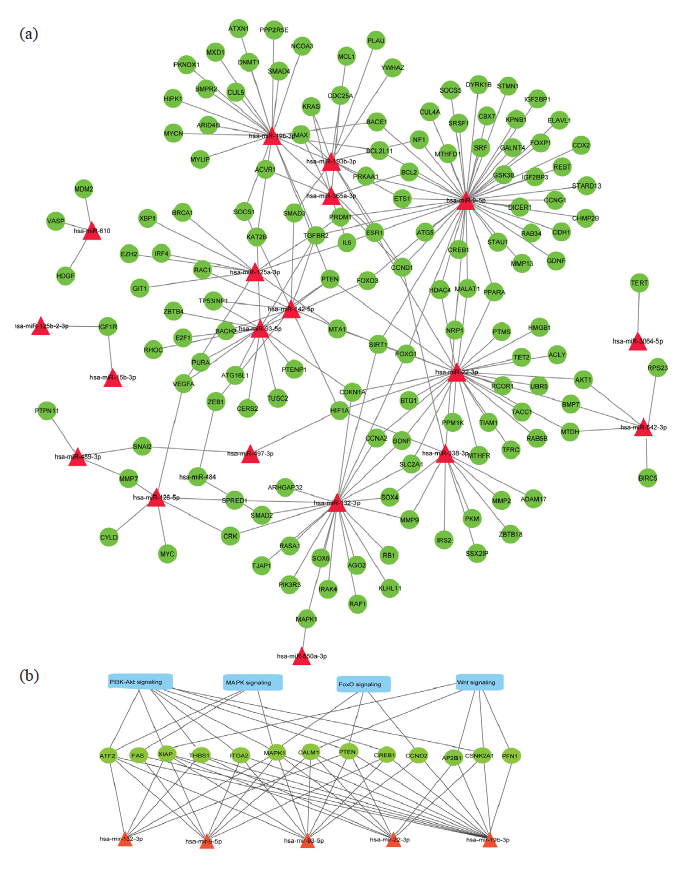
- (a) Predicted regulatory networks of differentially expressed miRNAs and their target genes associated with T2D. Red triangular shapes represent miRNAs, and green ellipses represent target genes associated with T2D. (b) Wnt, PI3K-Akt, MAPK, and FoxO signaling pathways are intricately linked to the target genes regulated by miRNAs.
3.3 Predicted functions and pathways of differentially expressed miRNAs
A functional enrichment analysis of the predicted target genes of dysregulated miRNAs was done for T2D patients treated with liraglutide. KEGG (Kyoto Encyclopedia of Genes and Genomes) and Biological pathway enrichment analyses were performed using DAVID (Database for Annotation, Visualization, and Integrated Discovery) to endorse the involvement of these miRNAs in ADPKD (autosomal dominant polycystic kidney disease). The results revealed that the targeted genes were predominantly enriched in several key pathways, including phosphoinositide 3-kinase/protein kinase B (PI3K/Akt), MAPK (mitogen-activated protein kinase), forkhead box protein O1 (FOXO1), p53, apoptosis, and Wnt signaling pathways (Fig. 3).
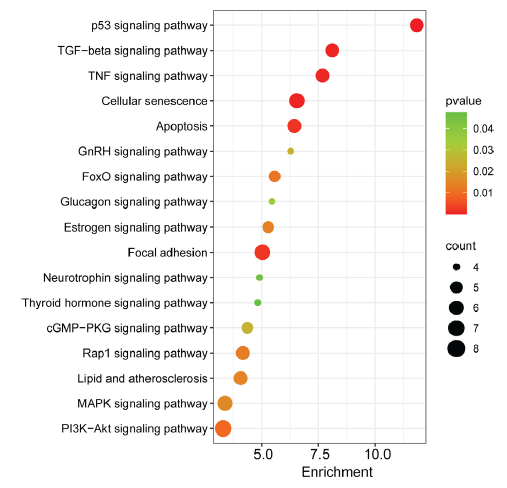
- Significantly enriched KEGG pathways of genes targeted by miRNAs. Dot size indicates the gene count, where “count” represents the number of genes associated with each pathway. The dot colour denotes the p-values of pathways, and the x-axis represents fold enrichment. KEGG: Kyoto encyclopedia of genes and genomes. TGF-beta: Transforming growth factor beta, TNF: Tumor necrosis factor, GnRH: Gonadotropin-releasing hormone, FOXO1: Forkhead box protein O1, cGMP: Cyclic guanosine monophosphate, PKG: Protein kinase G, Rap1: Ras-associated protein-1, MAPK: Mitogen-activated protein kinase, PI3K: Phosphoinositide 3-kinase, Akt: Protein kinase B.
