Translate this page into:
Malacitanolide, reissantin E and paclitaxel compounds as inhibitors of envelope, NS5 and NS2B/NS3 target proteins of dengue virus: Computational docking and molecular dynamics simulations studies
⁎Corresponding author at: Central Research Laboratory Meenakshi Academy of Higher Education and Research (Deemed to be University), Chennai 600 078, Tamil Nadu, India. n_arunagiri@yahoo.co.in (Arunagirinathan Narasingam)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Objective
The objective of this study is to find out the role of terpenoid compounds as potential inhibitors against certain protein targets of dengue virus.
Methods
The 2-dimensional structures of terpenoid compounds were retrieved from the PubChem database. They were analysed for their interactions with target proteins of dengue virus such as envelope (1OKE), NS5 (1R6A-RPV & SHA) and NS2B/NS3 (4M9K) by docking studies followed by molecular dynamics (MD) simulations using Schrödinger software, version 10.7.
Results
Out of 513 terpenoid compounds studied, malacitanolide showed the highest interaction energy values of −7.072 kcal/mol (hydrogen bond (HB) interactions with Thr280, Gln200, Gln271, Gln49 and Ala50), −5.295 kcal/mol (HB interactions with Ser150, Lys29, Ser214) and −4.030 kcal/mol (HB interactions with Glu1169 and Asn1119) against 1OKE, 1R6A-RPV and 4M9K targets, respectively. Paclitaxel had shown the interaction energy value of −9.334 kcal/mol (HB with Lys61, Asp146, Trp87, Gly148, and Arg84) with 1R6A-SAH. MD simulation studies revealed that the best interacting compound malacitanolide maintained a stable complex with 1OKE of dengue virus. Malacitanolide, reissantin E, and paclitaxel exhibited very good interactions with all three-protein targets of dengue virus and had also shown significant stability.
Conclusions
In the present study, it is concluded that the terpenoid compounds malacitanolide, reissantin E, and paclitaxel could act as potential inhibitors against all three target proteins of dengue virus.
Keywords
Dengue virus
Terpenoid
Malacitanolide
Molecular dynamics simulations
1 Introduction
Dengue is one of the important mosquito-borne diseases which is caused by four serotypes of dengue virus (DENV 1–4) (Bhatt et al., 2013). Aedes aegypti and Aedes albopictus are the major insect vectors that transmit dengue virus to human beings. Dengue haemorrhagic fever (DHF) is an important clinical symptom of dengue viral disease (Wang et al., 2020). World Health Organization estimated that around 390 million dengue cases occur annually and around 96 million cases have disease severity. The genome of dengue virus contains a positive sense long Open Reading Frame (ORF) encoding a polyprotein that contains structural proteins [C (capsid), prM (membrane), and E (envelope)] and non-structural proteins [NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5] (Nowak et al., 1989).
The E protein is involved in the attachment of virus to the host cell and fusion of virus to host cell membrane (Perera and Kuhn, 2008). NS2B/NS3 proteins process the polyprotein precursor prior to the assembly of viral replicase complex (Wahaab et al., 2021). The NS5 has RNA methyltransferase (MTase) and RNA-dependent RNA-polymerase (RdRp) in the N-terminal and C-terminal regions, respectively (Rodenhuis-Zybert et al., 2010). Based on the above vital functions, E, NS5 and NS2B/NS3 proteins are considered as potential targets for finding out antiviral therapeutics (Wahaab et al., 2021).
Target-based drug identification is mainly used for drug discovery (Lim et al., 2013). Phytocompounds are mainly targeted for drug development against many viral diseases. The important phytocompound groups are flavonoids, terpenoids, alkaloids, and steroids (Othman et al., 2019). Many studies reported that flavonoid derivatives are potential inhibitors against the dengue virus by inhibiting its replication (Allard et al., 2011; Frabasile et al., 2017). Phytocompounds belonging to the terpenoid group are studied extensively against the dengue virus. Therefore, in the present study, terpenoid compounds were studied for their ability as potential inhibitors against target proteins of dengue virus using in silico methods.
2 Materials and methods
2.1 Protein targets
Structural E protein (1OKE) and non-structural proteins (NS5 MTase [1R6A] and NS2B/NS3 Protease [4M9K]) of dengue virus were employed as target proteins for docking analysis. Three-dimensional crystal structures of target proteins were downloaded from PDB (Protein Data Bank) as.pdb format. The binding sites of the targets selected were BOG (1OKE), and SAH and RVP (1R6A). The binding region of 4M9K suggested by Schrödinger (Schrödinger, LLC, NY, USA, 2016) software was used as an active site for docking studies.
2.2 Retrieval of terpenoid compounds
A library of terpenoid compounds was prepared by retrieval from the databases PubChem (https://pubchem.ncbi.nlm.nih.gov/), NPACT (https://crdd.osdd.net/raghava/npact/) (Mangal et al., 2013) and KEGG (https://www.genome.jp/kegg/). Three-dimensional structures of terpenoid compounds were obtained from the PubChem and the structures were converted from.sdf to.mol format by Open Babel tool (O’Boyle et al., 2011). Ligand molecules were 3D optimized using ACD/ Chemsketch Freeware.
