Translate this page into:
Isolation, phenotypic and genotypic characterization of Escherichia coli from the bloodstream samples in Riyadh, Saudi Arabia
⁎Corresponding author. aalqasim@ksu.edu.sa (Abdulaziz Alqasim)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Bacteraemia is an international threat caused by extra-intestinal pathogenic Escherichia coli (ExPEC). Recently, the antimicrobial resistance of ExPEC has increased substantially, and this is attributed to the dissemination of E. coli ST131 clone. The present study explored the antimicrobial susceptibility patterns, extended spectrum β-lactamase (ESBL) carriage, virulence capacity and the prevalence of ST131 clone in Riyadh, Saudi Arabia. Thirty-one E. coli blood isolates, collected between January 2018 and March 2018, were used. The prevalence of ST131 clone was determined based on the PCR assays. Twenty-eight (90.3%) of all tested isolates were resistant to ampicillin, while only 1 isolate (3.2%) showed resistance to imipenem. Sixteen (51.6%) of the all isolates were ESBL producers, with CTX-M-15 being the predominant ESBL type. The virulence potential was higher among ESBL-producing isolates. Overall, seventeen (54.8%) isolates belonged to the ST131 clone. ST131 isolates were associated with increased antibiotic resistance; ESBL carriage and virulence capacity compared to non ST131isolates. This study reported the high levels of antimicrobial resistance and ESBL carriage among ExPEC blood isolates. It is concluded that the prevalence of ST131 clone among E. coli blood population is high locally, and found high virulence potential and antimicrobial resistance levels among ST131 isolates. These might drive ST131 success as a major cause of bacteraemia worldwide.
Keywords
E. coli
Bacteremia
ST131
Antimicrobial resistance
Virulence capacity
ESBL
1 Introduction
Bacteraemia is an international threat caused by extra-intestinal pathogenic Escherichia coli (ExPEC) (Rogers et al., 2011). It has been shown that ExPEC comprises 17–37% of all clinically important bacteria isolated from bloodstream infections (BSIs) globally (Russo and Johnson, 2003). Recently, the number of reported bacteraemia cases due to ExPEC has increased substantially, leading to high levels of morbidity and mortality worldwide. Over the past decades, ExPEC resistance to cephalosporins, fluoroquinolones, and trimethoprim–sulfamethoxazole, has increased markedly (Gupta et al., 1999). In the United Kingdom, the incidence of bacteraemia due to E. coli increased by approximately 70% between 1999 and 2011, and this was mostly driven by infections with antibiotic resistant isolates (Schlackow et al., 2012). More recently, resistance the antibiotics such as carbapenems and polymyxins, has increasingly been reported globally (Al-Dhabi and Ghilan, 2018:; Nordmann et al., 2011), which has complicated the management of patients.
Escherichia coli ST131 is a member of phylogroup B2, and belongs to serotype O25:H4 (Nicolas-Chanoine et al., 2008). It is often multidrug resistant (MDR), i.e. showing resistance to at least 1 agent in ≥ 3 antibiotic classes, and frequently carries a variety of extended-spectrum β-lactamases (ESBLs)such as CTX-M,OXA and TEM, as well as the aminoglycosides/fluoroquinolone acetyltransferase AAC (6′)-Ib-cr (Woodford et al., 2009). ST131 is commonly associated with carrying the blaCTX-M−15 gene, encoding the CTX-M-15 ESBL enzyme, on the IncFII plasmids (Coque et al., 2008). Many reports claimed that the high virulence potential among ST131 isolates has been identified (Coelho et al., 2010).
In Saudi Arabia, there is very limited information available on the antimicrobial resistance, virulence potential and molecular epidemiology of ExPEC blood isolates. We characterized the E. coli blood isolates at a molecular level and determined their antimicrobial susceptibility profiles, ESBL carriage and ST131 status.
2 Materials and methods
2.1 Bacterial isolates
Thirty one E. coli isolates were obtained from different infected patients from the tertiary hospital in Riyadh, Saudi Arabia. The clinical samples were collected from January 2018 to March 2018 and the isolated strains were initially identified using Vitek 2 identification system (Vitek2-ID-GNB, BioMerieux).
