Translate this page into:
Larvicidal and antiviral nature of phoenix dactylifera L. natural products by targeting dengue virus and Aedes aegypti L. Proteins through molecular docking
⁎Corresponding author at: Bioproducts Research Chair, Zoology Department, College of Science, King Saud University, P.O. Box 2455, Riyadh 11451, Saudi Arabia. falmekhalfi@ksu.edu.sa (Fahd A. AL-mekhlafi)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Mosquito-borne infections are a global health threat. Different species of mosquitoes transmit viruses and cause several human diseases. In this study, in silico molecular docking of 23 phytochemicals of Phoenix dactylifera was performed to look for potential hits that bind effectively at the active site of different protein targets of the dengue virus (2FOM, 3U1I, and 2BMF) and Aedes aegypti mosquito (1YIY, 1PZ4 and 3UQI). The docking results of coumestrol to 2FOM resulted in four hydrogen bonds and ten hydrophobic interactions with binding energy of −9.5 kcal/mol. Similarly, the docking simulation of 2FOM to pinoresinol formed seven covalent bonds resulting in − 9.5 kcal/ mol energy. There were also two hydrophobic (THR289 and THR450) and one π-cation (LYS515) interactions with amino acid residues. Similarly, isofucosterol exhibited the best binding conformations with the lowest binding energy values with the two target proteins 1YIY and 1PZ4 of Ae.egyptia. The docking simulation of 1YIY to isofucosterol resulted in one hydrogen bond with binding energies of – 10.3 kcal/mol and 16 hydrophobic interactions with different amino acid residues. A similar observation of target protein 1PZ4 was noted in isofucosterol resulting in −9.7 kcal/mol energy. From the docking studies reported in this paper, promising candidates can be further optimized and studied in vitro.
Keywords
Phoenix dactylifera
Dengue Virus
Aedes aegypti
Molecular docking
In silico
1 Introduction
Dengue fever is a severe disease with a diagnostic rate of 2 million infections per year worldwide. One-third of the world's population is at risk of dengue virus infection. This disease is caused by the dengue virus, which spreads to people through the bites of infected Aedes species mosquitoes. DENV-1, DENV-2, DENV-3 and DENV-4 and DENV-5 are different dengue virus serotypes (Mustafa et al., 2015). They cause the same disease despite their genomic variation. A long single-stranded RNA encodes the polypeptide of the dengue virus. It is further cleaved into three structural proteins, including membrane, envelope, capsid, and seven non-structural (NS) proteins, including NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5(Endy et al., 2011; Normile, 2013; Qaddir et al., 2017).
Targeting DENV multifunctional enzymes are targeted to design antiviral therapies. NS2B/NS3 protease and NS3 helicase were targeted in this study. S2B-NS3 protease is the second-largest protein of the DENV genome (Chandramouli et al., 2010) and responsible for cytoplasmic cleavages, including at junctions between NS4B/NS5, NS3/NS4A, NS2B/NS3, and NS2A/NS2B protein and within the capsid, NS4A and NS2A proteins (Phoo et al., 2016). Similarly, NS3 helicase unwinds dsRNA to release ssRNA used as a template for NS5 protein in replication (Bartelma and Padmanabhan, 2002). NS2B/NS3 protease and NS3 helicase are conserved within the four serotypes (Chandramouli et al., 2010; Li et al., 2005), which permit the design of the drugs that are promising against the different serotypes (Keller et al., 2006; Xu et al., 2005).
Sterol carrier protein-2 (SCP-2) and kynurenine aminotransferase (KAT) are expressed throughout the animal kingdom, including insects. Aedes aegypti kynurenine aminotransferase (AeKAT) is a multifunctional enzyme that catalyzes the transamination of several amino acids and uses α-keto acids as amino group acceptors (Han et al., 2008). AeKAT has 45–50 % sequence identity with mammalian KAT-Is (Fang et al., 2002). AeKAT is mainly expressed in adult heads, demonstrating its significant role in the central nervous system (Fang et al., 2002). Aedes aegypti sterol carrier protein-2 protein (AeSCP-2) is an intracellular lipid carrier found in the midgut (Krebs and Lan, 2003). AeSCP-2 was reported to be involved in the cholesterol delivery and uptake across the cellular barrier between the hemocoel and the midgut (Blitzer et al., 2005; Dyer et al., 2003; Krebs and Lan, 2003).