2.3 Molecular docking
Preliminary screening of phytocompounds was performed using iGEMDOCK and Molecular Docking Server. Reference ligand binding regions were selected as binding sites. The top hit three terpenoids were analysed by Glide module, Schrödinger. LigPrep and protein preparation wizard tools were used to prepare ligand and protein targets, respectively. Furthermore, optimized the H-bond networks and carried out the energy minimization using OPLS 2005 force filed. The active sites (Dscore > 1) of the target proteins were identified by SiteMap of Glide module in Mastero 10. The Grid for protein targets was generated using Grid generation panel. Induced Fit Docking (IFD) protocol was used to perform docking analysis where the protein and ligands were considered to be flexible to generate receptor conformations.
2.4 Post docking analysis - Molecular dynamics simulation
Finally, the screened compounds were further checked for interaction stability by performing Molecular Dynamics (MD) Simulations. Behaviours of receptors and ligands and physical movement of atoms in the real environment were also analysed. MD simulations were carried out for energy-minimised structure of the best compounds against the target proteins of the Dengue virus using Schrodinger’s Desmond module (Wang et al., 2013). The midpoint/Desmond methods were employed to perform high computational parallelism efficiently. The protein interactions were studied by OPLS-2005 and further solvated with a simple point charge water model. The protein–ligand complex quality was evaluated using Root Mean Square Deviation (RMSD) and Root Mean Square Fluctuation (RMSF) plots. The Nose-Hoover thermostat method was used to perform simulations with temperature of 300 K and coupling constant τt = 0.1 ps (Martyna et al., 1992). The short range of coulomb interactions was calculated and smooth particle mesh Ewald (PME) method with cut-off of 9.0 Å was used to calculate electrostatic interactions (Essmann et al., 1995). Simulation of prepared receptor-ligand complexes was performed for 5-ns times to investigate the protein–ligand complex stability.
2.5 ADMET profiling
The admetSAR tool was used to analyse the ADMET (Adsorption, distribution, metabolism, excretion and toxicity) properties of the screened phytocompounds (Cheng et al., 2012). Mutagenicity and carcinogenicity properties of the compounds were also studied, which helped to identify the potential drug candidate for animal model studies and clinical trials.
2.6 Density function theory (DFT) analysis
Screened terpenoid compounds were studied by DFT analysis. It analysed the orbital energies (HOMO and LUMO) followed by a detailed analysis of band gap energy and Dipole Movement using DMOL3/DFYL in DS V 4.0 (Lee et al., 1988).
3 Results
In the present study, a total of 513 terpenoid compounds were screened against three target proteins using iGEMDOCK. Top 50 compounds showing higher binding energy values were analysed for accurate docking and drug screening. Then, best ten terpenoid compounds such as malacitanolide, reissantin E, paclitaxel, gamma-tocopherol, saikosaponin B2, brunneogaleatoside, taiwaniaquinol A, englerin A, taxumairol, and saikosaponin D were docked with target proteins by Molecular Docking Server. It was observed that the phytocompounds paclitaxel, reissantin E, and malacitanolide exhibited docking scores of −6.43, −6.34 and −6.29 kcal/mol, respectively against 1OKE target (Fig. 1). The reference ligand BOG exhibited the interaction energy value of −5.70 kcal/mol against 1OKE. Likewise, the interaction energy values of −9.39, −7.56, −6.12 and −5.14 kcal/mol were obtained for paclitaxel, malacitanolide, reissantin E and SAH respectively against 1R6A-SAH, values of −7.37, −6.52, −6.19 and −5.90 kcal/mol for paclitaxel, malacitanolide, reissantin E and RVP respectively against 1R6A-RVP and values of −6.49, −6.43, −5.83 kcal/mol for paclitaxel, reissantin E and malacitanolide respectively against 4M9K. An inbuilt reference ligand was not reported for the 4M9K hence binding site region suggested by the MDS was adopted for docking analysis (Table 1).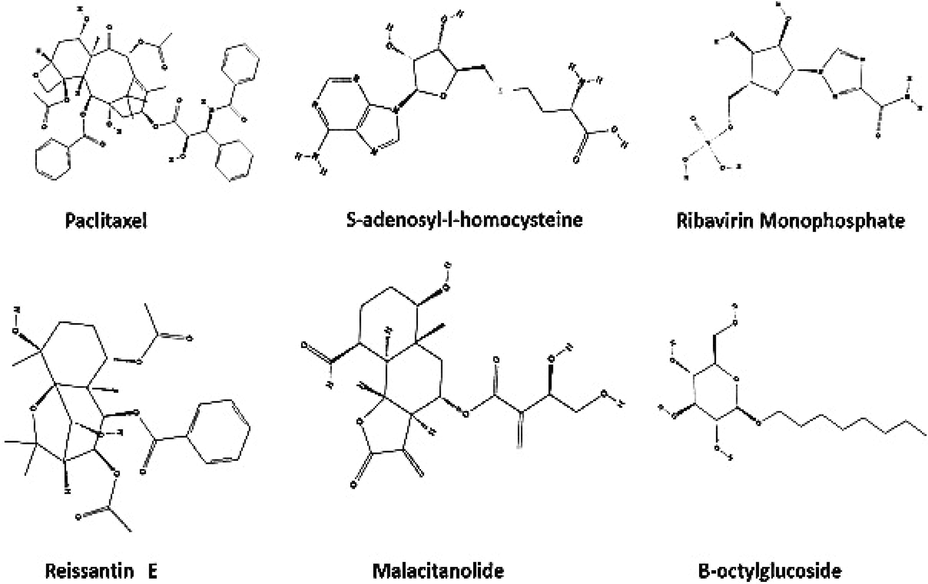
Chemical structures of reference ligands and terpenoid compounds.