2.2 Antimicrobial susceptibility testing
Antimicrobial susceptibility patterns of the isolates were determined as reported by the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2018). E. coli ATCC 25,922 was used as the positive control to compare the susceptibility using ten different antibiotics obtained from LIOFILCHEM, Italy (Table 1).
Antibiotic group
Antibiotic name (abbreviation)
Concentration (μg/disc)
Penicillins
Ampicillin (AM)
10 μg
β-lactam/β-lactamase inhibitors combination
Amoxicillin-clavulanic acid (AUG)
20/10 μg
Aminoglycosides
Gentamicin (GM)
10 μg
Second generation cephalosporins
Cefoxitin (FOX)
30 μg
Third generation cephalosporins
Ceftazidime (CAZ)
30 μg
Fourth generation cephalosporins
Cefepime (FEP)
30 μg
Tetracyclines
Tetracycline (T)
30 μg
Folate pathway inhibitors
Trimethoprim-Sulfamethoxazole (SXT)
1.25/23.75 μg
Carbapenems
Imipenem (IMP)
10 μg
Fluoroquinolones
Ciprofloxacin (CIP)
5 μg
2.3 Phenotypic and molecular detection of ESBL production
Initial screening and phenotypic confirmation of ESBL production were performed by CLSI guidelines. For molecular detection of ESBL encoding genes, gene based PCR was performed using different genes such as blaOXA,blaTEM,blaSHV, and blaCTX-M Groups 1, 2 and 9 using multiplex PCR primer sets and conditions previously described [15]. E. coli ATCC 25,922 acted as the positive and K. pneumoniae ATCC 700,603 acted as the negative control for the comparison.
2.4 Identification of CTX-M ESBL gene variants
The ESBL gene variants were performed by sequencing and then comparing using the NCBI gene bank BLAST search programme.
2.5 Screening for E. coli ST131 status
All isolates were screened for E. coli ST131 using the O25b and O16 ST131 clades primers by PCR amplification and sequencing (Clermont et al., 2009).
2.6 Identification of virulence-associated gene (VAG) carriage
PCR amplification methodology was implemented for the determination of VAG gene by following the methodology of the previous reports (Oteo et al., 2014).
2.7 Statistical analysis
Data were analysed using the SPSS software (version 19.0). Fisher’s exact test (FET) was used to compare between different groups.
3 Results
3.1 Antimicrobial susceptibility profiles and ESBL carriage of ExPEC isolates
Results indicated that ampicillin was the most ineffective antibiotic with 28 (90.3%) of isolates showing resistance to ampicillin (Table 2). Of all isolates, 16 (51.6%) were resistant to amoxicillin-clavulanic acid and 4 (12.9%) were resistant to gentamicin and cefoxitin, 14 (45.2%) to ceftazidime and cefepime, and 17 (54.8%) to trimethoprim-sulfamethoxazole and ciprofloxacin respectively. It was shown that 22 of 31 (71%) E. coli isolates were MDR (Fig. 1). Of these, 5 (16.1%) were resistant to 3 antibiotic groups, 4 (12.9%) to 4 antibiotic groups and 1 (3.2%) to 5 antibiotic groups. Additionally, 2 (6.5%), 6 (19.4%) and 4 (12.9%) were resistant to 6, 7 and 8 antibiotics, respectively.
Antibiotic
Number (%) of E. coli isolates
Sensitive isolates
Resistant isolates
Ampicillin
3 (9.7%)
28 (90.3%)
Amoxicillin-clavulanic acid
15 (48.4%)
16 (51.6%)
Gentamicin
27 (87.1%)
4 (12.9%)
Cefoxitin
27 (87.1%)
4 (12.9%)
Ceftazidime
17 (54.8%)
14 (45.2%)
Cefepime
17 (54.8%)
14 (45.2%)
Tetracycline
13 (41.9%)
18 (58.1%)
Trimethoprim-Sulfamethoxazole
14 (45.2%)
17 (54.8%)
Imipenem
30 (96.8%)
1 (3.2%)
Ciprofloxacin
14 (45.2%)
17 (54.8%)
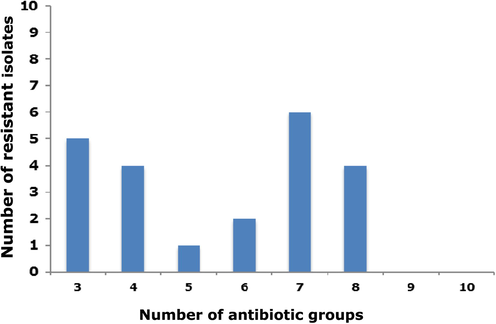
Number of clinical E. coli blood isolates show ing multi drug resistance (MDR) phenotype.