4. Discussions
The present study explored the differential expression of miRNAs in T2D patients treated with liraglutide and identified the key miRNA associated with therapeutic responses. Liraglutide treatment significantly influences the expression levels of 33 miRNAs, with 19 miRNAs showing increased levels and 14 miRNAs showing decreased expression levels. Network analysis revealed five miRNAs, hsa-miR-9-5p, hsa-miR-22-3p, hsa-miR-19b-3p, hsa-miR-132-3p and hsa-miR-93-5p were highly interconnected with other miRNAs and linked to 45, 28, 23, 22, and 16 target genes, respectively. These miRNAs and their target genes are associated with several key signaling pathways, such as PI3K/Akt (CCND2, THBS1, XIAP, ITGA2, MAPK1, CREB1, PTEN, and ATF2 genes), MAPK (MAPK1, FAS, and ATF2 genes), FOXO1 (MAPK1, CCND2, and PTEN genes), and Wnt (CSNK2A1, CALM1, AP2B1, XIAP, and PFN1 genes) signaling pathways. The PI3K/Akt, MAPK, and FOXO1 pathways are intricately connected to the target genes regulated by these miRNAs (Fig. 2b). These pathways play crucial roles in various cellular processes, for e.g., differentiation, proliferation, apoptosis, and stress responses, emphasizing the significance of miRNA-mediated intercellular communication in modulating these signaling networks.
The findings of this study are corroborated by several other in vivo or in vitro studies published in the recent past. A study demonstrated that liraglutide treatment enhanced the levels of PI3K, Akt, and mTOR in rate with acute myocardial injury (Abdel-Reheim et al. 2024) and promoted anti-apoptotic activity through activation of PI3K/Akt and MAPK signaling pathways (Zhu et al. 2016; Nizam et al. 2024). Similarly, an in vitro study reported that liraglutide treatment enhanced β-cell mass by activating the PI3K/Akt pathway and inhibiting FOXO1 (Fang et al. 2012). Likewise, another study reported that liraglutide activates the PI3K/Akt pathway in HaCaT cells (Nagae et al. 2018). Additionally, liraglutide was reported to exert neuroprotective action against ischemia-induced apoptosis by activating the PI3K/Akt and MAPK signaling pathways (Zhu et al. 2016). Western blot analysis in murine MC3T3-E1 preosteoblasts cells revealed that liraglutide activates the PI3K/AKT, ERK1/2, cAMP/PKA/β-cat-Ser675 signaling pathways (Wu et al. 2017).
Fig. 4 depicts various signaling pathways activated by liraglutide. Liraglutide exerts its effects through the GLP-1 receptor and activates PI3K/Akt signaling pathway (Wu et al. 2017). The activation of Akt inhibits FOXO1, thereby suppresses pro-apoptosis activity, promotes β-cell proliferation, and enhances insulin secretion (Zheng et al. 2024; Camaya et al. 2022). Moreover, Akt stimulates the mammalian target of rapamycin (mTOR) complex, which interacts with multiple downstream substrates (Camaya, Donnelly, and O’Brien 2022), leading to increased β-cell mass and improves insulin secretion (Halabitska et al. 2024). Akt activation also facilitates the translocation of GLUT4 (glucose transporter 4)-containing vesicles from intracellular compartments to the plasma membrane, enabling glucose uptake into cells. Additionally, liraglutide activates the MAPK pathways (Zhu et al. 2016), which is crucial for glucose-stimulated insulin secretion by pancreatic β-cells (Sidarala and Kowluru 2017). Liraglutide is also linked with Wnt/β-catenin signaling (Mali et al. 2022), a pathway indispensable for the transcription of genes involved in β-cell proliferation and insulin secretion (Nie et al. 2021).
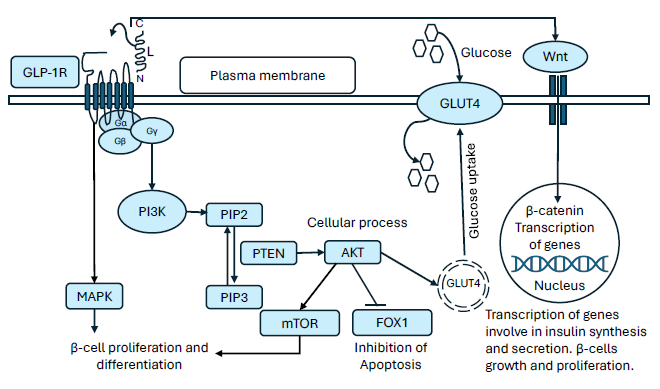
- Diagrammatic representation of intracellular signaling pathways activated by liraglutide via GLP-1 receptor in pancreatic β-cells. GLP-1R: Glucagon-like peptide-1 receptor, MAPK: Mitogen-activated protein kinase, PI3K: Phosphoinositide 3-kinase, PIP2: Phosphatidylinositol 4,5-biphosphate, PIP3: Phosphatidylinositol 3,4,5-triphosphate, PTEN: Phosphatase and tensin homologue, AKT: Protein kinase B, mTOR: Mechanistic target of rapamycin, FOXO1: Forkhead box protein O1, GLUT4: Glucose transporter 4.