Target
Ligand
Glide GScore
Kcal/mol
Glide Energy
XP HBond
Hydrogen bond interacting amino acids
No. of Hydrogen bond
Other non-bounded interactions
1OKE-BOG
BOG *
−8.354
−36.076
−3.125
OH – ALA 50;OH – THR 48; OH– THR 280
3
PRO 53, GLU 49, GLN 200, THR 268, LEU 191, GLY 190
Malacitanolide
−7.072
−37.387
−3.056
OH– THR 280;O- GLN 200
O-GLN271;OH-GLN 49O- ALA 505
LYS 47, GLY 275, PHE 193, LEU 135, PHE 279, THR 48, LEU277, LEU 198, PRO 53, VAL 130, ILE 270, ALA 205, SER 274
Paclitaxel
−6.895
−46.394
−2.125
OH – GLU 49;O-LYS 128
OH– ASP 203;OH– THR 48O- ALA 505
PRO53, GLN 52, LYS 47, LYS202, GLY 275, SER 274, LEU 135, LEU 198, VAL 130, PHE 193, LEU 207, PHE 279, THR 279,THR 280, LEU 277, ILE 270, GLN 200,GLN 271, ALA 205
Ressantin E
−4.426
−27.754
−1.220
2
PRO 53, LYS 128, GLN 52, THR 48, THR 280,ALA 50, LEU 135, GLU 49, VAL 130, LEU 198, ALA 205, GLN 271, ILE 270,LEU 277,LYS 47,GLY 275
1R6A-SAH
SAH*
−11.669
−63.258
−5.765
O- SER 56;O- GLY 86
OH– TRP 87;N- LYS 105
NH2– ASP 131;N- VAL 1326
PHE 133, VAL 130, LEU 103, THR 104, HIS110, GLY 106, GLU 111, GLY 83, GLY 148, CYS 82, ARG84, GLY 85
Paclitaxel
−9.334
−52.896
−3.367
OH– LYS 61;OH– ASP146
O-TRP 87;OH-GLY 148OH-ARG 845
ARG 160, PHE 133, ASP 131,VAL 132,ILE 147, THR 104, GLY 81,GLY 83,HIS 110,GLY 106,CYS 82, ASP 79, GLY 85,GLY 58,GLU 111, TYR 219, SER 56, LYS 181, GLU 217,GLU 149, ARG 38, SER 150,LYS 42,ARG 57,ARG 84
Malacitanolide
−6.130
−41.751
−2.055
O-LYS 105;O-GLY 106
OH– GLU 1493
GLY 148, HIS 110,GLU 111,GLY 83, THR 104, GLY 81,ASP 131, VAL 132,VAL 130,PHE 133,ILE 147,VAL 164,ARG 163, ARG 160 GLY 148
Ressantin E
−5.840
−39.204
−2.105
OH-SER 150, O-LYS-22
2
LYS 29, SER 214, PRO 152, LYS 14, LEU 17, ASN 18, GLY 21,PHE 25,LEU 20,GLN 26, SER 151
1R6A-RVP
RVP*
−8.004
−53.579
−3.169
OH-SER 150;OH-SER 214
OH-LYS14;OH-ASN 18O-LYS 22;NH2-LEU 206
LYS 29,THR 215,PRO 152,SER 151, LEU 17, GLY 21, PHE 25
Malacitanolide
−5.295
−39.257
−1.625
OH-SER 150;OH-LYS 14O-LYS 22
3
PHE 25,ALA 19, GLY 21,ASN 18, LEU 20,LEU 17,TRP13,PRO 152,THR 215, LYS 29
Paclitaxel
−4.561
−55.678
−1.862
O-LYS 22;O- LYS 29O-SER 214
3
