Translate this page into:
Isolation and identification of Leptospira species from bovines by rpoB and LipL41 genes based phylogenetic analysis
⁎Corresponding authors. shivachemist@gmail.com (Shiva Prasad Kollur), chandans@jssuni.edu.in (Chandan Shivamallu)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Isolation and Identification of Leptospira species from bovines by rpoB and LipL41 genes based phylogenetic analysis by Shivaraj Murag et al. 2020.
Abstract
Abstract
In this study, in the beginning 582 serum samples were subjected to Microscopic Agglutination Test (MAT) with eight different serovars prevalent in the region to know the seroprevalence of Leptospira in bovines in Karnataka, India. Based on the findings of the MAT, different samples like blood, urine, aborted materials and uterine discharge collected from the MAT positive animals were used for isolation and genomic detection by conventional PCR targeting two lipL32 and seqY genes using specific primers. Out of the 163 MAT positive samples screened 12 samples (including three isolates) were found positive in PCR. Subsequently, to identify the different species prevalent in the geographical region the PCR positive samples were subjected to rpoB and LipL41 gene amplification. and nucleotide sequence analysis of rpo B, it was found that all the samples were belonging to L. interrogans species with overlapping/superimposing L. interrogans and L.borgpetersenii species. Further, the LipL41 gene sequence phylogenetic analysis differentiated these two species clearly. Therefore, it can be concluded that LipL41 gene based phylogenic analysis besides rpoB gene can be effectively utilized to identify different Leptospira species in a geographical niche including the identification of intermediate species. This is first of its kind in India using LipL41 gene based phylogenetic analysis for Leptospira species identification in limited number of samples from bovines, hence the same can be explored on a larger geographical area with more number of samples and even to identify the prevalence or presence of intermediate species in different geographical locations.
Keywords
Leptospira
MAT
LipL41
rpoB
Phylogenic analysis
1 Introduction
Leptospirosis, a bacterial disease of zoonotic importance, prevalent globally and affects most of the mammalian species including both domestic and wild animals. Many leptospira serovars are isolated even from aquatic species. The disease spreads through multiple routes both directly or indirectly, however contanimnated soil or water plays a crucial role in transmission of the disease (Hartskeerl and Terpstra, 1996). According to World Health Organization (WHO, 1999), in India huge and diversified animal population with their close contact with farming community is playing an important role in the spread of the disease. Apart from this, poor sanitary conditions, poor animal management and close association between man and animals provide a favourable environment for the spread of the disease. Leptospirosis occurs round the year with a high incidence during rainy reason due to the survival of the Leptospira for a quite long time in the logged water conditions and slightly alkaline environment (Levett, 2001).
Among domestic animals, bovine leptospirosis has been associated with serious financial loss due to infertility, abortions, high culling rates, mastitis or decrease in milk yield. Most of the times the cattle remains as symptomless carrier of leptospires for long periods and pose a great threat to other healthy animals and working personnel. The abortions with leptospirosis are due to interference with the implantation of embryo or with other pregnancy events (Dhaliwal and Murray, 1996; Guitian et al., 1999).
Routinely, in the laboratory, the leptospirosis is diagnosed based on either antigen or antibody detection. However, detection of Leptospira serovar by serology like Microscopic Agglutination Test (MAT) and ELISA needs biosafety and biosecurity laboratories. Therefore, in identification of leptospirosis conventional Polymerase Chain Reaction (PCR) for antigen detection plays a significant role in detection of types of serovar/serogroup involved in the given outbreak or prevalent in that geographical area. In this context, various genes from 16S rRNA and outer membrane proteins (OMP)-like LipL41 have been targeted by different research workers to detect pathogenic leptospires by conventional PCR (Rapiphan et al., 2011; Raven, 2006; Senthilkumar et al., 2011). Considering the advantages of each gene over the other, two genes viz., partial secY (G1,G2) and LipL32 were targeted for identification of pathogenic leptospires.