Cholesterol is crucial for insects to grow, develop and reproduce (Perera and Wijerathna, 2019). AeSCP2 plays a vital role in cholesterol and fatty acid uptake in both larval and adult mosquitoes (Kumar et al., 2010). Knockdown of AeSCP-2 expression in larvae resulted in a reduction in cholesterol uptake, higher mortality, and decreased fecundity (Blitzer et al., 2005). In female adults, knockdown of AeSCP-2 expression leads to reduction of cholesterol uptake from the blood meal (Dyer et al., 2008). Few studies have focused on developing new mosquitocidal targeting AeSCP2 and AeKAT. Therefore, targeting AeSCP2 and AeKAT could be a substitute target for the discovery of promising mosquitocidal agents.
Medicinal plants have been used to treat various diseases since ancient times. Date palm, P. dactylifera L., a member of the Arecaceae family, is widely spread in the Arabian Peninsula. There are several potential health benefits of P. dactylifera (Al-Yahya, 1986; Allahyari et al., 2021; Demirci et al., 2013; Echegaray et al., 2020; Jassim and Naji, 2010). The crude extracts from this species have demonstrated antiviral (Allahyari et al., 2021) and insecticidal activities against Ae.aegypti (Demirci et al., 2011). A previous review on P. dactylifera reported more than 25 compounds, including phytoestrols, phytoestrogens, phenolic acid, and flavonoids (Al-Alawi et al., 2017). In the present study, these compounds will be used for docking against different target proteins of dengue virus and Ae. aegypti, and their interactions are observed to discover effective drugs.
Computational tools significantly impact drug discovery because of their fast and promising results. Docking studies predict the binding affinities of phytochemicals to the target enzymes. With our interest in searching for biological drug targets, we carried out this study to look for effective antiviral and mosquitocidal inhibitors in silico. The phytochemicals which show promising potential will be selected for further in vitro and in vivo testing in the future.
2 Materials and methods
2.1 Ligand preparation
The 23 phytochemical compounds of P. dactylifera comprised of 10 phytoestrols and phytoestrogens, 10 phenolic acid compounds and 3 flavonoids were downloaded from PubChem (https://pubchem.ncbi.nih.gov/). The files in ‘.sdf’ were converted to ‘.pdb’ format using the PyMol 1.1.0 tool. Torsion adjustment was carried out for the ligands. The Autodock 4.2 (Scripps Research Institute, La Jolla, CA, USA) tool was used to create the ‘pdbqt’ file.
2.2 Receptor preparation
The present study targets the protein from different dengue virus serotypes. The crystal structure of proteases (PDB ID: 2FOM, and 3U1I for dengue virus 2, and 3 respectively) and DENV-2 NS-3 helicase [PDB ID: 2BMF] were downloaded from RCSB databank (https://www.rcsb.org). Similarly, kynurenine aminotransferase [PDB ID: 1YIY], sterol carrier protein-2 [PDB ID: 1PZ4], and FKBP12 isomerase [PDB ID: 3UQI] of Aedes aegypti mosquito were also retrieved from the RCSB databank. The crystal structures for each protein were processed by deleting water molecules, adding missing atoms, and adding charges. The Autodock 4.2 tool was used to create the ‘pdbqt’ file.
2.3 Molecular docking and binding energy calculation
The protein and ligand preparation was carried out in AutoDock Tools. Once the grid box had been prepared, the docking was performed using AutoDock Vina 1.1.2 scoring function (Morris et al., 2009; Trott and Olson, 2010) to estimate binding energies. The complex formed with the least energy was selected and analyzed using PyMol (https://pymol.org), and Protein-Ligand Interaction Profiler (https://plip-tool.biotec.tu-dresden.de/plip-web/plip/index) software tools from the several docking poses.
3 Result
This evaluation of the bioactive compounds was based upon their binding parameters with the target proteins. The result of phytoestrols and phytoestrogens, phenolic acid, and flavonoid compounds against three different DENV enzymes (2FOM,3U1I, 2BMF) from Dengue Virus and three target proteins (1YIY, 1PZ4, 3UQI) from Ae. Aegypti (Table 1). The docked pose of 2FOM with the phytoesterol demonstrated the ligand's binding affinity with various targets of Dengue Virus (Table 2). The best potential binding sites of phytoestrols and phytoestrogens were −9.5 kcal/mol, −9.3 kcal/mol, and −8.9 kcal/mol for coumestrol, formononetin, and matairesinol, respectively. The best potential binding sites of phenolic acid and flavonoid compounds were 5-O-Caffeoylshikimic acid (-9.1 kcal/mol) and apigenin (-9.2 kcal/mol). In 2BMF, the potential binding sites among the different compounds docked were the best for pinoresinol (-9.5 kcal/mol) followed by luteolin (-9.0 kcal/mol) and chorogenic acid (-8.9 kcal/mol).
