Translate this page into:
Lactobacilli species diversity in gut microbiota of renal failure patients
⁎Corresponding author. kafilhs@tbzmed.ac.ir (Hossein Samadi Kafil)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Lactobacillus species and other acid producing bacteria could inhibit aerobic bacteria, normalize the intestinal microbiota and decrease the uremic toxins in patients with chronic kidney disease. Therefore, the aim of this study is assessment of abundance of lactobacillus spp. in fecal flora of End-stage kidney disease (ESKD) and kidney transplanted patients. DNA of 20 fecal samples of ESKD and kidney-transplanted as patients group and 20 fecal samples of non-ESKD and -kidney-transplanted as control group, which were admitted to hospitals, were extracted. Amplified DNA by 16srRNA illumina V3 and V4 primers were sequenced by MiSeq system. Total 651 strains, 37 (5.68%) strains were identified as order Lactobacillales. The strains were classified into six family, nine genus and 37 species. The most abundance genus in both groups are Streptococcus spp. and Lactobacillus spp. and the lowest abundance genus in both groups are Pediococcus and Leuconostoc. Comparing the abundance mean of the strains revealed that there is no significant association between control group and disease group, but the abundance mean of the strains in control group increase in compared to disease group. More diversity was exhibited in lactobacillus spp. in patients with chronic kidney disease compared to control group. Some of the species of Lactobacillaceae family such as L. acidophilus was not found in both patients and control groups. In addition, more of the species of Lactobacillaceae were decreased in patients group, whereas certain species were increased in patients group compared to control group.
Keywords
Lactobacillus
Chronic kidney disease
Renal failure patient
End-stage kidney disease (ESKD)
Fecal flora
1 Introduction
The gastrointestinal tract of human consists of various bacteria including aerobic and anaerobic bacteria. The entire of colon is filled by mostly commensal and obligatory anaerobic bacteria such as lactobacilli (Muyzer et al., 1993; Dahroud et al., 2016; Quagliariello et al., 2016). Lactobacillus genus is a gram-positive, non-sporulating, catalase negative and rod-shaped bacillis that contains more than ninety validly described species and subspecies (Jabbari et al., 2017). Lactobacillus species are included in the division of Firmicutes (Ludwig et al., 2010) and are belonged to the members of lactic acid bacteria, which are found in human and animals intestinal microbiota (Vaughan et al., 2005; Ludwig et al., 2010). As well as, Lactobacillus species are one of the most commonly used probiotic strains in the wide range of food components in preventing chronic inflammation disease and enhancing health (Pandey et al., 2015). The fermentative characteristics of lactobacillus spp. have classified them into three divisions including obligate heterofermentative lactobacilli (OHEL), obligate homofermentative lactobacilli (OHOL) and facultative heterofermentative lactobacilli (FHEL)[24]. The activity of these aerobic and anaerobic bacteria has a major impact on the host health and characteristics such as development of the immune system, host protection against pathogens, and positive effects on the host nutrition and colonic health (Falk et al., 1998; Hooper et al., 2001; Gholizadeh et al., 2019). The protein-bound uremic toxins such as p-cresyl sulphate (PCS), indoxyl sulphate (IS), p-cresylglucuronide, kynurenic acid and indole acetic acid (IAA), which are normally excreted into the urine, could be contributing factor to progression chronic renal disease that have deleterious effects (Vanholder and Glorieux, 2015). These uremic toxins could lead to damage endothelial cells or renal tubular cells by fibrotic changes, inflammatory responses or enhancing cellular oxidative by activation of Nicotinamide adenine dinucleotide (NADPH) oxidase, P53, nuclear factor-ĸB (NFĸB) (Shimizu et al., 2013; Watanabe et al., 2013; Vanholder et al., 2014; Vanholder and Glorieux, 2015). The precursors of these uremic toxins are produced by gut microbiota from tyrosine or tryptophan and absorbed from the colon. Alters in the absorption of the uremic toxins from the colon may affect renal function (Vanholder and Glorieux, 2015) (Fig. 1).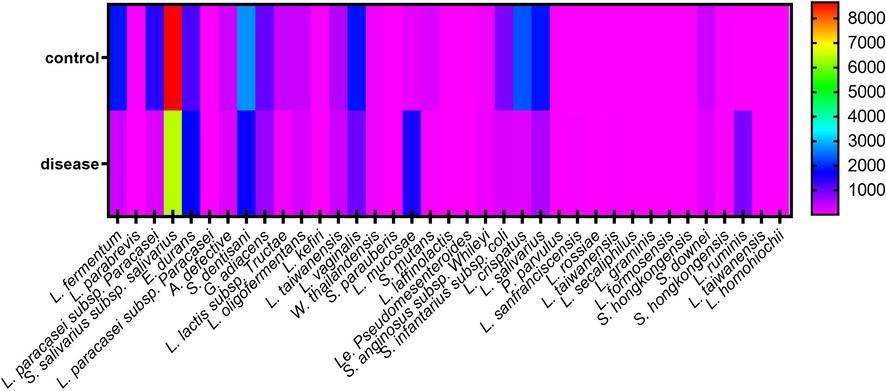
The heat map generated from abundance mean of various species.