Molecular analysis of the Leptospira genome especially nucleotides (nts) sequences of specific genes is paramount significant to determine the genetic variations and the evolutionary relatedness including the understanding of the new species or types evolution. Phylogenetic analysis of the isolates or the PCR amplified products using rpoB and LipL41 genes will give a clear picture on the presence of different species of leptospires, which can not be identified completely by MAT or by other PCR techniques. As per Venkatesha and Ramadass (2001), Bevans et al. (2020), Chandan et al. (2016), rpoB gene can be effectively used for phylogenetic analysis and detection of Leptospira species from field materials. The outer membarane proteins like LipL41 plays a crucial role in obligatory bacteria like leptospires and sequence variability can be ascertained effectively in OMPs (Lascola et al., 2006; Marjo and Windell, 2019). The phylogenetic analysis will also help us to understand the prevalence of Leptospira intermediate species which are playing a very significant role in the disease control strategy in India as per the earlier findings (Balamurugan et al., 2013). The phylogenetic analysis targeting two different genes will further help us to know the molecular epidemiology of Leptospria species in the geographical area.
2 Methods
Collection of samples: Cows and Buffaloes with clinical signs and history of suspected leptospiral infections like abortion, stillbirth, infertility problems (repeat breeder, pyometra and cystic ovaries), blood in milk and icterus with dullness, haematuria were chosen for study purpose and samples like blood, serum, urine, milk, aborted materials and tissue samples were collected from these animals.
Isolation of leptospires: All the samples collected from MAT positive animals were used for isolation i.e., sediments in liquid samples and liquid suspension just above the sediment for tissue samples (3–4 drops) were inoculated into the liquid EMJH media using 0.45 µm syringe filter. The tubes were incubated at 30 °C for 4–6 weeks with a weekly observation and sub culturing as per the requirement.
2.1 Polymerase chain reaction (PCR)
DNA extraction: The DNA extraction from the suspected materials was carried out using QIAmp DNA mini kit – (Qiagen, Cat. No: 51306) following the manufacturer’s instructions.
Primers: The following sets of primers were used for the gene G1 F – 5′ CTG AAT CGC TGT ATA AAA GT 3′ and G2 R – 5′ GGA AAA CAA ATG GTC GGA AG-3′ [4]. LipL32-F 5′ CATATGGGTCTGCCAAGCCTAAA 3′ and LipL32-R 5′ CTCGAGTTACTTAGTCGCGTCAGAA 3′ [3]. The PCR reaction was done in 25 ul as previously described (Bal et al., 1994; Meenambigai et al., 2011). The PCR was performed in the total volume of 25 µl reaction mixture as follows:
Master Mix12.5 μl, Forward Primer (10 pmole/μl) 1.0 μl, Reverse Primer (10 pmole /μl) 1.0 μl, Template DNA 4.0 μl, Nuclease free water 6.5 μl, Total 25.0 μl, For positive control four µl of DNA template (L. interogans serovar Hardjo) and for Negative control known negative DNA (Staphylococcus species) was used.
For secY PCR, using primer G1 and G2, one cycle of 94 °C for 5 min for initial denaturation followed by 32 cycles of 94 °C for 1 min-denaturation, 55 °C for 1 min- Annealing, 72 °C for 2 min for extension and final extension of one additional cycle of 72 °C for 6 min.
Agarose gel electrophoresis:
Following electrophoresis, the gel was visualized at 300 nm wave length using a UV transilluminator for the presence of specific amplicons along with DNA molecular weight marker.
2.2 Phylogenetic analysis
The PCR positive samples and the isolates were amplified for sequencing and subsequent phylogenetic analysis using partial rpoB and LipL41 gene sequences using the primers rpoB-F: 5′CCTCATGGGTTCCAACATGCA 3′ and rpoB-R :5′CGCATCCTCRAAGTTGTAWCCTT 3′ and LipL41-F 5′ TAGGAAATTGCGCAGCTACA 3′ and LipL41-R: 5′GCATCGAGAGGAATTAACATCA 3′ and PCR conditions as described earlier (Balamurugan et al., 2013; Bal et al., 1994; Ahmed et al., 2006) along with positive and negative controls. For partial rpoB first cycle of 94 °C for 3 min for initial denaturation, followed by 39 cycles of 94 °C for 90 sec denaturation, 55 °C for 1 min Annealing, 72 °C for 45 sec for extension and final extension of final extension of 72 °C for 20 min. Similarly, for LipL41, first cycle 95 °C for 5 min for initial denaturation and then 34 cycles of 94 °C for 30 sec denaturation, 58 °C for 30 sec Annealing, 72 °C for 1 min for extension and final extension of one cycle of 72 °C for 6 min.