No.
Protein
PDB ID
1
DEN2 NS2B/NS3 serine protease
2FOM
2
DENV3 NS2B-NS3 protease
3U1I
3
DENV-2 NS-3 helicase
2BMF
4
Ae. aegypti kynurenine aminotransferase
1YIY
5
Ae. aegypti sterol carrier protein-2
1PZ4
6
Ae. aegypti FKBP12 Isomerase
3UQI
Sr. No.
Compound ID
Name
2FOM
3U1I
2BMF
Phytoestrols and phytoestrogens
1
Compound CID: 173183
Campesterol
−8.7
−6.6
−7.5
2
Compound CID: 5281326
Isofucosterol
−6.2
−8.0
−6.7
3
Compound CID: 5280961
Genistein
−8.8
−4.7
−9.0
4
Compound CID: 187808
Glycitin
−7.7
−7.4
−8.7
5
Compound CID: 5281812
Pformonetin
−9.3
−4.5
−7.3
6
Compound CID: 119205
Matairesinol
−8.9
−7.5
−8.3
7
Compound CID: 332427
Lariciresinol
−8.8
−7.1
−8.7
8
Compound CID: 73399
Pinoresinol
−8.7
−7.9
−9.5
9
Compound CID: 65373
Secoisolariciresinol
−6.6
−6.4
−7.0
10
Compound CID: 5281707
Coumestrol
−9.5
−8.1
−9.2
Phenolic acid compounds
11
Compound CID: 135
p-hydroxybenzoic acid
12
Compound CID: 72
Protocatechuic Acid
−7.2
−6.6
−8.4
13
Compound CID: 8468
Vanillic Acid
−6.4
−5.6
−6.0
14
Compound CID: 5372020
Cinamic Acid
−7.2
−5.6
−6.2
15
Compound CID: 689043
Caffeic Acid
−7.0
−6.0
−6.5
16
Compound CID: 445858
Ferulic Acid
−7.0
−5.9
−6.4
17
Compound CID: 637775
Sinapic Acid
−5.7
−6.1
−7.7
18
Compound CID: 5281762
5-O-Caffeoylshikimic Acid
−9.1
−7.7
−8.7
19
Compound CID: 1794427
Chlorogenic Acid
−8.8
−7.5
−8.9
20
Compound CID: 441772
Pelargonin
−6.2
−8.1
−8.7
Flavonoids
21
Compound CID: 5280443
Apigenin
−9.2
−7.9
−8.6
22
Compound CID: 5280445
Luteolin
−8.8
−8.2
−9.0
23
Compound CID: 5280804
Isoquercitrin
−6.6
7.0
−7.8
The target proteins of Ae. aegypti, namely 1YIY, 1PZ4, and 3UQI were docked with phytoestrols and phytoestrogens, phenolic acid, and flavonoid compounds by Autodock Vina. The energy values and the binding affinities are presented in Table 3. The energy values obtained of the drug targets of the most promising compound were recorded for 1YIY and 1PZ4 target proteins. The best potential binding sites of all compounds tested were recorded for isofucosterol (-10.3, kcal/mol) with 1YIY target proteins. This is followed by coumestrol (-9.3 kcal/mol) and campesterol (-8.9 kcal/mol) for the same target protein (1YIY). In 1PZ4, the best potential binding sites among the different compounds docked was isofucosterol (-9.7 kcal/mol) followed by coumestrol (-9.3 kcal/mol), p-fotmonrtin (-9.2 kcal/mol), and genistein (-8.9 kcal/mol). The best potential binding sites of phenolic acid and flavonoid compounds were apigenin (-9.2 kcal/mol) for 1YIY target proteins and chlorogenic acid (-8.6 kcal/mol) for 1PZ4 target proteins, respectively.
Sr. No.