SER 151,ARG 160,GLU 149, GLY 149, SER 150, LEU 183, LYS 181,THR 215, SER 211, GLU217, ARG 212,LYS 61,ARG 57,PRO 152, PHE 25,GLY 21, LEU 20, ASN 19,GLN 26
−3.363
−38.770
−1.438
O-LYS 105,O-GLY 148, OH-GLU 149
3
ARG 160, ARG 163, PHE 133, VAL 130, VAL 132, ASP 131, GLY 81, GLY 106,HIS 110, GLU 111,GLY 83,THR 104,ILE 147
4
M9K
Malacitanolide
−4.030
–33.592
−1.145
OH-GLU 1169O- ASN 1119
2
THR 1118,VAL 1154,LYS 1073,LYS 1074,ASN 1152,ALA 1166,ILE 1123,VAL 1155,ALA 1164
Ressantin E
−3.943
−36.509
−1.460
O-ASN 1119
1
ARG 1157, LYS 1117,VAL 1154, THR 1156, GLY 1153,VAL 1155, LYS 1073, ILE 1123,THR 1118,THR 1120, SER 1171,GLU 1169
Paclitaxel
−2.034
−20.342
−1.008
OH-ARG 1157
1
LYS 1117,VAL 1154, THR 1156, GLY 1153,VAL 1155, LYS 1073, ILE 1123,THR 1118,THR 1120, SER 1171,GLU 1169
3.1 Induced Fit docking Maestro 10.7
The screened compounds were further subjected to Maestro 10.7. Against 1OKE, malacitanolide showed Glide GScore of −7.02 kcal/mol and XP HBond of −3.056 kcal/mol and formed HB interactions with Thr280, Gln200, Gln271, Gln49 and Ala50 (Figs. 1 & 2), while paclitaxel showed Glide GScore of −6.895 kcal/mol and XP HBond of −2.125 kcal/mol and formed HB interactions with Glu49, Lys128, Asp203, Thr48 and Ala50 and reissantin E showed Glide GScore of −4.426 kcal/mol and XP HBond of −1.220 kcal/mol and formed HB interactions with Ser274 and Gln200. Against 1R6A-SAH, paclitaxel showed Glide GScore of −9.334 kcal/mol and XP HBond of −3.367 kcal/mol and HB interactions formed with Lys61, Asp146, Trp87, Gly148, and Arg84, while Malacitanolide showed Glide GScore of −6.130 kcal/mol and XP HBond of −2.055 kcal/mol and formed HB interactions with Lys105, Gly106, Glu149, and reissantin E showed Glide GScore of −5.840 kcal/mol and XP HBond of −2.105 kcal/mol and formed HB interactions with Ser150 and Lys22 (Figs. 2–4).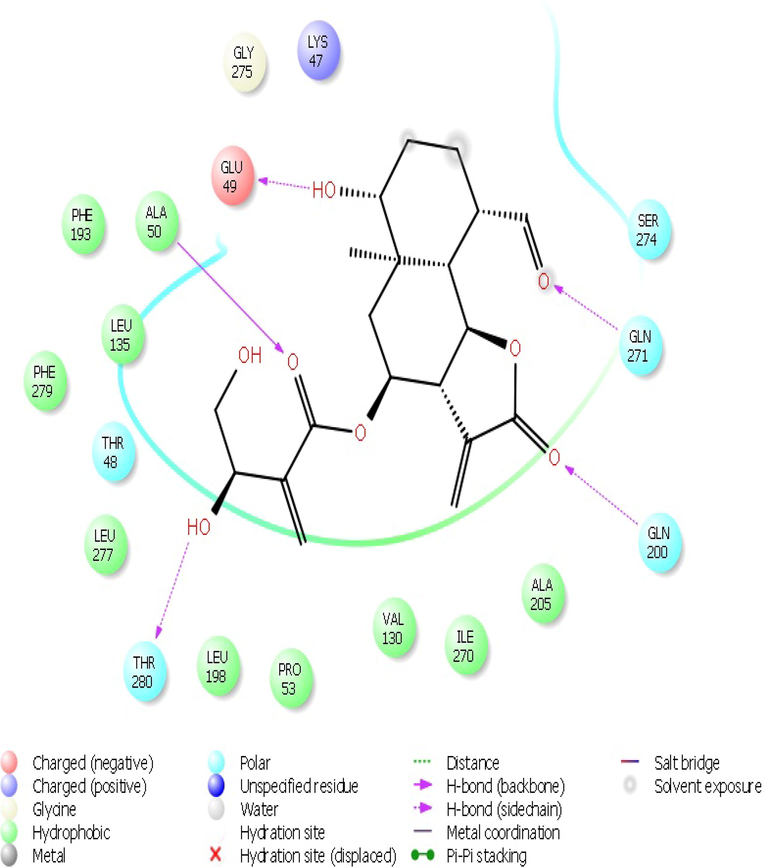
The 2D interaction plots of compound Malacitanolide with the BOG site of envelop protein of dengue virus.
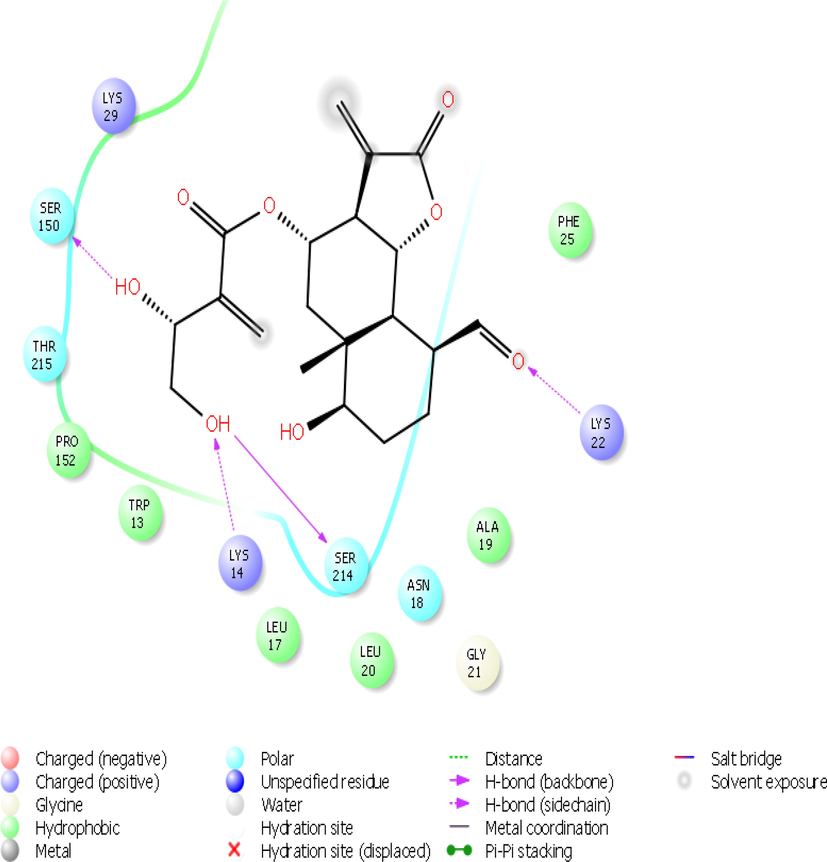
The 2D interaction plots of compound Malacitanolide with the 1R6A-RVP site of dengue virus.