Recent studies suggested that two third of individuals with abnormalities and a disequilibrium in the gastrointestinal and ecosystem have uremia (Kang, 1993). The majority of these abnormalities and disequilibrium happen in the colon and ileum that intestinal microbiota plays critical roles. The increase in aerobic bacteria such as Escherichia coli results in a decreasing in the anaerobic bacteria such as bifidobacteria and lactobacilli, as well as, generating toxic substrates, called uremic toxins (Simenhoff and Dunn, 1996; Vaziri et al., 2015). Several studies demonstrated that there is a deteriorated intestinal barrier in individuals with uremia, which is due to the abnormalities and disequilibrium of gut microbiota caused by the increase of pathogenic bacteria (Lenoir-Wijnkoop et al., 2007; Nakabayashi et al., 2010; Vaziri, 2012). Higher concentrations of urea and, consequently, increased ammonium are demonstrated in chronic renal diseases, which resulted in an increase pH that could promotes aerobic bacteria in the gastrointestinal and, subsequently, produce uremic toxins. Conversely, carbohydrates fermentation and acetic and lactic acid production by bifidobacteria and lactobacilli acidify the intestinal that could inhibit aerobic bacteria and normalize the intestinal microbiota in patients with chronic renal failure. In uremic individuals the fecal urease activity is increased at high concentrations of plasma urea. Therefore, a beneficial and consideration factor in uremic patients is the increase in colon microbial urease (Chow et al., 2003). Chronic renal disease is a health problem that can lead to end-stage kidney disease (ESKD), which is required kidney transplantation therapy. The modulation of interrelationship between the gut and kidney may be a potential role to slow progression of chronic renal disease (Ritz, 2011). Several studies suggested that alters in the gut microbiota affects the gut environment, which could correlate with kidney disease (Chow et al., 2003; Anders et al., 2013; Ramezani and Raj, 2014). Rossi et al. (2012) demonstrated that the pre- and probiotic therapy could significantly reduce IS and PCS in the chronic renal disease population. In addition, Yoshifuji et al. (2015) demonstrated that Lactobacillus spp. supplementation are improved intestinal changes and renal damage through activation of Lactobacillus recognized TLR (toll-like receptor). In another study, Vaziri et al. (2013) demonstrated that the abundance of Lactobacillaceae and Prevotellaceae in the intestinal microbiota flora are significantly decreased in the uremic animals compared to the control animals. Therefore, the aim of this study is assessment of abundance of lactobacillus spp. in fecal flora of ESKD and kidney transplanted patients.
2 Methods and materials
2.1 Sample collection
Fresh fecal samples were directly collected from the anus of 20 ESKD and kidney-transplanted patients, which were admitted to kidney transplantation ward of Emam-Reza teaching and treatmenet hospital, Tabriz, Iran. 20 fecal samples of admitted patients who had not been admitted for ESKD and kidney-transplantation as a control group. The underlying causes of ESKD in the study population included chronic pyelonephritis in one patient, polycystic kidney disease in one, post renal and urolithiasis in two, chronic kidney disease of unknown etiology in three, glomerulonephritis in five and hypertensive nephrosclerosis in eight patients. Patients with gastrointestinal individuals, malignancy, individuals who had been treated with antibiotics within 3 months before the enrolment in the study, infections, active inflammatory disorders, diabetes were excluded. The fecal samples were immediately stored at −80 °C until DNA extraction. In addition, all patients were evaluated for blood creatinine test, blood urea-nitrogen test and blood uric-acid test.