Compound ID
Name
1YIY
1PZ4
3UQI
Phytoestrols and phytoestrogens
1
Compound CID: 173183
campesterol 10
−9.3
−9.5
−6.3
2
Compound CID: 5281326
Isofucosterol 4
−10.3
−9.7
−6.9
3
Compound CID: 5280961
Genistein 2
−8.5
−8.9
−6.0
4
Compound CID: 187808
Glycitin 3
−8.6
−6.2
−7.0
5
Compound CID: 5281812
Pformonetin 1
−8.3
−9.2
−5.8
6
Compound CID: 119205
Matairesinol 6
−8.1
−8.8
−6.1
7
Compound CID: 332427
Lariciresinol 5
−8.0
−8.8
−6.1
8
Compound CID: 73399
Pinoresinol 7
−8.2
−6.4
−6.9
9
Compound CID: 65373
Secoisolariciresinol 8
−8.1
−8.2
−5.7
10
Compound CID: 5281707
Coumestrol 9
−9.4
−6.7
−7.2
Phenolic acid compounds
11
Compound CID: 135
p-hydroxybenzoic acid
12
Compound CID: 72
protocatechuic acid
−7.3
−7.6
−5.5
13
Compound CID: 8468
Vanillic Acid
−6.5
−5.8
−5.1
14
Compound CID: 5372020
Cinamic acid
−7.2
−7.1
−5.4
15
Compound CID: 689043
Caffeic Acid
−7.3
−6.5
−5.4
16
Compound CID: 445858
Ferulic Acid
−7.1
−7.0
−5.4
17
Compound CID: 637775
Sinapic Acid
−7.5
−5.1
−5.7
18
Compound CID: 5281762
5-O-Caffeoylshikimic Acid
−7.9
−8.5
−6.3
19
Compound CID: 1794427
Chlorogenic Acid
−8.2
−8.6
−6.4
20
Compound CID: 441772
Pelargonin
−8.1
−6.4
−6.4
Flavonoids
21
Compound CID: 5280443
Apigenin
−8.9
−8.5
−5.9
22
Compound CID: 5280445
Luteolin
−8.6
−8.4
−6.1
23
Compound CID: 5280804
Isoquercitrin
−8.1
−5.7
−6.9
The docking poses and the 2D docking images of the best compounds with various Dengue virus and Ae. aegypti targets were analyzed with Protein-Ligand Interaction Profiler and PyMol software tools (Figs. 1-4). The docking poses were explored, and the amino acid residues involved in the various interactions were evaluated.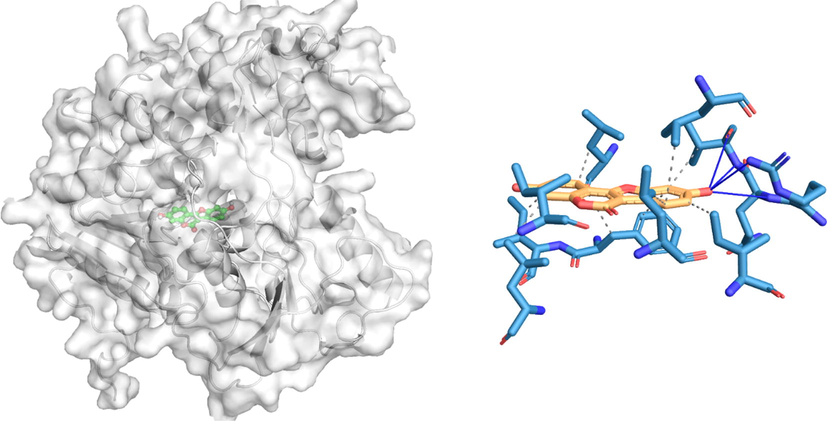
In silico molecular docking of the binding interaction of coumestrol compound with virus target protein (2FOM) based on the binding energy generated by AutoDock program. (A) A close-up view of the surface structure of 2FOM with coumestrol binding at the active site. (B) 2D structure of coumestrol interacting with 2FOM active site residues. …. Hydrophobic interaction
 Hydrogen bond.