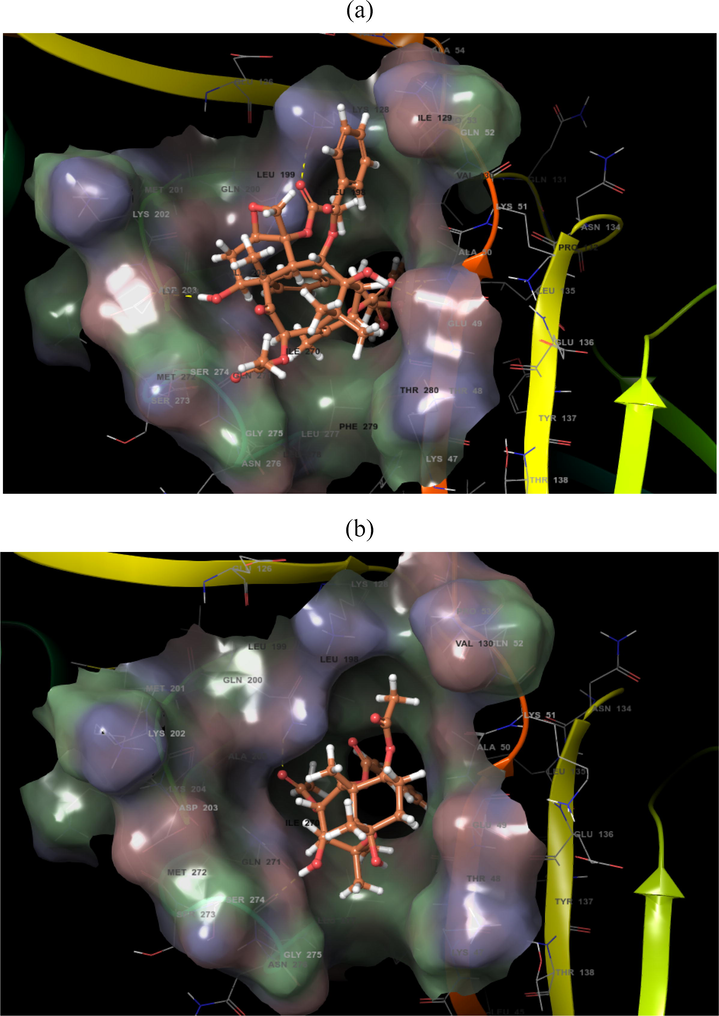
Docking pose of compounds (a) Paclitaxel and (b) Reissantin E with the 1OKE target protein of dengue virus.
While malacitanolide showed Glide GScore of −5.295 kcal/mol and XP HBond of −1.625 kcal/mol and formed HB interactions with Ser150, Lys14 and Lys22, Paclitaxel showed Glide GScore of −4.561 kcal/mol and XP HBond of −1.862 kcal/mol and formed HB interactions with Lys22, Lys29 and Ser214 and reissantin E showed Glide GScore of −3.363 kcal/mol and XP HBond of −1.438 kcal/mol and formed HB interactions with Lys105, Gly148 and Glu149 against 1R6A-RVP. Against 4M9K, the malacitanolide showed Glide GScore of −4.030 kcal/mol and XP HBond of −1.145 kcal/mol and formed HB interactions with Glu1169 and Asn1119, whereas reissantin E showed Glide GScore of −3.943 kcal/mol and XP HBond of −1.460 kcal/mol and formed HB interaction with Asn 1119 and paclitaxel showed Glide GScore of −2.034 kcal/mol and XP HBond of −1.008 kcal/mol and formed HB interaction with Arg1157 (Figs. 2 & 3). The Glide Energy and the interactions of other non-bonded amino acids of the above three compounds with protein targets using Maestro 10.7 (Schrödinger) are given in Table 1.