Overall, activation of the PI3K/Akt pathway following liraglutide treatment appears to play a crucial role in insulin signaling, glucose uptake, glycogen synthesis, and maintaining glucose homeostasis (Taheri et al. 2024; Singh et al. 2024; Feng et al. 2024).
Earlier studies have suggested that variation in miRNA expression contributes to the modulation of various biological processes. Herein, miR-365a-3p and miR-125b-2-3p were found to be downregulated following liraglutide treatment in T2D patients. However, these miRNAs have previously been reported to be upregulated under hyperglycemic condition (Satake et al. 2018; Cheung et al. 2022). Similarly, miR-542-3p, which was reported to be downregulated in diabetic cardiomyopathy (Chavali et al. 2014) and ischemic stroke (Li, Tan, et al. 2019), was found upregulated in the present study. Additionally, other studies have linked the upregulation of miR-550a-3p to polyneuropathies (Pellegrino et al. 2023), damaged vascular smooth muscles (Chen et al. 2022), severe acute pancreatitis and acute lung injury (Lu et al. 2017; Satoh et al. 2015). Whereas, in the present study, miR-550a-3p was found downregulated following liraglutide treatment in T2D patients. These results were found consistent with the earlier study, wherein miR-550a-3p downregulation in plasma samples of T2D patients treated with sitagliptin was reported (Catanzaro et al. 2018).
Herein, we noticed that miR-19b-3p was upregulated following liraglutide treatment in T2D patients. However, a previous study reported miR-19b-3p downregulation in conditions of diabetic neuropathy (Rajabinejad et al. 2022), muscular dystrophy, and diabetic myopathy (Copier et al. 2017). Another study informed overexpression of miR-19b-3p in human skeletal muscle cells after aerobic exercise that enhances insulin signaling, increases glucose uptake, and improves maximal oxygen consumption (Massart et al. 2021). Likewise, miR-19b-3p overexpression has been shown to enhance insulin-sensitive signaling by upregulation of Akt phosphorylation (Liu et al. 2017).
A reduced expression of miR-338-3p was found associated with adverse effects on the Akt/glycogen synthase kinase 3 beta signaling pathway, impairing glycogen synthesis and inducing insulin resistance (Dou et al. 2017). In contrast, upregulation of miR-338-3p observed in this study following liraglutide treatment may contribute to metabolic control via the PI3K/Akt signaling pathway (Nagae et al. 2018).
Further, miR-93-5p was found to be downregulated in T2D patients treated with liraglutide. This finding aligns with the previous research study, which reported liraglutide inhibits miR-93-5p expression in the aortas of diabetic rats (Zhang et al. 2019). Lately, a study reported an upregulation of miR-93-5p in adipose tissue of T2D patients, which was inversely correlated with GLUT4 expression (Yan et al. 2022). Whereas, another study reported that the overexpression of miR-93-5p enhances insulin resistance and contributes to the progression of T2D in HepG2 cells (Zhou et al. 2021).
The second-highest hub miRNA, i.e., miR-22-3p was found to be downregulated in T2D patients treated with liraglutide. In contrast, earlier study reported overexpression of miR-22-3p in the liver of diabetic mice is related to impaired gluconeogenesis and increased insulin resistance (Kaur et al. 2015). Also, the elevated hepatic miR-22-3p expression in diabetic db/db mice is linked with the silencing of Wnt-responsive transcription factor Tcf7 (Kaur et al. 2015). Moreover, the overexpression of miR-22 obstructed stimulation of the PTEN/Akt/mTOR signaling pathway in myocardial tissues of myocardial infarction rats (Li et al. 2024). Furthermore, the elevated levels of miR-22-3p, miR-122, miR-192, miR-27a-3p, and miR-27b-3p in the plasma of obese mice have been found linked with insulin resistance, glucose intolerance, and dyslipidemia (Castaño et al. 2018). Conversely, studies have demonstrated that silencing miR-22-3p with an antagomir drug improves glucose tolerance, enhances insulin sensitivity, reduces body weight, and alleviates cholesterol levels (Thibonnier et al. 2020; Hu et al. 2020). The downregulation of miR-22-3p observed in the present study following liraglutide treatment in T2D patients may yield similar metabolic benefits.