It was found that 16 (51.6%) of the all isolates were ESBL producers (Table 3). The ESBL gene types detected in these E. coli isolates belonged to the CTX-M-Group 1, CTX-M-Group 9, TEM and OXA ESBLs, while the SHV type and CTX-M-Group 2 were not detected in any of the tested isolates. Among all ESBL genes, CTX-M-Group 1 dominated with 11 of 16 (68.8%) isolates carrying this ESBL type. With respect to the CTX-M gene variants, the CTX-M-15 was found in all the CTX-M-Group 1 producing isolates, while CTX-M-14 variant was the only detected variant among CTX-M-Group 9 producers (Table 3). 1 NA: Non-applicable.
Isolate ID
β-lactamase type(s)
CTX-M variant
CTX-M G1
CTX-M G2
CTX-M G9
TEM
OXA
SHV
B1
–
–
–
+
–
–
NA1
B7
+
–
–
–
+
–
CTX-M-15
B8
+
–
–
+
–
–
CTX-M-15
B9
–
–
–
–
+
–
NA
B10
+
–
–
–
–
–
CTX-M-15
B16
+
–
–
–
+
–
CTX-M-15
B20
+
–
–
–
+
–
CTX-M-15
B21
–
–
+
–
–
–
CTX-M-14
B22
+
–
–
–
+
–
CTX-M-15
B23
–
–
+
+
–
–
CTX-M-14
B24
+
–
–
+
–
–
CTX-M-15
B26
–
–
–
+
–
–
NA
B28
+
–
–
+
–
–
CTX-M-15
B29
+
–
–
+
–
–
CTX-M-15
B30
+
–
–
+
–
–
CTX-M-15
B31
+
–
–
–
+
–
CTX-M-15
3.2 Characteristics of ESBL-producing and non ESBL-producing E. coli isolates
The ESBL producing isolates were reported in table 4. Results indicated that 17 of 31 (54.8%) ExPEC isolates belonged to ST131 clone. Of these, 15 (93.8%) were ESBL-producing while only 2 (13.3%) were non ESBL-producing. ESBL-producers were significantly more capable of displaying resistance to these five antibiotics such as ceftazidime, amoxicillin-clavulanic acid, trimethoprim-sulfamethoxazole cefepime, and ciprofloxacin (Table 4). All the 16 (100%) ESBL-producing isolates were MDR, while this phenotype was found in only 6 of 15 (40%) (Table 4).
Category
Specific trait
Number of isolates (%)
ESBL-producing (n = 16)
Non ESBL-producing (n = 15)
Total (n = 31)
P valuea
ST131 isolates
ST131
15 (93.8%)
2 (13.3%)
17 (54.8%)
<0.001
ST131 O25b
14 (87.5%)
2 (13.3%)
16 (51.6%)
<0.001
ST131 O16
1 (6.3%)
0 (0%)
1 (3.2%)
1.000
Antimicrobial resistance
AM
16 (100%)
12 (80%)
28 (90.3%)
0.10
AUG
14 (87.5%)
2 (13.3%)
16 (51.6%)
<0.001
GM
3 (18.8%)
1 (6.7%)
4 (12.9%)
0.59
FOX
4 (25%)
0 (0%)
4 (12.9%)
0.10
CAZ
13 (81.3%)
1 (6.7%)
14 (45.2%)
<0.001
FEP
13 (81.3%)
1 (6.7%)
14 (45.2%)
<0.001
T
10 (62.5%)
8 (53.3%)
18 (58.1%)
0.72
SXT
14 (87.5%)
3 (20%)
17 (54.8%)
0.002
IMP
0 (0%)
1 (6.7%)
1 (3.2%)
0.48
CIP
15 (93.8%)
2 (13.3%)
17(54.8%)
<0.001
FQ phenotype
FQ Rb
15 (93.8%)
2 (13.3%)
17 (54.8%)
<0.001
MDR phenotype
MDR
16 (100%)
6 (40%)
22 (71%)
0.002
Adhesions