Hydrogen bond.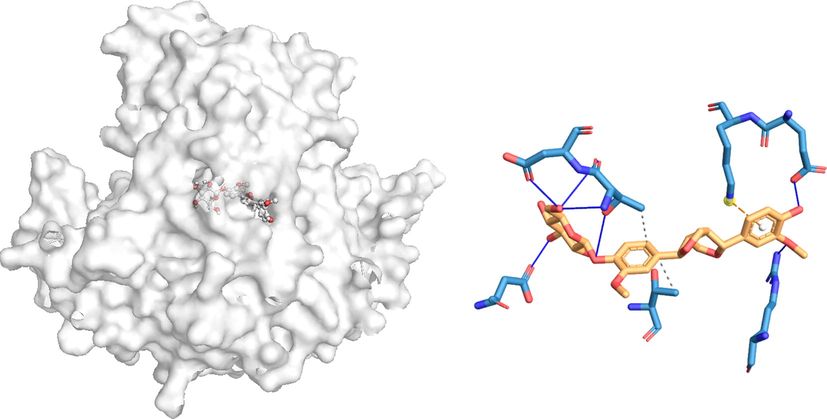
In silico molecular docking of the binding interaction of pinoresinol compound with virus target protein (2BMF) based on the binding energy generated by AutoDock program. (A) A close-up view of the surface structure of 2BMF with pinoresinol binding at the active site. (B) 2D structure of pinoresinol interacting with 2BMF active site residues. …. Hydrophobic interaction
 Hydrogen bond.
Hydrogen bond.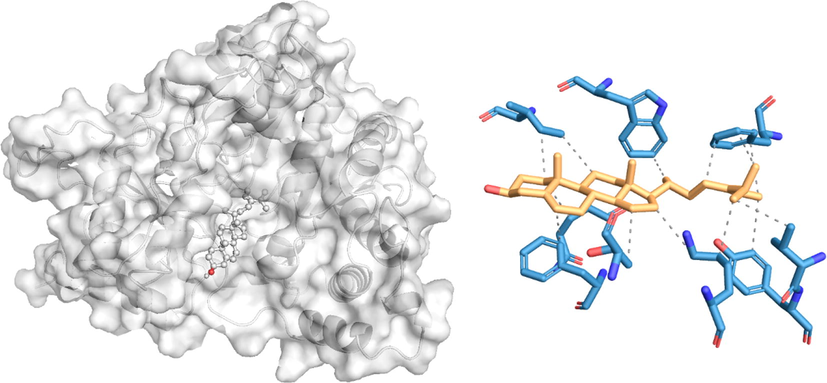
In silico molecular docking of the binding interaction of isofucosterol compound with the mosquito target protein (1YIY) based on the binding energy generated by the AutoDock program. (A) A close-up view of the surface structure of 1YIY with isofucosterol binding at the active site. (B) 2D structure of isofucosterol interacting with 1YIY active site residues. - - - - - Hydrophobic interaction.
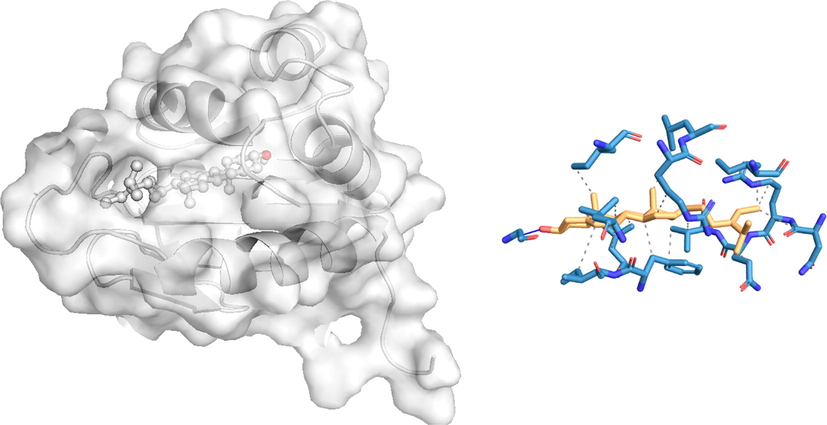
In silico molecular docking of the binding interaction of isofucosterol compound with mosquito target protein (1PZ4) based on the binding energy generated by AutoDock program. (A) A close-up view of the surface structure of 1PZ4with isofucosterol binding at the active site. (B) 2D structure of isofucosterol interacting with 1PZ4 active site residues. …. Hydrophobic interaction
 Hydrogen bond.
Hydrogen bond.