In this study, miR-542-5p, miR-497-3p, miR-125a-3p, and miR-9-5p were upregulated, while miR-193b-3p was downregulated in T2D patients treated with liraglutide. These miRNAs are associated with FOXO signaling pathway (Favaro et al. 2021; Tian et al. 2020; Hu et al. 2022). The upregulation of miR-542-5p, miR-497-3p, and miR-125a-3p have been reported to inhibit FOXO1 expression via activation of the PI3K/Akt pathway (Tian et al. 2020). Notably, the overexpression of miR-542-5p was shown to reverse elevated blood lipid and glucose levels (Tian et al. 2020). Whereas, the downregulation of miR-193b-3p has been reported to reverse impaired glucose metabolism by inhibiting FOXO1 in the PI3K/Akt pathway in T2D patients (Hu et al. 2022). Additionally, the downregulation of miR-9-5p in diabetic retinopathy was connected with greater expression of both, SIRT1 and FOXOs (Li et al. 2019), thereby contributing to diabetes progression through the regulation of genes involved in glycolytic pathways (Zhang and Zhu 2018; Yu et al. 2021). The upregulation of miR-9-5p elicits a protective response against renal fibrosis and chronic kidney injury (Fierro-Fernández et al. 2020). Similarly, the overexpression of miR-193b-3p under hyperglycemic conditions was associated with the collection of intracellular lipid droplets in human hepatocyte-derived cells (Mollet and Macedo 2023). Based on these findings, the upregulation of mir-9-5p and downregulation of miR-193b-3p observed in this study following liraglutide treatment in T2D patients may contribute to reducing the progression of diabetes related chronic kidney disease (CKD) and liver fibrosis.
In the present study, miR-125a-5p was upregulated in T2D patients treated with liraglutide. Previous studies have reported that overexpression of miR-125a-5p is associated with reduced blood glucose and lipids levels in diabetic mice (Xu et al. 2018; Ji et al. 2014). Also, miR-125a-5p overexpression increased hepatic glycogen content, but decreased lipid droplet accumulation in the liver of diabetic mice (Xu et al. 2018; Ji et al. 2014).
In the present finding, miR-132-3p was downregulated in T2D patients treated with liraglutide. A recent study reported that miR-132 is drastically upregulated in circulating microvesicles as well as plasma of diabetic dyslipidemia patients (Nemecz et al. 2023). Whereas, another study reported that antagomir-132 effectively downregulates miR-132 under in vivo (BALB/c mice) conditions, thereby, improving insulin secretion and reducing blood glucose levels (Bijkerk et al. 2019). Additionally, the inhibition of miR-132–3p induced by fluorosis has been shown to potentiate the activation of the MAPK pathway, while its overexpression exhibits the opposite effect (He et al. 2024).
Overall, the miRNA expression data produced from this study revealed that liraglutide activates several critical signaling pathways, including PI3K/Akt, MAPK, FOXO1, and Wnt. These pathways collectively enhance β-cell function, improve insulin secretion, and increase glucose uptake in peripheral tissues, presenting liraglutide’s potential as a comprehensive therapeutic agent for T2D management.
Study limitations: Despite several important findings, this study suffers from some inherent limitations. The small sample size may lower the statistical power of the results, as miRNA studies typically require larger samples to account for individual variability in miRNA expression. The findings of this study are based on bioinformatics analysis of publicly available datasets supported with in vivo studies in animal models, the lack of in vivo experimental validation in T2D patients may weaken the strength of the findings. Furthermore, individual genetic, environmental, and lifestyle factors that may influence miRNA expression and possibly affect the reproducibility and generalizability of the results across different populations were not covered in the scope of this study.
5. Conclusion
The miRNA expression data revealed that liraglutide activates PI3K/Akt along with other signaling pathways, which subsequently enhances β-cell function, improves insulin secretion, and increases glucose uptake in T2D patients. This study emphasizes the significance of key miRNAs, i.e., hsa-miR-9-5p, hsa-miR-22-3p, hsa-miR-19b-3p, hsa-miR-132-3p, and hsa-miR-93-5p as potential predictive biomarkers for assessing the therapeutic response to liraglutide in T2D patients. Our findings indicate that these miRNAs offer vital insights into the mechanisms by which liraglutide mitigates T2D risk, opening possibilities for the development of novel biomarkers, therapeutic approaches, and precision health strategies. These findings provide a solid foundation for further experimental validation of the key signaling pathways involved in liraglutide therapy.
Acknowledgments
The authors gratefully acknowledge the funding of the Deanship of Graduate Studies and Scientific Research, Jazan University, Saudi Arabia, through project number: RG24-L06. The authors would like to acknowledge their respective institutes for providing the necessary support for the work.