3.2 Molecular dynamics (MD) simulation
Structural dynamics of malacitanolide bound to 1OKE were analysed by performing molecular dynamics simulations for 5 ns. The top-ranked compound with IOKE of dengue virus was chosen for MD simulations. All five properties, like Energy fluctuations, RMSD, Root Mean Square Fluctuation (RMSF), ROG (radius of gyration), and H-bonds were analysed for assessing the protein–ligand complex stability. RMSD of protein structure backbone was simulated by plotting a graph with nanosecond verse angstrom units. The protein fluctuation was greatly observed until 1.5 Å at the initial stage, where slight conformational changes occurred after which the system attained stability. The Fig. 5 explains the stability of the complex and the degree of fluctuation. The RMSF of carbon α residues of the protein chain fluctuated at specific range of amino acids viz. 53–90, 101–110, and 325–329. These fluctuating sites mainly formed turns and coils, not the alpha helix or beta sheets. The protein residues involved in ligand contacts were highlighted in green colour. The ligand had maintained a stable confirmation at 1.5 to 2.0 Å. Initially, the fluctuation was seen in ROG for the ligand until 1 ns, then acquired and showed a regular conformation throughout the simulations. The radius of gyration ranged from 4.0 Å to 4.16 Å throughout the 5 ns simulations, and while looking into the interaction profile of ligand during simulation we observed that Thy280 had formed two hydrogen bonds with malacitanolide with 95% and 67% stability and Ala50 formed one hydrogen bond with 81% stability. A few other amino acids such as Glu49, Lys128, Thr48 and Lys47 also contributed to interactions supported by water bridges and this finding supports that water molecules maintaining the receptor–ligand complex (Fig. 4).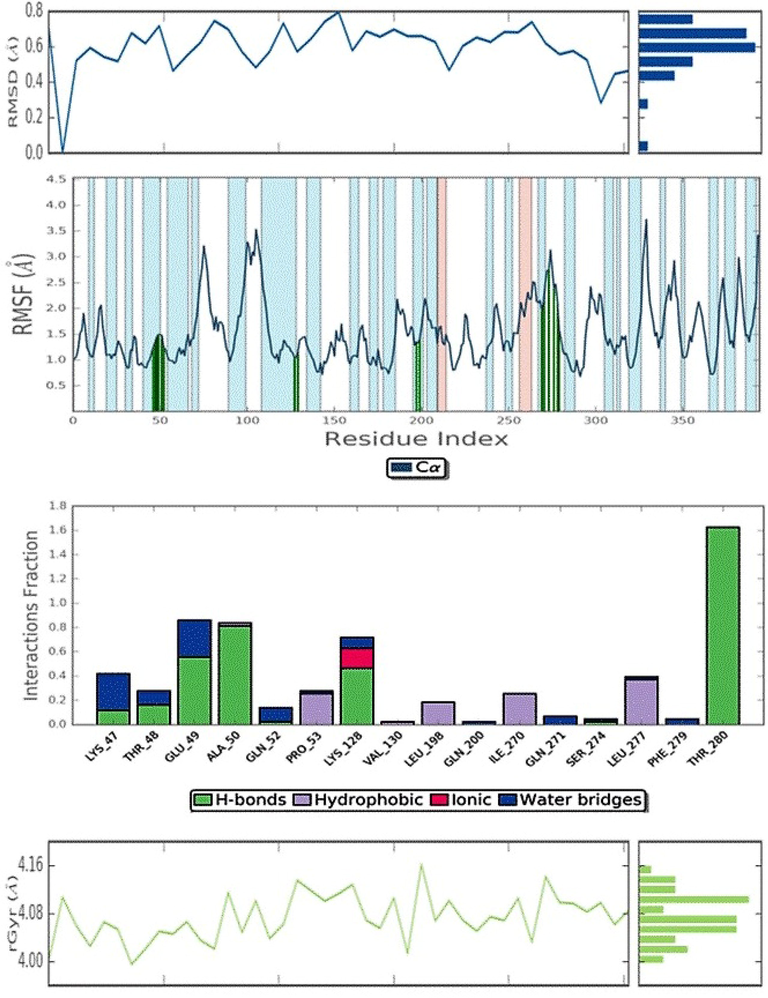
Molecular dynamic simulation of Dengue virus 1OKE complexed with malacitanolide.
3.3 ADMET properties
In this study, the ADMET analysis of malacitanolide, reissantin E, and paclitaxel revealed that all the three compounds were non-toxic and non-carcinogenic in nature and exhibited good human intestinal absorption properties. Other ADMET properties of the compounds are provided in Table 2.
S.
No.
Parameter
Paclitaxel
Malacitanolide
Reissantins E
Standard
BOG
B-Octylglucoside
SAH-Adenosyl-L-Homocysteine
RVP- Ribavirin monophosphate
1
BBB
Negative
Positive
Positive
Negative
Negative
Positive
2
HIA
Positive
Positive
Positive
Negative
Positive
Positive
3
CaCo2 PermeabilityCm/S
0.4145
0.1357
0.4588
0.3330
0.2404
0.7457
4
AMES
Non Toxic
Non Toxic
Non Toxic
Non Toxic
Non Toxic
Non Toxic
5
Carcinogenes
Non Carcinogen
Non Carcinogen
Non Carcinogen
Non Carcinogen
Non Carcinogen
Non Carcinogen
6
Biodegradation
Not readily Degradable
Not readily Degradable
Not readily Degradable
readily Degradable
Not readily Degradable
Not readily Degradable
7
Aqueous SolubilityLogS
−3.8728
−3.8155
−3.2918
−1.4197
−2.0627
−2.1749
8
Rat Acute ToxicityMol/Kg
2.4391
2.8626
2.9419
1.4828
2.3223
2.3307
9
Fish Oral ToxicityMg/L
0.6419
0.5096
2.8626
2.4039
1.6400
1.6538
3.4 Density function theory
The DFT analysis was performed for the lead molecules and the binding energy was correlated with the band gap energy. All the three lead molecules possess lower band gap energy. When a comparison was made with the three lead molecules, it was found that the compound paclitaxel showed lower band gap energy (0.0989602 kcal/mol) than malacitanolide and reissantin E (0.104356).
4 Discussion
The unavailability of effective antiviral drug in treating dengue infections complicates the clinical management of dengue patients. This study analyses the terpenoid compounds against dengue viral target proteins such as structural E protein and nonstructural proteins like NS5 and NS2B-NS3 using computational approaches. Many studies have suggested that the dengue E, NS2B/NS3 and NS5 proteins are potential targets to identify antiviral molecules (Modis et al., 2003; Hrobowski et al., 2005; Tomlinson et al., 2009; Murugesan and Manoharan, 2020). Non-structural proteins were also the most important targets due to their involvement in the dengue virus replication. They may have two domains of conserved nature, namely, methytransferase and RdRp (Rawlinson et al., 2006). It was found that among the terpenoid compounds screened in this study, three compounds contributed to the effective interactions with the structural and non-structural protein targets of the dengue virus. A study reported that the compounds doxycycline and rolitetracycline have shown an effective interaction with the envelope protein and exhibited hydrogen bonds with amino acids such as Thr48, Gln49, Ala50, Lys51, Glu52, Gln271 and Gln200 in the BOG site and also showed antiviral activity at 67.1 µM and 55.6 µM, respectively (Yang et al., 2007). In this study, malacitanolide formed the hydrogen bond with amino acids such as Thr280, Gln200, Gln271, Gln49 and Ala50 in the BOG site and it was hypothesised that malacitanolide compound would have significant antiviral properties against dengue virus owing to similar interaction profile with that of doxycycline and rolitetracycline.