The amplicons were commercially sequences and edited nts sequences by mMEGA 5 software was analyzed by BLAST and Clustal W methodology using the published sequences (Tables 1 and 2).
Sl. No.
Place of Origin
GenBank accession no.
Sl.No.
Place of Origin
GenBank accession no.
1
India
JN388642
21
India
HM046990
2
India
JN388657
22
Brazil
EU747301
3
India
HM046991
23
India
HM046995
4
India
JN388617
24
India
HM046989
5
India
JN388655
25
Brazil
EU747315
6
India
HM046992
26
France
DQ296139
7
India
HM046993
27
India
HM046994
8
India
JN388654
28
India
HM046996
9
India
JN388632
29
India
HM046997
10
India
JN388656
30
India
JN388636
11
India
JN388629
31
India
JN388633
12
Brazil
EU747299
32
India
JN388625
13
Brazil
EU747311
33
France
DQ296138
14
Brazil
EU747304
34
France
DQ296143
15
France
DQ296133
35
France
DQ296136
16
India
JN388624
36
France
EU747316
17
India
JN388631
37
France
DQ296142
18
Brazil
EU747310
38
Brazil
EU747309
19
India
JN388644
39
Brazil
EU 747,303
20
Brazil
EU747307
40
India
EU388638
Sl. No.
Place of Origin
GenBank accession no.
Sl. No.
Place of Origin
GenBank accession no.
1
China
AY622686
21
China
AY776298
2
Brazil
GQ204270
22
USA
AY461952
3
India
DQ132992
23
Mayotte
JN683923
4
USA
AY461945
24
USA
AY461957
5
USA
AY461946
25
USA
AY461956
6
Argentina
KF184576
26
USA
AY461950
7
China
AY622673
27
USA
JN461949
8
China
AY622677
28
India
JN683923
9
China
AY622679
29
USA
AY461953
10
Japan
AY240677
30
USA
AY461954
11
India
AY642286
31
USA
AY461958
12
China
AY622687
32
China
AY622674
13
China
AY622678
33
China
AY622676
14
Japan
AB240674
34
China
AY622684
15
China
AY622683
35
USA
AY461937
16
China
AY776300
36
Mayotte
JN683912
17
China
AY776299
37
Mayotte
JN683918
18
Japan
AB240676
38
Japan
AB240677
19
India
AY642287
39
USA
AY461947
20
China
AY622682
3 Results and discussion
3.1 Isolation of leptospires in EMJH medium
The different samples collected from the MAT positive animals were subjected for isolation, out of which three samples (1.84%) yielded growth of leptospires (Fig. 1) during the second week of incubation. Eventhough, 12 samples were positive by PCR , we could isolate leptospires only from three samples (25%) indicating that less sensitivity of the isolation method or it may be due to the contamination of the other organism while employing the isolation. However, inspite of repeated sub culture and following strict biosecurity and sterility measures the growth was observed in only three samples.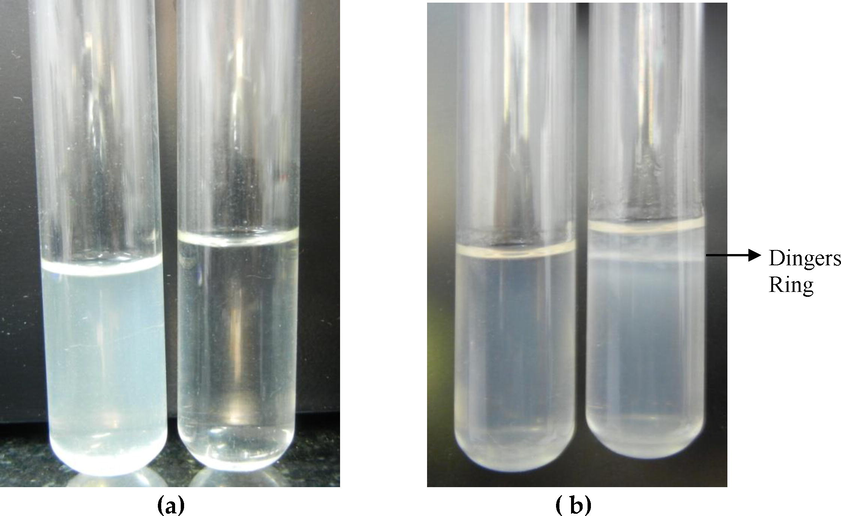
Growth of leptospires in EMJH liquid (a) growth &control and EMJH semisolid medium (b) control &growth.