2.2 DNA extraction
Approximately 4 g of Fecal samples, which mixed vigorously with a spoon was homogenized aseptically and vortexed for more than 5 min. DNA of the mixture were extracted by the QIAamp Stool Mini Kit (Qia gene, Germany), according to the manufacturer’s instruction.
2.3 PCR amplification and next generation sequencing
The PCR amplification for each sample was performed by the V3 and V4 hyper variable region of bacterial 16 s rRNA. The primer sequences of two universal bacterial 16 s rRNA gene were Illumina V3 F: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG and Illumina V4 R: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC (Klindworth et al., 2013). The amplification reaction was set up as follows: 2X KAPA HiFi Hot Start Ready Mix 25 µl, 2 µl aliquots of both amplicon forward and reverse primer (10 µM), and template DNA 5 µl in a total 50 µl volume. Polymerase chain reaction (PCR) reaction was performed in a T100TM thermal (Bio-Rad, USA) using following program: 1 cycle of initial denaturing at 95 °C for 5 min, followed by 35 cycles of denaturing at 95 °C for 1 min, annealing at 55 °C for 45 s and extension at 72 °C for 1 min; and a final extension at 72 °C for 1 min. The PCR products were assessed using electrophoresis in 1% agarose gel in Tris-boric acid-EDTA buffer, stained with ethidium-bromide and visualized under UV light. The PCR products were sent to Omega Bioservices company and sequencing of the PCR products was performed on a MiSeq system (100 k 2 × 300 bp paired-end reads) (Illumina, USA). Bioinformatics analyses were performed via Illumina’s BaseSpace in parallel with Illumina’s in-house QIIme 2 pipeline.
2.4 Statistical analysis
Statistical analysis was performed in GraphPad Prism 8. The study data was analyzed by using descriptive statistics including mean and standard deviation (STDEV) and Mann-Whitney nonparametric test and unpaired t test with Welch's correction were applied to compare mean of data.
3 Results
The characteristics of participants between control group did not differ with disease group in age (59.3 ± 7.89 vs. 53.20 ± 12.03), and gender (male: 10, female: 10 vs. male: 14 vs. female: 6). Total 651 strains from 40 fecal samples were identified, which 37 (5.68%) strains were identified as order Lactobacillales. The strains were classified into six family including Lactobacillaceae (18/37), Streptococcaceae (14/37), Enterococcaceae (1/37), Aerococcaceae (1/37), Carnobacteriaceae (1/370 and Leuconostocaceae (2/37). In addition, the strains were classified into nine genus and 37 species including Lactobacillus (17/37), Straptococcus (10/37), Lactococcus (4/37), Abiotrophia (1/37), Granulicatella (1/37), Enterotoccoccus (1/37), Weisella (1/37), Leuconostoc (1/37), and Pediococcus (1/37). The most abundance genus in both groups are Streptococcus (4.47% ± 3.67% in control group and 3.12% ± 2.47% in disease group) and Lactobacillus (3.41% ± 4.15% in control group and 1.78% ± 2.88% in disease group) and the lowest abundance genus in both groups are Pediococcus (0.00005% ± 0.00009% in control group and 0.0006% ± 0.0009% in disease group) and Leuconostoc (0.004% ± 0.003% in control group and 0.002% ± 0.001% in disease group). S. dentisiani (2711.9 ± 4067.4) and L. crispatus (2289.65 ± 6280) were the most abundant species in control group and P. parvulus (0.15 ± 0.63) and L. graminis (2.45 ± 5.44) were the lowest species. In disease group, S. salivarius subsp. salivarius (6436.2 ± 9572.81) and E. durans (1756.85 ± 3674.53) were the most abundant species and L. secaliphilus (1 ± 3.54) and S. hongkongensis (1.65 ± 4.47) were the lowest abundant. The abundance of various species are shown in Table 1. S. salivarius subsp. salivarius, S. dentisiani, G. adiacens, and L. oligofermentans were found in all control group. As well as, S. salivarius subsp. salivarius, E. durans and S. dentisiani were found in all disease group. Comparing the abundance mean of the strains revealed that there is no significant association between control group and disease group (P-value > 0.05), but the abundance mean of the strains in control group increase in compared to disease group (719.9 ± 1534 vs. 462.8 ± 1115, P = 0.275). In addition, the presence of order Lactobacillales in the individuals of control group was not significant difference in compared to the individuals of disease group (10.49 ± 1.071 vs. 10.27 ± 1.044, P = 0.848).