CRediT authorship contribution statement
Mohammad Irshad: Conceived and designed the study and experiments, experimental execution, manuscript writing and critical reviewing. Soniya Yadav: Conceived and designed the study and experiments, experimental execution, manuscript writing and reviewing. Darin Mansor Mathkor: Conceived and designed the study and experiments, contributed materials and analysis tools, manuscript writing and reviewing. Ashjan Saeed Babegi: Conceived and designed the study and experiments, contributed materials and analysis tools, manuscript writing and reviewing. Shafiul Haque: Conceived and designed the study and experiments, contributed materials and analysis tools, manuscript writing and critical reviewing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Declaration of Generative AI and AI-assisted technologies in the writing process
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
References
- Unlocking the miRNA-34a-5p/TGF-β and HMGB1/PI3K/Akt/mTOR crosstalk participate in the enhanced cardiac protection of liraglutide against isoproterenol-induced acute myocardial injury rat model. Int. Immunopharmacol.. 2024;127:111369. https://doi.org/10.1016/j.intimp.2023.111369
- [CrossRef] [PubMed] [Google Scholar]
- Network centrality approaches used to uncover and classify most influential nodes with their related miRNAs in cardiovascular diseases. Gene Reports. 2022;27:101555. https://doi.org/10.1016/j.genrep.2022.101555
- [CrossRef] [Google Scholar]
- Liver fibrosis and liver stiffness in patients with obesity and type 1 diabetes. Diabetes. Obes. Metab.. 2024;26:4052-4059. https://doi.org/10.1111/dom.15760
- [CrossRef] [PubMed] [Google Scholar]
- Glucagon-like peptide-1 agonists combined with sodium-glucose cotransporter-2 inhibitors reduce weight in type 1 diabetes. Obesity (Silver Spring). 2023;31:716-723. https://doi.org/10.1002/oby.23677
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of obesity in people with and without type 1 diabetes across Belgium, Kuwait, and Mexico: An IMI2 SOPHIA study. eClinicalMedicine. 2024;77:102869. https://doi.org/10.1016/j.eclinm.2024.102869
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Dysregulated urinary extracellular vesicle small RNAs in diabetic nephropathy: Implications for diagnosis and therapy. J. Endocr. Soc.. 2024;8:bvae114. https://doi.org/10.1210/jendso/bvae114
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Global analysis of urinary extracellular vesicle small RNAs in autosomal dominant polycystic kidney disease. J. Gene Med.. 2024;26:e3674. https://doi.org/10.1002/jgm.3674
- [CrossRef] [PubMed] [Google Scholar]
- In vivo silencing of microRNA-132 reduces blood glucose and improves insulin secretion. Nucleic Acid. Ther.. 2019;29:67-72. https://doi.org/10.1089/nat.2018.0763
- [CrossRef] [PubMed] [Google Scholar]
- The long-acting glucagon-like peptide-1 analogue, liraglutide, inhibits beta-cell apoptosis in vitro. Biochem. Biophys. Res. Commun.. 2005;330:577-584. https://doi.org/10.1016/j.bbrc.2005.03.013
- [CrossRef] [PubMed] [Google Scholar]
- Targeting the PI3K/Akt signaling pathway in pancreatic β-cells to enhance their survival and function: An emerging therapeutic strategy for type 1 diabetes. J. Diabetes. 2022;14:247-260. https://doi.org/10.1111/1753-0407.13252
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proc. Natl. Acad. Sci. U S A. 2018;115:12158-12163. https://doi.org/10.1073/pnas.1808855115
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Circulating microRNAs in elderly type 2 diabetic patients. Int. J. Endocrinol.. 2018;2018:6872635. https://doi.org/10.1155/2018/6872635
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Therapeutic miRNA and siRNA: Moving from bench to clinic as next generation medicine. Mol. Ther. Nucleic Acids.. 2017;8:132-143. https://doi.org/10.1016/j.omtn.2017.06.005
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res.. 2020;48:W244-W251. https://doi.org/10.1093/nar/gkaa467
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Differential expression of dicer, miRNAs, and inflammatory markers in diabetic Ins2+/- akita hearts. Cell Biochem. Biophys.. 2014;68:25-35. https://doi.org/10.1007/s12013-013-9679-4
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- MiR-550a-3p restores damaged vascular smooth muscle cells by inhibiting thrombomodulin in an in vitro atherosclerosis model. Eur. 2022 J. Histochem. 66. https://doi.org/10.4081/ejh.2022.3429
- [Google Scholar]
- miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res.. 2020;48:D127-D131. https://doi.org/10.1093/nar/gkz757
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Glucose-dependent miR-125b is a negative regulator of β-cell function. Diabetes. 2022;71:1525-1545. https://doi.org/10.2337/db21-0803
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8 Suppl. 2014;4:S11. https://doi.org/10.1186/1752-0509-8-S4-S11
- [CrossRef] [Google Scholar]
- Circulating miR-19b and miR-181b are potential biomarkers for diabetic cardiomyopathy. Sci. Rep.. 2017;7:13514. https://doi.org/10.1038/s41598-017-13875-2