3.2 Polymerase Chain reaction (PCR)
The different samples collected from MAT positive animals were initially screened using G1 and G2 primers (Gravekamp et al., 1993) could detect 12 (7.36%) leptospiral DNA with an amplicon size of 285 bp. Subsequently, when all the samples were subjected to LipL32 specific PCR the same 12 samples were found positive with an amplicon size of 756 bp (Figs. 2 and 3). The identification of Leptospira is a fool-proof mechanism to determine the active infection in any given animal, hence genomic detection by PCR was employed to confirm the disease in the seropositive animals as PCR being more specific, rapid, less cumbersome. The detection of same 12 samples as positive by both primer sets has shown the efficacy of both the primer sets and the results were in agreement (Cheema et al., 2007), as they also reported that the above primers were similar in their efficacy for diagnosis of leptospirosis. Further, it was found that all the 12 samples were positive by both primers indicating the absence of Grippotyphosa serogroup/serovar. Moreover, all the PCR positive cases were from the animals with aborted history especially from various tissues of the aborted foetus has resulted in specific amplicons. This finding clearly indicates that targeting more aborted animals with repeated collections could help in finding more positive cases by PCR.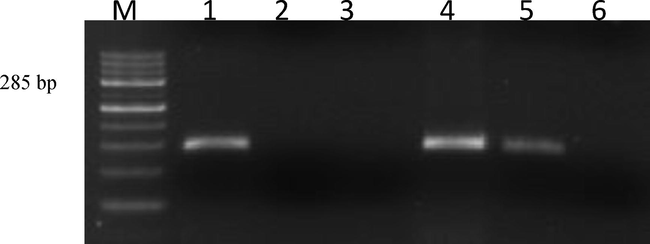
Agarose gel showing PCR amplifications with G1G2 Primers. Lane M: 100 bp ladder; Lane 1: Positive control (Reference sejroe strain); Lane 2: Negative control (Staphylococcus Spp); Lane 3: No template control; Lane 4: Positive isolate (KVAFSU_BANG_KAR I); Lane 5: Positive isolate (KVAFSU_BANG_KAR II); Lane 6: Negative sample.
Agarose gel showing PCR amplifications with LipL32 Primers. Lane M: 100 bp ladder; Lane 1: Positive sample (KVAFSU_BANG_KAR 1); Lane 2: Positive isolate (KVAFSU_BANG_KAR I); Lane 3: Positive control (Reference sejroe strain); Lane 4: Negative control (Staphylococcus Spp); Lane 5: Negative template control; Lane 6: Negative sample.