Species
Control group mean
Individuals collected
Min
Max
STDEV
Disease group mean
Individuals collected
Min
Max
STDEV
p-value
L. fermentum
1861.5
19
0
28,623
6158.6
334.05
15
0
4887
1079.9
0.497
L. parabrevis
59.95
6
0
1109
235.76
1.7
4
0
12
3.78
0.330
L. paracasei subsp. paracasei
1427.8
18
0
27,153
5775.4
234.75
16
0
3303
742.79
0.350
S. salivarius subsp. salivarius
8650
20
237
29,245
8977.9
6436.2
20
517
32,920
9572.81
0.242
E. durans
1204.2
19
0
15,560
3412.4
1756.85
20
3
13,984
3674.53
0.337
L. paracasei subsp. paracasei
38.1
10
0
425
90.69
27.2
7
0
285
74.97
0.328
A. defective
316.4
15
0
3012
695.88
220.6
15
0
3474
770.99
0.690
S. dentisani
2711.9
20
14
17,221
4067.4
1642.05
20
37
16,661
3750.48
0.059
G. adiacens
1031
20
3
7570
1819.1
681.55
19
0
10,394
2291.94
0.139
Lc. lactis subsp. tructae
381.35
10
0
7102
1509.1
119.15
7
0
1814
403.56
0.438
L. oligofermentans
378.45
20
2
2479
633.57
278.45
19
0
1585
473.20
0.147
L. kefiri
60.8
5
0
999
213.78
3.15
4
0
36
8.59
0.511
L. taiwanensis
500.5
17
0
3667
976.43
341.7
17
0
5605
1242.55
0.541
L. vaginalis
1847.35
16
0
15,259
3800.3
977.6
19
0
15,009
3330.58
0.984
W. thailandensis
85.85
7
0
1663
354.06
97.45
5
0
1910
426.66
0.551
S. parauberis
9.1
9
0
119
25.14
6.05
7
0
44
11.98
0.667
L. mucosae
173.35
11
0
3286
698.2
1536.85
10
0
29,149
6506.20
0.837
S. mutans
253.8
12
0
3199
681.86
67.1
14
0
547
136.17
0.810
Lc. raffinolactis
4.15
2
0
79
3479.1
5.25
6
0
40
10.82
0.173
Le. Pseudomesenteroides
12.95
6
0
73
24.34
6.8
9
0
37
11.55
0.624
S. anginosus subsp. whileyi
120.85
18
0
685
6225
135.75
16
0
953
243.39
0.909
S. infantarius subsp. coli
865.25
15
0
15,643
3328.1
254.7
13
0
3724
858.44
0.303
L. crispatus
2289.65
19
0
28,128
6280
199.3
19
0
1614
350.88
0.920
L. salivarius
1859.05
16
0
13,385
3879.2
549.6
18
0
3717
1041.07
0.920
P. parvulus
0.15
1
0
3
0.63
1.85
4
0
28
6.27
0.164
L. sanfranciscensis
26.8
5
0
488
103.53
34
8
0
528
117.25
0.324
L. rossiae
4.35
6
0
45
10.90
3.35
5
0
26
6.90
0.915
L. taiwanensis
15.7
7
0
168
38.55
49.2
5
0
972
217.20
0.399
L. secaliphilus
22.6
5
0
375
79.69
1
2
0
16
3.64
0.181
L. graminis
2.45
4
0
20
5.44
7.3
6
0
78
18.75
0.513
Lc. formosensis
11.9
4
0
205
45.59
4.35
5
0
74
16.45
0.978
S. hongkongensis
9.95
6
0
141
30
1.65
4
0
19
4.47
0.416
S. downei
348.6
7
0
4686
1068
222.4
8
0
4205
937.68
0.771
S. hongkongensis
33.75
7
0
305
78.82
7.4
6
0
101
22.63
0.530
L. ruminis
6.55
4
0
114
24.23
872.35
6
0
17,298
3866.24
0.408
Lc. taiwanensis
7.25
1
0
145
74.57
4.45
1
0
89
19.90
1
L. homohiochii
3.35
1
0
67
14.28
1.5
1
0
30
6.70
1
4 Discussion
This study describes the abundance and diversity of order Lactobacillales in two groups including control group and disease group described as ESKD and kidney transplanted patients. In control group, the abundance of Lactobacillales were lower than disease group, and their species composition was different in comprised to disease group.