The docking results of coumestrol to 2FOM resulted in four hydrogen bonds (ARG24, GLN25, VAL26) and ten hydrophobic interactions (ILE12, LEU16, ILE19, VAL26, LEU48, LEU102, PHE105, ILE106, LEU109) with binding energies of −9.5 kcal/mol. Similarly, the docking simulation of 2BMF to pinoresinol resulted in seven hydrogen bonds to THR289, ASP290, ARG322, ASP409, and GLU514, resulting in the energy of − 9.5 kcal/ mol. There were also two hydrophobic (THR289 and THR450) and one π-cation (LYS515) interaction with amino acid residues.
Among the 23 phytocompounds, isofucosterol exhibited the best binding conformations with the lowest binding energy values with the two target proteins 1YIY and 1PZ4 of Ae.egyptia. The docking simulation of 1PZ4 to isofucosterol resulted in one hydrogen bond (GLY17) with − 9.7 kcal/mol binding energies. Also, formation of 16 hydrophobic interactions with ILE12, ARG15, LEU16, ILE19, ASN23, ARG24,GLN25, VAL26, LEU102, PHE105, ILE106, and LEU109. A similar observation of target protein 1YIY was noted with isofucosterol, with –10.3 kcal/mol energy. No hydrogen bond interaction was observed with isofucosterol, but there were 13 hydrophobic interactions with TRP27, ILE31, GLN44, PHE46, PHE135, VAL223, TYR224, LYS255, and THR260.
4 Discussion
Computational advances have a significant influence on the process of drug development. Virtual screening is widely used to reduce the time and cost of drug discovery. Molecular docking is an approach used to discover novel ligands for a target protein and plays an essential role in structure-based drug design (Meng et al., 2011). The relationship between the receptor and ligand plays a vital role in drug formulations. Several drugs were isolated from the natural products based on bioactivity–guided fractionation. Natural products can help cure many human diseases.
Natural products-derived antiviral compounds such as alkaloids (Acridone, Aporphine, β-Carboltne), carbohydrates (Glucosamine, Sulphated polysaccharides, γ-carragenan), chromones (Khellin, Visnagin, Psoralen), furanocoumarins and flavonoids (Cyanidin, Pelargodin), phenolics (Benzoic acid and Caffeic acid derivatives), terpene (Scopadulcic acid β, Scopadulin), triterpene steroids (Nigranoic acid, Buxamine E, Cyclobuxamine H) and many more have been published in several review articles (Che, 1991; El Sayed, 2000; Meng et al., 2011; Rinehart et al, 1993). Similarly, Several compounds have been published dealing with natural products-derived larvicidal compounds such as octacosane isolated from Moschosma polystachyum, falcarinol and falcarindiol (Cryptotaenia Canadensis), trans-asarone (Daucus carota), 3-n-butyl-4,5- dihydrophthalide (Apium graveolens), geranial, and neral (Magnolia salicifolia), β-selinene (Apium graveolens), β-selinene (graveolens, neoduline), 4-methoxyneoduline, and nepseudin ((Neorautanenia mitis) and many more (Kishore et al., 2014).
This study investigated the binding capability of bioactive compounds from P. dactylifera with key antiviral and larvicidal protein targets since various biological activities had been reported from this plant. Our study shows that the selected compounds can efficiently bind to the target protein and that molecular docking can be successfully used to find inhibitors from P. dactylifera.
Dengue protease plays an important role in dengue virus replication (Geiss et al., 2009). Therefore, blocking the interaction between its subunits or the active site disturb viral replication in the host. Reports revealed that disulphide cyclic peptides have inhibition potential against dengue protease(Tambunan et al., 2011; Trott and Olson, 2010). Therefore, pinoresinol and coumestrol reported in this study may obstruct the activity of the targeted protein via affecting the interactions with the active site or blocking the binding of subunits necessary for complex formation.
Mosquito control is carried out either on adults or immature larvae. The use of synthetic insecticides in controlling mosquito's adults and the larval population is harmful to the environment. Therefore, this necessitates the search for eco-friendly mosquitocides for controlling the mosquitoes' larvae.
AeKATis a multifunctional aminotransferase that catalyzes the transamination of several amino acids and uses α-keto acids as amino group acceptors (Han et al., 2008). It has a vital role in neuroactive activity (Han and Li, 2004). AeKAT is mainly expressed in adult heads of mosquitoes, demonstrating its significant role in the CNS (Fang et al., 2002). Cholesterol uptake is essential for the larval population and is carried out through the AeSCP-2. Several compounds were screened to block the target protein. Currently, isofucosterol was a promising analog to dock with cholesterol carrier protein AeSCP-2.