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Type 2 diabetes and myocardial infarction: Recent clinical evidence and perspective. Front Cardiovasc. Med.. 2021;8:644189. https://doi.org/10.3389/fcvm.2021.644189
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Management of hyperglycemia in type 2 diabetes, 2022 A consensus report by the american diabetes association (ADA) and the European association for the study of diabetes (EASD) Diabetes Care. 2022;45:2753-2786. https://doi.org/10.2337/dci22-0034
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Mir-338-3p mediates tnf-a-induced hepatic insulin resistance by targeting PP4r1 to regulate PP4 expression. Cell. Physiol. Biochem.. 2017;41:2419-2431. https://doi.org/10.1159/000475912
- [CrossRef] [PubMed] [Google Scholar]
- The akt/FoxO1/p27 pathway mediates the proliferative action of liraglutide in β cells. Mol. Med. Rep.. 2012;5:233-238. https://doi.org/10.3892/mmr.2011.607
- [CrossRef] [PubMed] [Google Scholar]
- Influence of high glucose in the expression of miRNAs and IGF1R signaling pathway in human myometrial explants. Arch. Gynecol. Obstet.. 2021;303:1513-1522. https://doi.org/10.1007/s00404-020-05940-5
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Metabolites of traditional chinese medicine targeting PI3K/AKT signaling pathway for hypoglycemic effect in type 2 diabetes. Front. Pharmacol.. 2024;15:1373711. https://doi.org/10.3389/fphar.2024.1373711
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- MiR-9-5p protects from kidney fibrosis by metabolic reprogramming. FASEB J.. 2020;34:410-431. https://doi.org/10.1096/fj.201901599RR
- [CrossRef] [PubMed] [Google Scholar]
- miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res.. 2012;40:37-52. https://doi.org/10.1093/nar/gkr688
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Liraglutide increases serum levels of microRNA-27b, -130a and -210 in patients with type 2 diabetes mellitus: A novel epigenetic effect. Metabolites. 2020;10:391. https://doi.org/10.3390/metabo10100391
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Explore the effect and target of liraglutide on islet function in type 2 diabetic rats by miRNA omics technology. Diabetes Metab. Syndr. Obes.. 2021;14:3795-3807. https://doi.org/10.2147/DMSO.S325030
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Diabetes and osteoarthritis: Exploring the interactions and therapeutic implications of insulin, metformin, and GLP-1-based interventions. Biomedicines. 2024;12:1630. https://doi.org/10.3390/biomedicines12081630
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The potential for microRNA therapeutics and clinical research. Front Genet.. 2019;10:478. https://doi.org/10.3389/fgene.2019.00478
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Regulating effect of miR-132–3p on the changes of MAPK pathway in rat brains and SH-SY5Y cells exposed to excessive fluoride by targeting expression of MAPK1. Ecotoxicol. Environ. Saf.. 2024;279:116467. https://doi.org/10.1016/j.ecoenv.2024.116467
- [CrossRef] [PubMed] [Google Scholar]
- Clinical potential of liraglutide in cardiovascular risk reduction in patients with type 2 diabetes: Evidence to date. Diabetes Metab. Syndr. Obes. 2019;12:505-512. https://doi.org/10.2147/DMSO.S174568
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Plasma miR-193b-3p is elevated in type 2 diabetes and could impair glucose metabolism. Front Endocrinol (Lausanne). 2022;13:814347. https://doi.org/10.3389/fendo.2022.814347
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- miR-22 inhibition reduces hepatic steatosis via FGF21 and FGFR1 induction. JHEP Rep. 2020;2:100093. https://doi.org/10.1016/j.jhepr.2020.100093
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- miRTarBase 2020: Updates to the experimentally validated microRNA–target interaction database. Nucleic Acids Res. 2019;48 (D1):D148-D54.
- [Google Scholar]
- Network-based identification of miRNAs and transcription factors and in silico drug screening targeting δ-secretase involved in alzheimer’s disease. Heliyon. 2021;7:e08502. https://doi.org/10.1016/j.heliyon.2021.e08502
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- miR-125a inhibits porcine preadipocytes differentiation by targeting ERRα. Mol. Cell. Biochem.. 2014;395:155-165. https://doi.org/10.1007/s11010-014-2121-4
- [CrossRef] [PubMed] [Google Scholar]
- Elevated hepatic miR-22-3p expression impairs gluconeogenesis by silencing the wnt-responsive transcription factor Tcf7. Diabetes. 2015;64:3659-3669. https://doi.org/10.2337/db14-1924
- [CrossRef] [PubMed] [Google Scholar]
- Toward the blood-borne miRNome of human diseases. Nat. Methods. 2011;8:841-843. https://doi.org/10.1038/nmeth.1682
- [CrossRef] [PubMed] [Google Scholar]
- Epidemiology of type 2 diabetes - global burden of disease and forecasted trends. J. Epidemiol. Glob. Health. 2020;10:107-111. https://doi.org/10.2991/jegh.k.191028.001
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Identification of microRNA and gene interactions through bioinformatic integrative analysis for revealing candidate signatures in prostate cancer. Gene Reports. 2022;27:101607. https://doi.org/10.1016/j.genrep.2022.101607
- [CrossRef] [Google Scholar]