Marteau et al. (2001) demonstrated that the rRNA of Lactobacillus-Enterococcus group in healthy human is 6.6% of fecal bacterial rRNA, which is more than our study that found 3.8% in control group and 2.38% in disease group. In addition, Stsepetova et al. detected lactobacilli in all fecal samples by real-time PCR and 90% by the culture method in the samples, and we found lactobacilli in all fecal samples, which could suggested that the NGS method is the best for assessment of fecal bacteria.
This study is one of the few research concerning abundance of lactobacilli and their effects on chronic kidney disease patients, specifically comparing in the two groups as control and chronic kidney disease patients groups. However, the importance of this type of study in kidney disease patients lies in the benefits that could be obtained if the decrease of lactobacilli promoted symptoms in chronic kidney disease patients.
It was reported that gut microbiota may be a key role that maintains the gut in patients with chronic kidney disease (Kotanko et al., 2006). In addition, progression of chronic kidney disease may promoted by uremic toxins, which are made by certain gut microbiota (Satoh et al., 2003). Several studies suggested that early probiotic therapy to remove various uremic toxins could be delayed the onset of ESKD in individuals with progressive chronic kidney disease. Based on the previous study by Alatriste et al. (2014), Yoshifuji et al. (2015) and Vaziri et al. (2013), a decreased of lactobacilli abundance are shown in chronic kidney disease. In addition, the population of butyrate-producing enzymes bacterial families such as Lactobacillaceae and Prevotellaceae are lower in patients with uremia, whereas the population of indole- or p-cresol-producing bacterial families such as Enterobacteriaceae, Clostridiaceae, and Verrocomicrobiaceae are higher compared to health individuals (Wong et al., 2014). Similar to previous study, Lactobacillaceae is lower in patients group compared to healthy individuals in the present study. Ranganathan et al. suggested that probiotics with combination of S. thermophiles, B. longum and L. acidophilus are significantly reduced BUN and enhanced well-being in patients with chronic kidney disease (Ranganathan et al., 2010). Whereas, we have not found L. acidophilus in both control and patients groups. While, L. fermentum, L. paracasei, L. kefiri, L. vaginalis, L. crispatus, L. salivarius and other families of order Lactobacillales were reduced two or more folds in patients group compared to control group. In addition, L. mucosa, L. taiwanensis and L. ruminis were increased two or more folds in patients group compared to control group. Therefore, it has been suggested that future studies are necessary to elucidate more precise mechanisms of different lactobacilli strains in chronic kidney disease, subsequently develop more efficient therapeutic strategies. Our data could provide relevant information for these future studies.
5 Conclusion
More diversity was exhibited in lactobacillus spp. in patients with chronic kidney disease compared to control group. Some of the species of Lactobacillaceae family such as L. acidophilus was not found in both patients and control groups. In addition, more of the species of Lactobacillaceae were decreased in patients group, whereas certain species were increased in patients group compared to control group. This diversity could suppose a novel mechanisms and therapeutic strategies to slow the progression of renal damage and delay the onset of ESKD in patients with chronic kidney disease.
Acknowledgment
This study was done as dissertation of GR Hanifi for PhD of Microbiology in Islamic azad University. Project was approved and got local ethic committee.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
References
- Alatriste, P.V.M., Arronte, R.U., Espinosa, C.O.G., Cuevas, M.d.l.Á.E., 2014. Effect of probiotics on human blood urea levels in patients with chronic renal failure. Nutricion hospitalaria 29, 582–590.
- The intestinal microbiota, a leaky gut, and abnormal immunity in kidney disease. Kidney Int.. 2013;83:1010-1016.
- [Google Scholar]
- Free and microencapsulated Lactobacillus and effects of metabolic induction on urea removal. Artificial Cells Blood Substitutes Biotechnol.. 2003;31:425-434.
- [Google Scholar]
- Low intensity ultrasound increases the fermentation efficiency of Lactobacillus casei subsp.casei ATTC 39392. Int. J. Biol. Macromol.. 2016;86:462-467.
- [Google Scholar]
- Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol. Mol. Biol. Rev.. 1998;62:1157-1170.