papA
7 (43.8%)
2 (13.3%)
9 (29%)
0.11
papC
7 (43.8%)
2 (13.3%)
9 (29%)
0.11
papGallele I
0 (0%)
0 (0%)
0 (0%)
1.000
papGallele II
6 (37.5%)
1 (6.7%)
7 (22.6%)
0.08
sfa/focDE
1 (6.3%)
0 (0%)
1 (3.2%)
1.000
afa/draBC
2 (12.5%)
0 (0%)
2 (6.5%)
0.48
fimH
15 (93.8%)
14 (93.3%)
29 (93.5%)
1.000
iha
8 (50%)
1 (6.7%)
9 (29%)
0.02
Siderophores
iutA
12 (75%)
7 (46.7%)
19 (61.3%)
0.15
iroNE. coli
3 (18.8%)
9 (60%)
12 (38.7%)
0.03
Toxins
hlyA
5 (31.3%)
0 (0%)
5 (16.1%)
0.04
cnf1
2 (12.5%)
0 (0%)
2 (6.5%)
0.48
Polysaccharide coatings
kpsMTII
6 (37.5%)
5 (33.3%)
11 (35.5%)
1.000
kpsMTIII
0 (0%)
1 (6.7%)
1 (3.2%)
0.48
Miscellaneous
PAI
10 (62.5%)
3 (20%)
13 (41.9%)
0.03
cvaC
2 (12.5%)
6 (40%)
8 (25.8%)
0.11
traT
10 (62.5%)
11 (73.3%)
21 (67.7%)
0.70
ompT
11 (73.3%)
9 (60%)
20 (64.5%)
0.72
usp
10 (62.5%)
7 (46.7%)
17 (54.8%)
0.48
sat
10 (62.5%)
1 (6.7%)
11 (35.5%)
0.002
Virulence scoresc
8 (1–13)
5.3 (1–12)
6.7 (1–13)
0.03
Testing the VAG carriage of all ExPEC isolates showed that 5 VAGs: fimH, iutA, PAI, traT and usp were expressed by more than 50% of isolates, while they failed to express the papG allele I. ESBL-producing isolates had higher VAG carriage than non ESBL-producing isolates, and they were significantly associated with carrying iha, hlyA, PAI and sat VAGs. However, iroNE. coli was highly found among non ESBL-producing isolates(Table 4). The median virulence scores (ranges) were 8 (1–13) for ESBL-producing isolates and 5.3 (1–12) for non ESBL-producing isolates, and this difference was found to be significant (P = 0.03) (Table 4).
3.3 Characteristics of E. coliST131 and non ST131 isolates
The data of this research found increased resistance of ST131 compared to non ST131 isolates, and there was a significant association between ST131 isolates and showing resistance to 5 agents. All the 17 (100%) ST131 isolates were able to show MDR phenotype, while it was exhibited by only 5 of 14 (35.7%) non ST131 isolates (P < 0.001) (Table 5). E. coli ST131 had a higher VAG carriage in comparison to non ST131 isolates, and ST131 isolates were significantly associated with carrying 3 VAGs: iha, hlyA and sat (Table 5). The median virulence scores (ranges) were 7.5 (1–13) for ST131isolatesand 4 (1–7) for non ST131 isolates, and this difference was significant (P = 0.03) (Table 5). a P values (by Fisher’s exact test) are for 2-group comparison: ST131 and non ST131. Conflict of Interest statement. The authors of the manuscript entitled declared no conflict in this manuscript and publications.