- The profiling and role of miRNAs in diabetes mellitus. J. Diabetes Clin. Res.. 2019;1:5-23. https://doi.org/10.33696/diabetes.1.003
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The discovery and development of liraglutide and semaglutide. Front Endocrinol. (Lausanne). 2019;10:155. https://doi.org/10.3389/fendo.2019.00155
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- miRBase: From microRNA sequences to function. Nucleic Acids Res.. 2019;47:D155-D162. https://doi.org/10.1093/nar/gky1141
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- MiR-22 inhibits myocardial fibrosis in rats with myocardial infarction by targeting PTEN/Akt/mTOR signaling pathway. Cell. Mol. Biol. (Noisy-le-grand).. 2024;70:28-33. https://doi.org/10.14715/cmb/2024.70.1.4
- [Google Scholar]
- Downregulation of miR-139-5p contributes to the antiapoptotic effect of liraglutide on the diabetic rat pancreas and INS-1 cells by targeting IRS1. PLoS One. 2017;12:e0173576. https://doi.org/10.1371/journal.pone.0173576
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Identification of microRNAs and genes as biomarkers of atrial fibrillation using a bioinformatics approach. J. Int. Med. Res.. 2019;47:3580-3589. https://doi.org/10.1177/0300060519852235
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- RNA-seq revealed novel non-proliferative retinopathy specific circulating miRNAs in T2DM patients. Front Genet.. 2019;10:531. https://doi.org/10.3389/fgene.2019.00531
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- MIENTURNET: An interactive web tool for microRNA-target enrichment and network-based analysis. BMC Bioinformatics. 2019;20 https://doi.org/10.1186/s12859-019-3105-x
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Retracted: effects of microRNA‐19b on the proliferation, apoptosis, and migration of wilms’ tumor cells via the PTEN/PI3K/AKT signaling pathway. J. Cell. Biochem.. 2017;118:3424-3434. https://doi.org/10.1002/jcb.25999
- [CrossRef] [PubMed] [Google Scholar]
- ‘Differential analysis of count data–the DESeq2 package’. Genome Biol. 2014;15 (550):10-1186.
- [Google Scholar]
- Circulating miRNAs as biomarkers for severe acute pancreatitis associated with acute lung injury. World J. Gastroenterol.. 2017;23:7440-7449. https://doi.org/10.3748/wjg.v23.i41.7440
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Efficacy of liraglutide in patients with diabetic nephropathy: A meta-analysis of randomized controlled trials. BMC Endocr. Disord.. 2022;22 https://doi.org/10.1186/s12902-022-01006-6
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Microvascular and macrovascular complications of type 2 diabetes mellitus: Exome wide association analyses. Front Endocrinol (Lausanne). 2023;14:1143067. https://doi.org/10.3389/fendo.2023.1143067
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Metabolic and energetic benefits of microRNA-22 inhibition. BMJ Open Diabetes Res. Care. 2020;8:e001478. https://doi.org/10.1136/bmjdrc-2020-001478
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Endurance exercise training-responsive miR-19b-3p improves skeletal muscle glucose metabolism. Nat. Commun.. 2021;12:5948. https://doi.org/10.1038/s41467-021-26095-0
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Pre-diabetes-linked miRNA miR-193b-3p targets PPARGC1A, disrupts metabolic gene expression profile and increases lipid accumulation in hepatocytes: Relevance for MAFLD. Int. J. Mol. Sci.. 2023;24:3875. https://doi.org/10.3390/ijms24043875
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Glucagon-like peptide-1 analogue liraglutide facilitates wound healing by activating PI3K/Akt pathway in keratinocytes. Diabetes Res. Clin. Pract.. 2018;146:155-161. https://doi.org/10.1016/j.diabres.2018.10.013
- [CrossRef] [PubMed] [Google Scholar]
- Microvesicle-associated and circulating microRNAs in diabetic dyslipidemia: MiR-218, miR-132, miR-143, and miR-21, miR-122, miR-155 have biomarker potential. Cardiovasc. Diabetol.. 2023;22:260. https://doi.org/10.1186/s12933-023-01988-0
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The complex role of wnt ligands in type 2 diabetes mellitus and related complications. J. Cell. Mol. Med.. 2021;25:6479-6495. https://doi.org/10.1111/jcmm.16663
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Circulating hsa-miR-320a and its regulatory network in type 1 diabetes mellitus. Front. Immunol.. 2024;15:1376416. https://doi.org/10.3389/fimmu.2024.1376416
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Differential expression of microRNAs in serum of patients with chronic painful polyneuropathy and healthy age-matched controls. Biomedicines. 2023;11:764. https://doi.org/10.3390/biomedicines11030764
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Significance of circulating microRNAs in diabetes mellitus type 2 and platelet reactivity: Bioinformatic analysis and review. Cardiovasc. Diabetol.. 2019;18:113. https://doi.org/10.1186/s12933-019-0918-x
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The MALAT1-H19/miR-19b-3p axis can be a fingerprint for diabetic neuropathy. Immunol. Lett.. 2022;245:69-78. https://doi.org/10.1016/j.imlet.2022.03.004
- [CrossRef] [PubMed] [Google Scholar]
- limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res.. 2015;43:e47. https://doi.org/10.1093/nar/gkv007
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- MicroRNAs in cardiovascular disease: An introduction for clinicians. Heart. 2015;101:921-928. https://doi.org/10.1136/heartjnl-2013-305402
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Effects of liraglutide on weight loss, fat distribution, and β-cell function in obese subjects with prediabetes or early type 2 diabetes. Diabetes Care. 2017;40:1556-1564. https://doi.org/10.2337/dc17-0589
- [CrossRef] [PubMed] [Google Scholar]
- Circulating miRNA profiles associated with hyperglycemia in patients with type 1 diabetes. Diabetes. 2018;67:1013-1023. https://doi.org/10.2337/db17-1207
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- MicroRNA-seq data analysis pipeline to identify blood biomarkers for alzheimer’s disease from public data. Biomark. Insights. 2015;10:BMI.S25132. https://doi.org/10.4137/bmi.s25132
- [Google Scholar]
- Glimepiride compared to liraglutide increases plasma levels of miR-206, miR-182-5p, and miR-766-3p in type 2 diabetes mellitus: A randomized controlled trial. Diabetes Metab. J.. 2023;47:668-681. https://doi.org/10.4093/dmj.2022.0342
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The regulatory roles of mitogen-activated protein kinase (MAPK) pathways in health and diabetes: Lessons learned from the pancreatic β-cell. Recent Pat. Endocr. Metab. Immune Drug Discov. 2017;10:76-84. https://doi.org/10.2174/1872214810666161020154905
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- A network pharmacology approach with experimental validation to discover protective mechanism of poly herbal extract on diabetes mellitus. J. King Saud Univ. Sci.. 2024;36:103138. https://doi.org/10.1016/j.jksus.2024.103138
- [CrossRef] [Google Scholar]
- Molecular crosstalk: Notch can manipulate Hes1 and miR-9 behavior. J. Theor. Biol.. 2020;504:110404. https://doi.org/10.1016/j.jtbi.2020.110404
- [CrossRef] [PubMed] [Google Scholar]
- Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431-432. https://doi.org/10.1093/bioinformatics/btq675
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The PI3K/Akt signaling axis and type 2 diabetes mellitus (T2DM): From mechanistic insights into possible therapeutic targets. Cell Biol. Int.. 2024;48:1049-1068. https://doi.org/10.1002/cbin.12189
- [CrossRef] [PubMed] [Google Scholar]
- Metabolic and energetic benefits of microRNA-22 inhibition. BMJ Open Diabetes Res. Care. 2020;8:e001478. https://doi.org/10.1136/bmjdrc-2020-001478
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- MiR-542-5p inhibits hyperglycemia and hyperlipoidemia by targeting FOXO1 in the liver. Yonsei Med. J.. 2020;61:780-788. https://doi.org/10.3349/ymj.2020.61.9.780
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- ggplot2. WIREs Computational Stats. 2011;3:180-185. https://doi.org/10.1002/wics.147
- [CrossRef] [Google Scholar]
- Liraglutide, a glucagon-like peptide-1 receptor agonist, facilitates osteogenic proliferation and differentiation in MC3T3-E1 cells through phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT), extracellular signal-related kinase (ERK)1/2, and cAMP/protein kinase A (PKA) signaling pathways involving β-catenin. Exp. Cell Res.. 2017;360:281-291. https://doi.org/10.1016/j.yexcr.2017.09.018
- [CrossRef] [PubMed] [Google Scholar]
- miR-125a-5p ameliorates hepatic glycolipid metabolism disorder in type 2 diabetes mellitus through targeting of STAT3. Theranostics. 2018;8:5593-5609. https://doi.org/10.7150/thno.27425
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- CircRNA hsa_circ_0071336 is associated with type 2 diabetes through targeting the miR-93-5p/GLUT4 axis. FASEB J.. 2022;36:e22324. https://doi.org/10.1096/fj.202200149RR
- [CrossRef] [PubMed] [Google Scholar]
- miR-96-5p: A potential diagnostic marker for gestational diabetes mellitus. Medicine (Baltimore). 2021;100:e25808. https://doi.org/10.1097/MD.0000000000025808
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- ‘miR-9-5p plays an important role in gestational diabetes mellitus (GDM) progression by targeting HK-2’. Int. J. Clin. Exp. Med. 2018;11:6694-701.
- [Google Scholar]
- A glucagon-like peptide-1 analog, liraglutide, ameliorates endothelial dysfunction through miRNAs to inhibit apoptosis in rats. Peer J.. 2019;7:e6567. https://doi.org/10.7717/peerj.6567
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Glucagon-like peptide-1 receptor: Mechanisms and advances in therapy. Signal Transduct. Target Ther.. 2024;9:234. https://doi.org/10.1038/s41392-024-01931-z
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- miR-93-5p promotes insulin resistance to regulate type 2 diabetes progression in hepG2 cells by targeting HGF. Mol. Med. Rep.. 2021;23:329. https://doi.org/10.3892/mmr.2021.11968
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- The neuroprotection of liraglutide against ischaemia-induced apoptosis through the activation of the PI3K/AKT and MAPK pathways. Sci. Rep.. 2016;6:26859. https://doi.org/10.1038/srep26859
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]