- [Google Scholar]
- Microbial balance in the intestinal microbiota and its association with diabetes, obesity and allergic disease. Microb. Pathog.. 2019;127:48-55.
- [Google Scholar]
- Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881-884.
- [Google Scholar]
- Lactobacillus plantarum as a probiotic potential from Kouzeh Cheese (Traditional Iranian Cheese) and its antimicrobial activity. Probiotics Antimicrob. Proteins. 2017;9:189-193.
- [Google Scholar]
- Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M. and Glöckner, F.O., 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1-e1.
- Intestinal bacterial microflora—a potential source of chronic inflammation in patients with chronic kidney disease. Nephrol. Dial. Transplant.. 2006;21:2057-2060.
- [Google Scholar]
- Probiotic and prebiotic influence beyond the intestinal tract. Nutr. Rev.. 2007;65:469-489.
- [Google Scholar]
- Taxonomic Outline of the Prokaryotes. New York, NY: Bergey’s Manual of Systematic Bacteriology; 2010.
- Comparative study of bacterial groups within the human cecal and fecal microbiota. Appl. Environ. Microbiol.. 2001;67:4939-4942.
- [Google Scholar]
- Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol.. 1993;59:695-700.
- [Google Scholar]
- Effects of synbiotic treatment on serum level of p-cresol in haemodialysis patients: a preliminary study. Nephrol. Dial. Transplant.. 2010;26:1094-1098.
- [Google Scholar]
- Probiotics, prebiotics and synbiotics-a review. J. Food Sci. Technol.. 2015;52:7577-7587.
- [Google Scholar]
- Effect of Bifidobacterium breve on the intestinal microbiota of coeliac children on a gluten free diet: a pilot study. Nutrients. 2016;8
- [Google Scholar]
- The gut microbiome, kidney disease, and targeted interventions. J. Am. Soc. Nephrol.. 2014;25:657-670.
- [Google Scholar]
- Pilot study of probiotic dietary supplementation for promoting healthy kidney function in patients with chronic kidney disease. Adv. Therapy. 2010;27:634-647.
- [Google Scholar]
- Rossi, M., Klein, K., Johnson, D.W., Campbell, K.L., 2012. Pre-, pro-, and synbiotics: do they have a role in reducing uremic toxins? A systematic review and meta-analysis. Int. J. Nephrol.
- Uremic toxins overload accelerates renal damage in a rat model of chronic renal failure. Nephron Exp. Nephrol.. 2003;95:e111-e118.
- [Google Scholar]
- Indoxyl sulfate upregulates renal expression of ICAM-1 via production of ROS and activation of NF-κB and p53 in proximal tubular cells. Life Sci.. 2013;92:143-148.
- [Google Scholar]
- The intestine and the kidneys: a bad marriage can be hazardous. Clin. Kidney J.. 2015;8:168-179.
- [Google Scholar]
- The uremic toxicity of indoxyl sulfate and p-cresyl sulfate: a systematic review. J. Am. Soc. Nephrol.. 2014;25:1897-1907.
- [Google Scholar]
- Diversity, vitality and activities of intestinal lactic acid bacteria and bifidobacteria assessed by molecular approaches. FEMS Microbiol. Rev.. 2005;29:477-490.
- [Google Scholar]
- CKD impairs barrier function and alters microbial flora of the intestine: a major link to inflammation and uremic toxicity. Curr. Opin. Nephrol. Hypertens.. 2012;21:587.
- [Google Scholar]
- Chronic kidney disease alters intestinal microbial flora. Kidney Int.. 2013;83:308-315.
- [Google Scholar]
- Altered intestinal microbial flora and impaired epithelial barrier structure and function in CKD: the nature, mechanisms, consequences and potential treatment. Nephrol. Dial. Transplant.. 2015;31:737-746.
- [Google Scholar]
- p-Cresyl sulfate causes renal tubular cell damage by inducing oxidative stress by activation of NADPH oxidase. Kidney Int.. 2013;83:582-592.
- [Google Scholar]
- Expansion of urease-and uricase-containing, indole-and p-cresol-forming and contraction of short-chain fatty acid-producing intestinal microbiota in ESRD. Am. J. Nephrol.. 2014;39:230-237.
- [Google Scholar]
- Gut Lactobacillus protects against the progression of renal damage by modulating the gut environment in rats. Nephrol. Dial. Transplant.. 2015;31:401-412.
- [Google Scholar]