Category
Specific trait
Number of isolates (%)
ST131 (n = 17)
Non-ST131 (n = 14)
Total (n = 31)
Pvaluea
Antimicrobial resistance
AM
17 (100%)
11 (78.6%)
28 (90.3%)
0.08
AUG
14 (82.4%)
2 (14.3%)
16 (51.6%)
0.002
GM
3 (17.6%)
1 (7.1%)
4 (12.9%)
0.61
FOX
3 (17.6%)
1 (7.1%)
4 (12.9%)
0.61
CAZ
12 (70.6%)
2 (14.3%)
14 (45.2%)
0.003
FEP
12 (70.6%)
2 (14.3%)
14 (45.2%)
0.003
T
12 (70.6%)
6 (42.9%)
18 (58.1%)
0.16
SXT
13 (76.5%)
4 (28.6%)
17 (54.8%)
0.01
IMP
1 (5.9%)
0 (0%)
1 (3.2%)
1.000
CIP
17 (100%)
0 (0%)
17 (54.8%)
<0.001
FQ phenotype
FQ R
17 (100%)
0 (0%)
17 (54.8%)
<0.001
MDR phenotype
MDR
17 (100%)
5 (35.7%)
22 (71%)
<0.001
ESBL type(s)
ESBL
15 (88.2%)
1 (7.1%)
16 (51.6%)
<0.001
TEM
2 (11.8%)
0 (0%)
2 (6.5%)
0.49
OXA
1 (5.9%)
0 (0%)
1 (3.2%)
1.000
SHV
0 (0%)
0 (0%)
0 (0%)
1.000
CTX-M-15
1 (5.9%)
0 (0%)
1 (3.2%)
1.000
CTX-M-14
0 (0%)
1 (7.1%)
1 (3.2%)
0.45
CTX-M-15 + TEM
5 (29.4%)
0 (0%)
5 (16.1%)
0.05
CTX-M-15 + OXA
5 (29.4%)
0 (0%)
5 (16.1%)
0.05
CTX-M-14 + TEM
1 (5.9%)
0 (0%)
1 (3.2%)
1.000
Non-ESBL
2 (11.8%)
13 (92.9%)
15 (48.4%)
<0.001
Adhesions
papA
7 (41.2%)
2 (14.3%)
9 (29%)
0.13
papC
7 (41.2%)
2 (14.3%)
9 (29%)
0.13
papGallele I
0 (0%)
0 (0%)
0 (0%)
1.000
papGallele II
6 (35.3%)
1 (7.1%)
7 (22.6%)
0.09
sfa/focDE
1 (5.9%)
0 (0%)
1 (3.2%)
1.000
afa/draBC
2 (11.8%)
0 (0%)
2 (6.5%)
0.49
fimH
16 (94.1%)
13 (92.9%)
29 (93.5%)
1.000
iha
8 (47.1%)
1 (7.1%)
9 (29%)
0.02
Siderophores
iutA
13 (76.5%)
6 (42.9%)
19 (61.3%)
0.08
iroNE. coli
3 (17.6%)
9 (64.3%)
12 (38.7%)
0.01
Toxins
hlyA
5 (29.4%)
0 (0%)
5 (16.1%)
0.05
cnf1
2 (11.8%)
0 (0%)
2 (6.5%)
0.49
Polysaccharide coatings
kpsMTII
5 (29.4%)
6 (42.9%)
11 (35.5%)
0.48
kpsMTIII
1 (5.9%)
0 (0%)
1 (3.2%)
1.000
Miscellaneous
PAI
10 (58.8%)
3 (21.4%)
13 (41.9%)
0.07
cvaC
3 (17.6%)
5 (35.7%)
8 (25.8%)
0.41
traT
13 (76.5%)
8 (57.1%)
21 (67.7%)
0.44
ompT
11(64.7%)
9(64.3%)
20 (64.5%)
1.000
usp
10 (58.8%)
7 (50%)
17 (54.8%)
0.72
sat
10 (58.8%)
1(7.1%)
11 (35.5%)
0.007
Virulence scores
7.9 (1–13)
5.1 (1–12)
6.7 (1–13)
0.03
4 Discussion
The incidence of bacteremia due to ExPEC has recently increased globally (Rogers et al., 2011), and this is driven by a substantial rise in the MDR plasmids carriage among ExPEC isolates (Alhashash et al., 2015). In Saudi Arabia, information on the phenotypic and molecular traits, such as antimicrobial resistance, ESBL carriage, virulence capacity and ST131 status, of ExPEC bloodstream isolates is very scarce. Here we determined the antibiotic resistance levels and ESBL carriage of ExPEC blood isolates from Riyadh city, and found high resistance levels of these isolates to antibiotics commonly used for empiric treatment for extraintestinal intestinal infections, particularly amoxicillin-clavulanic acid, cephalosporins, trimethoprim-sulfamethoxazole and ciprofloxacin. These resistance levels were higher than those reported in many countries such as Turkey (Bozcal et al., 2018) and Mozambique (Mandomando et al., 2010).