Many studies have attempted to look for different sources of compounds that inhibit the action of SCP-2 via competitive binding. Some are natural compounds (Anstrom et al., 2012; Ee et al., 2006; Larson et al., 2014), and some are synthetic compounds (Kim et al., 2005),. Kim et al. reported several sterol carrier protein inhibitors. They found that these inhibitors showed physiological effects on cholesterol metabolism in cell culture, similar to the impact of AeSCP-2 knockdown (Kim et al., 2005). Several studies revealed that sterol carrier protein inhibitors have toxicity on Cx. pipiens, Cx. quinquefasciatus, and Anopheles gambiae (Larson et al., 2014; Li et al., 2009). Furthermore, it is reported that there is a synergistic activity of AeSCP-2 inhibitors when used with permethrin (Li et al., 2009). Furthermore, the AeSCP-2-mediated cholesterol uptake pathway is required for dengue virus production in Aedes mosquitoes, and the inhibition of AeSCP-2 activity resulted in repressed production of the virus within mosquito cells in vitro (Fu et al., 2015). However, further studies are required to elucidate the interaction mechanism between the promising compounds, the targeted proteins, and the feasibility of using these inhibitors in the fields.
Acknowledgements
Princess Nourah bint Abdulrahman University Researchers, Supporting Project number (PNURSP2022R82), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Date palm tree (Phoenix dactylifera L.): natural products and therapeutic options. Front. Plant Sci.. 2017;8:845.
- [Google Scholar]
- Antiviral activity of Phoenix dactylifera extracts against herpes simplex virus type 1: an animal study. Comp. Clin. Pathol.. 2021;30(6):945-951.
- [Google Scholar]
- Phytochemical and biological studies on Saudi medicinal plants. 11. The major componentt of the spathe oil of Phoenix dactylifera. Fitoterapia. 1986;57:284-285.
- [Google Scholar]
- Mosquitocidal properties of natural product compounds isolated from Chinese herbs and synthetic analogs of curcumin. J. Med. Entomol.. 2012;49(2):350-355.
- [Google Scholar]
- Expression, purification, and characterization of the RNA 5′-triphosphatase activity of dengue virus type 2 nonstructural protein 3. Virology. 2002;299(1):122-132.
- [Google Scholar]
- Functional analysis of AeSCP-2 using gene expression knockdown in the yellow fever mosquito, Aedes aegypti. Insect Mol. Biol.. 2005;14(3):301-307.
- [Google Scholar]
- Serotype-specific structural differences in the protease-cofactor complexes of the dengue virus family. J. Virol.. 2010;84(6):3059-3067.
- [Google Scholar]
- Marine products as a source of antiviral drug leads. Drug Dev. Res.. 1991;23(3):201-218.
- [Google Scholar]
- Phoenix dactylifera L. essential oil: Chemical composition, antimicrobial and insecticidal activities. Planta Med.. 2011;77(12)
- [Google Scholar]
- Phoenix dactylifera L. spathe essential oil: Chemical composition and repellent activity against the yellow fever mosquito. Acta Trop.. 2013;128(3):557-560.
- [Google Scholar]
- The structural determination of an insect sterol carrier protein-2 with a ligand-bound C16 fatty acid at 1.35-Å resolution. J. Biol. Chem.. 2003;278(40):39085-39091.
- [Google Scholar]
- Three-dimensional structure/function analysis of SCP-2-like2 reveals differences among SCP-2 family memberss. J. Lipid Res.. 2008;49(3):644-653.
- [Google Scholar]
- Phoenix dactylifera products in human health–A review. Trends Food Sci. Technol.. 2020;105:238-250.
- [Google Scholar]
- Xanthones from Garcinia mangostana (Guttiferae) Nat. Prod. Res.. 2006;20(12):1067-1073.
- [Google Scholar]
- Natural products as antiviral agents. Stud. Nat. Products Chem. Elsevier 2000:473-572.
- [Google Scholar]
- Determinants of inapparent and symptomatic dengue infection in a prospective study of primary school children in Kamphaeng Phet, Thailand. PLoS Negl. Trop. Dis.. 2011;5(3)
- [Google Scholar]
- Isolation, characterization, and functional expression of kynurenine aminotransferase cDNA from the yellow fever mosquito, Aedes aegypti. Insect Biochem. Mol. Biol.. 2002;32(8):943-950.