It was noted that malacitanolide formed five HB interactions with both hydroxyl and ketone moieties. The amino acids Thr280, Glu49, Aln50 and Gln200 formed the HB interactions. Likewise, another lead compounds, namely, paclitaxel formed 5 HB interactions and reissantin E formed 2 HB interactions with E protein target. HB interactions of the screened lead compounds against E protein observed in this study were higher than the flavonoid compounds reported by Mir et al. who found that baicalein exhibited HB interaction with Thr48, fisetin with Thr48, Ile270 and Thr280, quercetin with Thr280 and naringenin did not form any HB interaction (Mir et al., 2016).
The docking results revealed that malacitanolide exhibited high interaction energy (-5.295 kcal/mol) with NS5 protein and formed HB interactions with hydroxyl and ketone moieties. Lim et al. (2013) found that the compound SPH1-103–799 formed HB interactions with Lys22, Leu20 and Asn18 of NS5 and the compound SPH1-101–102 formed HB interactions with Lys14, Ser150 and Lys22 of NS5. In this study, the amino acids such as Lys-14, Leu-17, Ser-150, Ser-151, Asn-18, Leu-20, Phe-25, Lys-29, Pro-152, Glu-408, and Gly-409 were significantly involved in the ligand binding with NS5. All the above mentioned amino acid interactions were agreed with the findings of Benarroch et al. (2004) who reported that the same amino acid interactions in RNA cap site except Glu-408 and Gly-409.
Another study identified few significant interactions of SAH with the SAM site of NS5 protein. The amino acids such as Lys105, His110, Asp131 and Ile147 were involved in the interactions and methylation process (Dong et al., 2010). We found that malacitanolide developed hydrogen bond interactions with Lys105, Gly106, and Glu149 and other interactions with Ile147, Asp131, and His110 of NS5 protein. The paclitaxel formed a hydrogen bond with four hydroxyl moieties in NS5 (1R6A-SAH) and the two ketone moieties contributed two hydrogen bonds. Paclitaxel also formed a pi-pi interaction with Arg57 and Arg84 and a hydrogen displacement site was seen at Lys105.
Lim et al. (2013) also reported in another study that the compounds SH1-007–088 and SPH1-111–460 formed hydrogen bond interactions with Ser56, Gly86, Asp146 and Lys105. Another study by Singh et al. (2016) reported HB interactions of analog 1 with Lys181 and Arg57 analog 15 with Ser150 and analog 17 with Lys181 and Arg84.
Against NS2B/NS3 (M49K), the malacitanolide showed docking score of −4.327 kcal/mol. The amino acids such as Arg157, Lys117, Val154, Thr156, Gly153, Val155, Lys73, Ile123, Thr118, Thr120, Ser171 and Glu169 were involved in the above interactions. The other two molecules reissantin E and paclitaxel failed to form sufficient binding energy. It was noted that all three compounds interacted with the different active sites. However, the hydroxyl and ketone groups of the terpenoid compounds contributed to the hydrogen bond interaction. Existence of oxygen functional group increases the biological activities of terpenoid compounds (Wu et al., 2015). Similarly, the hit compound in this study also possesses the hydroxyl and ketone moieties at different positions.
Molecular dynamics simulations have played a significant role in developing antiviral drugs by showing the importance of residual motion in the binding of ligand molecules. Molecular dynamics simulations could provide the function of the time of motion of an individual particle (Karplus and McCammon, 2002). In the MD simulations studies, the fluctuations are required for protein–ligand interaction and conformational changes in the protein. MD simulation was reported for less than 10 ns in earlier studies and now which has increased to 100 ns due to the availability of advanced computers. The temperature of the protein and the solvent used as either 180 K or 300 K. In a study, various bioflavonoid compounds were studied for interactions with dengue-RdRp target. Protein-ligand interaction of hesperidin was confirmed in simulation analysis for 5 ns and identified as potent RdRp inhibitor (Fatriansyah et al., 2022). In another study, mulberroside A chebulic acid, mulberroside C and curcumin were identified as potent anti-dengue virus phytocompounds by MD simulations studies (Vora et al., 2019). Similarly, in our study, we performed the MD simulations at temperature of 300 K and at time of 5 ns. MD simulations proved effective in visualising the migration of water molecules and stabilising the protein–ligand interaction. Finally, while looking into the quantum chemistry aspect of the studied compounds, all three compounds had very low band gap energy (ΔE) between both HOMO and LUMO, favouring the transfer of energy and exchange of electrons and would relatively result in increased activity of the compounds.
5 Conclusions
The screened terpenoid compounds, viz. malacitanolide, reissantin E, and paclitaxel were identified as potential inhibitors against envelope, NS5 and NS2B/NS3 target proteins of dengue virus. Moreover, these compounds possess good pharmacokinetic properties that would support their usage as therapeutics for dengue. Both in-vitro and in-vivo studies have to be conducted to further confirm the anti-dengue virus activity of the above three screened terpenoid compounds.