We also demonstrated that 16 (51.6%) of all isolates were ESBL-producing, with CTX-M-15 being the most predominant ESBL variant. This prevalence of ESBL carriage among ExPEC blood isolates was found to be high compared to that demonstrated by many studies across the world (Koksal et al., 2009), although Alhashash and colleagues have found that 59.3% of ExPEC blood isolates were ESBL producers (Alhashash et al., 2013), which is higher than that detected here. However, the dominance of CTX-M-15 ESBL was consistent with many reports showing CTX-M-15 as the most prevalent ESBL among ExPEC blood isolates globally (Guiral et al., 2018). Additionally, the MDR phenotype was displayed by 71% of all tested isolates, and this was higher than that reported among ExPEC blood population in the United Kingdom where 50.7% of isolates were MDR (Alhashash et al., 2013). Taken together, our findings highlight the urgent need to revise the current local guidelines used for optimal treatment regimens for bacteraemia patients to combat the increasing resistance issue.
We compared the antibiotic resistance and virulence traits of ESBL-producing isolates, and found that ESBL-producing isolates were highly resistance to other antibiotics. This is not surprising given the strong association between ESBL carriage and multidrug resistance in ExPEC reported previously (Alqasim et al., 2018). Interestingly, we also showed the significant correlation between virulence potential and ESBL carriage, and this is in contrary to many previous comparative studies showing that ESBL-producing ExPEC isolates were not associated with higher virulence compared to non ESBL-producers (Karisik et al., 2008). However, the finding of this study concurs with a previous report demonstrating high virulence capacity among ESBL-producing ExPEC isolates (Pitout et al., 2005). These conflicting reports merit further investigation at a genetic level to elucidate the relationship between resistance and virulence of ExPEC isolates.
Our data showed that 17 (54.8%) of ExPEC isolates were members of the ST131 clone, and this is higher compared to many previous reports throughout the world demonstrating the prevalence of ST131 among ExPEC blood population to be between 5% and 30% (Adams-Sapper et al., 2013; Hung et al., 2019). Additionally, our ST131 isolates were higher in their antimicrobial resistance, ESBL carriage and multidrug resistance compared to non-ST131 isolates, and this is in agreement with many studies showing similar observations for ST131 globally (López-Cerero et al., 2013). The prevalence of ST131 reported here is alarming and might provide an explanation of the increased levels of antimicrobial resistance, ESBL carriage and multidrug resistance that has currently been described locally.
With regard to virulence capacity, our finding showing the significant correlation between ST131 and iha, hlyA and sat was in agreement with a recent study (Hung et al., 2019). However, our isolates were not assoacited with significat traT carriage which is in contrary to the finding by Hung and collaegues.
5 Conclusion
In conclusion, this study demonstrated high levels of antimicrobial resistance and ESBL carriage among clinical E. coli blood isolates in Saudi Arabia, and this highlights the need to revise the current guidelines of the empiric therapy for bloodstream infections. It also showed the high prevalence of ST131 isolates among bacteraemia isolates locally, and these ST131 isolates were found to be highly antimicrobial resistant and virulent compared to non ST131 isolates.
Acknowledgements
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through research group No (RG-1440-053).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Characterization of Silver Nanomaterials Derived from Marine Streptomyces sp. Al-Dhabi-87 and Its In Vitro Application against Multidrug Resistant and Extended-Spectrum Beta-Lactamase Clinical Pathogens. Nanomaterials. 2018;8(5)
- [Google Scholar]
- Prevalence of Multidrug Resistance and Extended-Spectrum β-Lactamase Carriage of Clinical Uropathogenic Escherichia coli Isolates in Riyadh, Saudi Arabia. Int. J. Microbiol.. 2018;2018:1-9.
- [Google Scholar]
- Clonal composition and community clustering of drug-susceptible and-resistant Escherichia coli isolates from bloodstream infections. Antimicrob. Agents Chemother. 2013;57(1):490-497.