- [Google Scholar]
- Sterol carrier protein 2, a critical host factor for dengue virus infection, alters the cholesterol distribution in mosquito Aag2 cells. J. Med. Entomol.. 2015;52(5):1124-1134.
- [Google Scholar]
- Focus on flaviviruses: current and future drug targets. Future Med. Chem.. 2009;1(2):327-344.
- [Google Scholar]
- Cysteine and keto acids modulate mosquito kynurenine aminotransferase catalyzed kynurenic acid production. FEBS Lett.. 2004;577(3):381-385.
- [Google Scholar]
- Structural insight into the mechanism of substrate specificity of Aedes kynurenine aminotransferase. Biochemistry. 2008;47(6):1622-1630.
- [Google Scholar]
- In vitro evaluation of the antiviral activity of an extract of date palm (Phoenix dactylifera L.) pits on a Pseudomonas phage. Evid.-Based Complement. Alternat. Med.. 2010;7(1):57-62.
- [Google Scholar]
- Finding new medicines for flaviviral targets, Novartis Foundation Symposium. Chichester; New York: John Wiley; 2006. p. :102. 1999
- Identification of mosquito sterol carrier protein-2 inhibitors. J. Lipid Res.. 2005;46(4):650-657.
- [Google Scholar]
- Natural products as leads to potential mosquitocides. Phytochem. Rev.. 2014;13(3):587-627.
- [Google Scholar]
- Isolation and expression of a sterol carrier protein-2 gene from the yellow fever mosquito, Aedes aegypti. Insect Mol. Biol.. 2003;12(1):51-60.
- [Google Scholar]
- A search for mosquito larvicidal compounds by blocking the sterol carrying protein, AeSCP-2, through computational screening and docking strategies. Pharmacog. Res.. 2010;2(4):247.
- [Google Scholar]
- Larvicidal activity of sterol carrier protein-2 inhibitor in four species of mosquitoes. J. Med. Entomol.. 2014;45(3):439-444.
- [Google Scholar]
- Larvicidal activity of mosquito sterol carrier protein-2 inhibitors to the insecticide-resistant mosquito Culex quinquefasciatus (Diptera: Culicidae) J. Med. Entomol.. 2009;46(6):1430-1435.
- [Google Scholar]
- Functional profiling of recombinant NS3 proteases from all four serotypes of dengue virus using tetrapeptide and octapeptide substrate libraries. J. Biol. Chem.. 2005;280(31):28766-28774.
- [Google Scholar]
- Molecular docking: a powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug Des.. 2011;7(2):146-157.
- [Google Scholar]
- AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem.. 2009;30(16):2785-2791.
- [Google Scholar]
- Discovery of fifth serotype of dengue virus (DENV-5): A new public health dilemma in dengue control. Med. J. Armed Forces India. 2015;71(1):67-70.
- [Google Scholar]
- Surprising New Dengue Virus Throws A Spanner In Disease Control Efforts. American Association for the Advancement of Science; 2013.
- Perera, H., Wijerathna, T., 2019. Sterol carrier protein inhibition-based control of mosquito vectors: current knowledge and future perspectives. Canadian Journal of Infectious Diseases and Medical Microbiology 2019.
- Structure of the NS2B-NS3 protease from Zika virus after self-cleavage. Nat. Commun.. 2016;7(1):1-8.
- [Google Scholar]
- Computer-aided analysis of phytochemicals as potential dengue virus inhibitors based on molecular docking, ADMET and DFT studies. J. Vector Borne Dis.. 2017;54(3):255.
- [Google Scholar]
- Antiviral substances. In: Attaway D.H., Zaborsky O.R., eds. Marine Biotechnology, Pharmaceutical and Bioactive Natural Products. Plenium Press; New York; 1993. p. :309-342.
- [Google Scholar]
- Computational design of disulfide cyclic peptide as potential inhibitor of complex NS2B-NS3 dengue virus protease. Afr. J. Biotechnol.. 2011;10(57):12281-12290.
- [Google Scholar]
- AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem.. 2010;31(2):455-461.
- [Google Scholar]
- Structure of the Dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 A. J. Virol.. 2005;79(16):10278-10288.
- [Google Scholar]