Acknowledgements
The authors would like to extend their sincere thanks to the Researchers Supporting Project (RSP2023R470), King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Alkylated Flavanones from the Bark of Cryptocarya chartacea As Dengue Virus NS5 Polymerase Inhibitors. J. Nat. Prod.. 2011;74:2446-2453.
- [CrossRef] [Google Scholar]
- A Structural Basis for the Inhibition of the NS5 Dengue Virus mRNA 2′-O-Methyltransferase Domain by Ribavirin 5′-Triphosphate. J. Biol. Chem.. 2004;279:35638-35643.
- [CrossRef] [Google Scholar]
- admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model.. 2012;52:3099-3105.
- [CrossRef] [Google Scholar]
- Biochemical and genetic characterization of dengue virus methyltransferase. Virology. 2010;405:568-578.
- [CrossRef] [Google Scholar]
- A smooth particle mesh Ewald method. J. Chem. Phys.. 1995;103:8577-8593.
- [CrossRef] [Google Scholar]
- Molecular docking and molecular dynamics simulation of fisetin, galangin, hesperetin, hesperidin, myricetin, and naringenin against polymerase of dengue virus. J. Trop. Med.. 2022;20:2022.
- [Google Scholar]
- The citrus flavanone naringenin impairs dengue virus replication in human cells. Sci. Rep.. 2017;7:41864.
- [CrossRef] [Google Scholar]
- Peptide inhibitors of dengue virus and West Nile virus infectivity. Virol. J.. 2005;2:49.
- [CrossRef] [Google Scholar]
- Molecular dynamics simulations of biomolecules. Nat. Struct. Biol.. 2002;9:646-652.
- [CrossRef] [Google Scholar]
- Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B. 1988;37:785-789.
- [CrossRef] [Google Scholar]
- Ten years of dengue drug discovery: progress and prospects. Antiviral Res.. 2013;100:500-519.
- [CrossRef] [Google Scholar]
- NPACT: naturally occurring plant-based anti-cancer compound-activity-target database. Nucleic Acids Res.. 2013;41:D1124-D1129.
- [CrossRef] [Google Scholar]
- Nosé-Hoover chains: the canonical ensemble via continuous dynamics. J. Chem. Phys.. 1992;97:2635-2643.
- [CrossRef] [Google Scholar]
- Identification of bioflavonoid as fusion inhibitor of dengue virus using molecular docking approach. Inf. Med. Unlocked. 2016;3:1-6.
- [CrossRef] [Google Scholar]
- A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci.. 2003;100:6986-6991.
- [CrossRef] [Google Scholar]
- Murugesan, A., Manoharan, M., 2020. Dengue Virus. In: Emerging and Reemerging Viral Pathogens. Elsevier, pp. 281–359. https://doi.org/10.1016/B978-0-12-819400-3.00016-8.
- Analyses of the terminal sequences of west nile virus structural proteins and of the in vitro translation of these proteins allow the proposal of a complete scheme of the proteolytic cleavages involved in their synthesis. Virology. 1989;169:365-376.
- [CrossRef] [Google Scholar]
- Antimicrobial activity of polyphenols and alkaloids in middle eastern plants. Front. Microbiol.. 2019;10:911.
- [CrossRef] [Google Scholar]
- Structural proteomics of dengue virus. Curr. Opin. Microbiol.. 2008;11:369-377.
- [CrossRef] [Google Scholar]
- Dengue virus RNA polymerase NS5: a potential therapeutic target? CDT. 2006;7:1623-1638.
- [CrossRef] [Google Scholar]
- Dengue virus life cycle: viral and host factors modulating infectivity. Cell. Mol. Life Sci.. 2010;67:2773-2786.
- [CrossRef] [Google Scholar]
- Inhibitor designing, virtual screening, and docking studies for methyltransferase: a potential target against dengue virus. J. Pharm. Bioall. Sci.. 2016;8:188.
- [CrossRef] [Google Scholar]
- Structure-based discovery of dengue virus protease inhibitors. Antiviral Res.. 2009;82:110-114.
- [CrossRef] [Google Scholar]
- Pharmacophore modeling, molecular docking and molecular dynamics simulation for screening and identifying anti-dengue phytocompounds. J. Biomol. Struct. Dyn.. 2019;38:1726-1740.
- [CrossRef] [Google Scholar]
- Potential role of flavivirus NS2B-NS3 proteases in viral pathogenesis and anti-flavivirus drug discovery employing animal cells and models: a review. Viruses. 2021;14:44.
- [CrossRef] [Google Scholar]
- Modeling Local Structural Rearrangements Using FEP/REST: application to Relative Binding Affinity Predictions of CDK2 Inhibitors. J. Chem. Theory Comput.. 2013;9:1282-1293.
- [CrossRef] [Google Scholar]
- Dengue hemorrhagic fever – A systemic literature review of current perspectives on pathogenesis, prevention and control. J. Microbiol. Immunol. Infect.. 2020;53:963-978.
- [CrossRef] [Google Scholar]
- Novel dengue virus NS2B/NS3 protease inhibitors. Antimicrob. Agents Chemother.. 2015;59:1100-1109.
- [CrossRef] [Google Scholar]
- Combinatorial computational approaches to identify tetracycline derivatives as flavivirus inhibitors. PLoS One. 2007;2:e428.
- [CrossRef] [Google Scholar]