- [Google Scholar]
- Increase in bacteraemia cases in the East Midlands region of the UK due to MDR Escherichia coli ST73: high levels of genomic and plasmid diversity in causative isolates. J. Antimicrob. Chemother.. 2015;71(2):339-343.
- [Google Scholar]
- Multidrug-Resistant Escherichia coli Bacteremia. Emerg. Infect Dis.. 2013;19(10):1699.
- [Google Scholar]
- The relationship between phylogenetic classification, virulence and antibiotic resistance of extraintestinal pathogenic Escherichia coli in İzmir province. Turkey. PeerJ. 2018;6:1-24.
- [Google Scholar]
- Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J. Antimicrob. Chemother.. 2009;64(2):274-277.
- [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing; 21st Informational Supplement. CLSI Document M100–S28. Clinical and Laboratory Standards. Wayne, PA: Institute; 2018.
- Spread of Escherichia coli O25b:H4-B2-ST131 producing CTX-M-15 and SHV-12 with high virulence gene content in Barcelona (Spain) J. Antimicrob. Chemother.. 2010;66:1-10.
- [Google Scholar]
- Dissemination of clonally related Escherichia coli strains expressing extended-spectrum -lactamase CTX-M-15. Emerg. Infect Dis.. 2008;14(2):195-200.
- [Google Scholar]
- Epidemiology and molecular characterization of multidrug-resistant Escherichia coli isolates harboring blaCTX-M group 1 extended-spectrum β-lactamases causing bacteremia and urinary tract infection in Manhiça. Mozambique. Infect Drug Resist. 2018;11:927-936.
- [Google Scholar]
- Increasing prevalence of antimicrobial resistance among uropathogens causing acute uncomplicated cystitis in women. JAMA. 1999;281(8):736-738.
- [Google Scholar]
- Lee SS-J, Chang T-H, Lin H-H, Hung C-H, Wang J-L: Bloodstream Infection with Extended-spectrum Beta-lactamase–producing Escherichia coli: the role of virulence genes. J. Microbiol. Immunol. Infect.. 2019;3:1-9.
- [Google Scholar]
- Virulence factors in Escherichia coli with CTX-M-15 and other extended-spectrum β-lactamases in the UK. J. Antimicrob. Chemother.. 2008;61(1):54-58.
- [Google Scholar]
- Prevalence and antimicrobial resistance patterns of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolated from blood cultures in an Istanbul University Hospital. Chemotherapy. 2009;55(4):293-297.
- [Google Scholar]
- Escherichia coli belonging to the worldwide emerging epidemic clonal group O25b/ST131: risk factors and clinical implications. J. Antimicrob. Chemother.. 2013;69:809-814.
- [Google Scholar]
- Antimicrobial drug resistance trends of bacteremia isolates in a rural hospital in southern Mozambique. Am. J. Trop Med. Hyg.. 2010;83(1):152-157.
- [Google Scholar]
- Intercontinental emergence of Escherichia coli clone O25: H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother.. 2008;61(2):273-281.
- [Google Scholar]
- Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect Dis.. 2011;17(10):1791-1798.
- [Google Scholar]
- Inhibitor-resistant TEM-and OXA-1-producing Escherichia coli isolates resistant to amoxicillin-clavulanate are more clonal and possess lower virulence gene content than susceptible clinical isolates. Antimicrob. Agents. Chemother.. 2014;58(7):3874-3881.
- [Google Scholar]
- Virulence factors of Escherichia coli isolates that produce CTX-M-type extended-spectrum β-lactamases. Antimicrob. Agents Chemother.. 2005;49(11):4667-4670.
- [Google Scholar]
- Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother.. 2011;66(1):1-14.
- [Google Scholar]
- Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect.. 2003;5(5):449-456.
- [Google Scholar]
- Increasing incidence of Escherichia coli bacteraemia is driven by an increase in antibiotic-resistant isolates: electronic database study in Oxfordshire 1999–2011. J. Antimicrob. Chemother.. 2012;67(6):1514-1524.
- [Google Scholar]
- Complete nucleotide sequences of plasmids pEK204, pEK499 and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25: H4-ST131 clone. Antimicrob. Agents Chemother.. 2009;53:4472-4482.
- [Google Scholar]