3.3 Nucleotide sequencing of PCR products
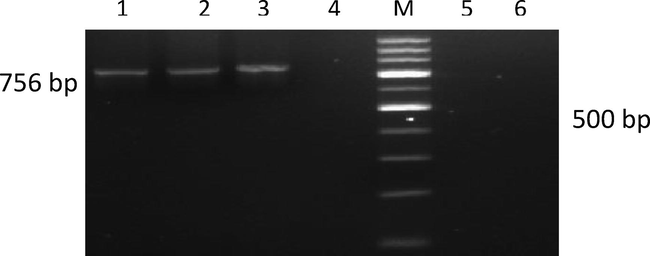
Direct sequencing of the purified PCR amplified partial rpoB and LipL41 gene products (Figs. 4, 5) was done using the gene specific primer pairs. The partial rpoB gene primer included Lept 1900F and Lept 2500R bordering a hypervariable region between the position 1900 and 2500. The LipL41 gene primer included the polymorphic sequence size of 518 coating for outer membrane lipoprotein with a PCR product size of 520 bp (Ahmed et al., 2006). There is an exponentially high sequencing data of biological genomes is available now for molecular biological or genetic investigations (Bansal, 2005). The genetic analysis of leptospires is to be established to show the genetic relatedness and evolutionary variation among the different Leptospira species prevalent in a geographical area. The study of nucleotide sequence is important to understand the evolution of new types or species, its importance in the development of diagnostic and molecular epidemiological investigations. Further, this technique is beneficial in identification of even single nucleotide deletion / addition or substitution, which cannot be detected in any other methods.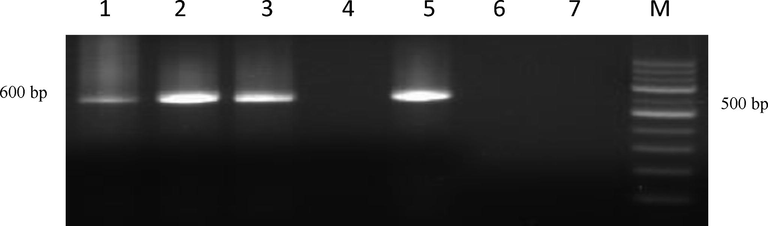
Agarose gel showing PCR amplification with rpoB Primers. Lane M: 100 bp ladder; Lane 1: Positive isolate (KVAFSU_BANG_KAR I); Lane 2: Positive isolate (KVAFSU_BANG_KAR II); Lane 3: Positive isolate (KVAFSU_BANG_KAR III); Lane 4: Negative control (Staphylococcus Spp); Lane 5: Positive control (Reference canicola strain); Lane 6: No template control; Lane 7: Negative sample.
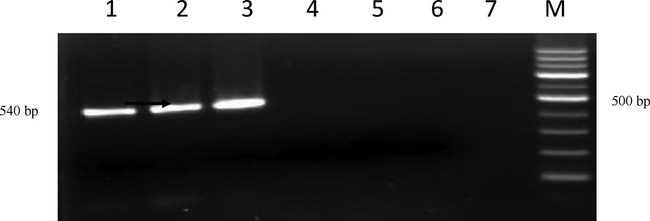
Agarose gel showing PCR amplification with LipL41 Primers Lane M:100 bp ladder; Lane 1: Positive isolate (KVAFSU_BANG_KAR I); Lane 2: Positive isolate (KVAFSU_BANG_KAR II); Lane 3: Positive control (Reference canicola strain); Lane 4: Negative control (Staphylococcus Spp); Lane 5: No template control; Lane 6 and 7: Negative sample.
In principle, the phylogenetic analysis based on alignment of nucleotide sequence of specific genes provide an insight into the genetic makeup of the leptospires and their sharing with the other members of the group. This approach is perhaps the most realistic and presents a quick bird’s eye view of the complex inter-relatedness.
3.4 Phylogenetic analysis
The Phylogenetic trees were constructed and the evolutionary history was inferred using the Neighbour-Joining method (Saitou and Nei, 1997) using corresponding sequences from both published reference rpoB and LipL41 sequences from different parts of the world and nucleotide sequences obtained in this study. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) were shown next to the branches (Felsenstein, 1985). The evolutionary distances were computed using the Neighbour-Joining tree method (Tamura et al., 2004) and are in the units of the number of base substitutions per site. The analysis involved nucleotide sequences from reference strains as well as amplified products sequenced in this study. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2007) The details are shown as per the cluster formation in (Figs. 6, 7).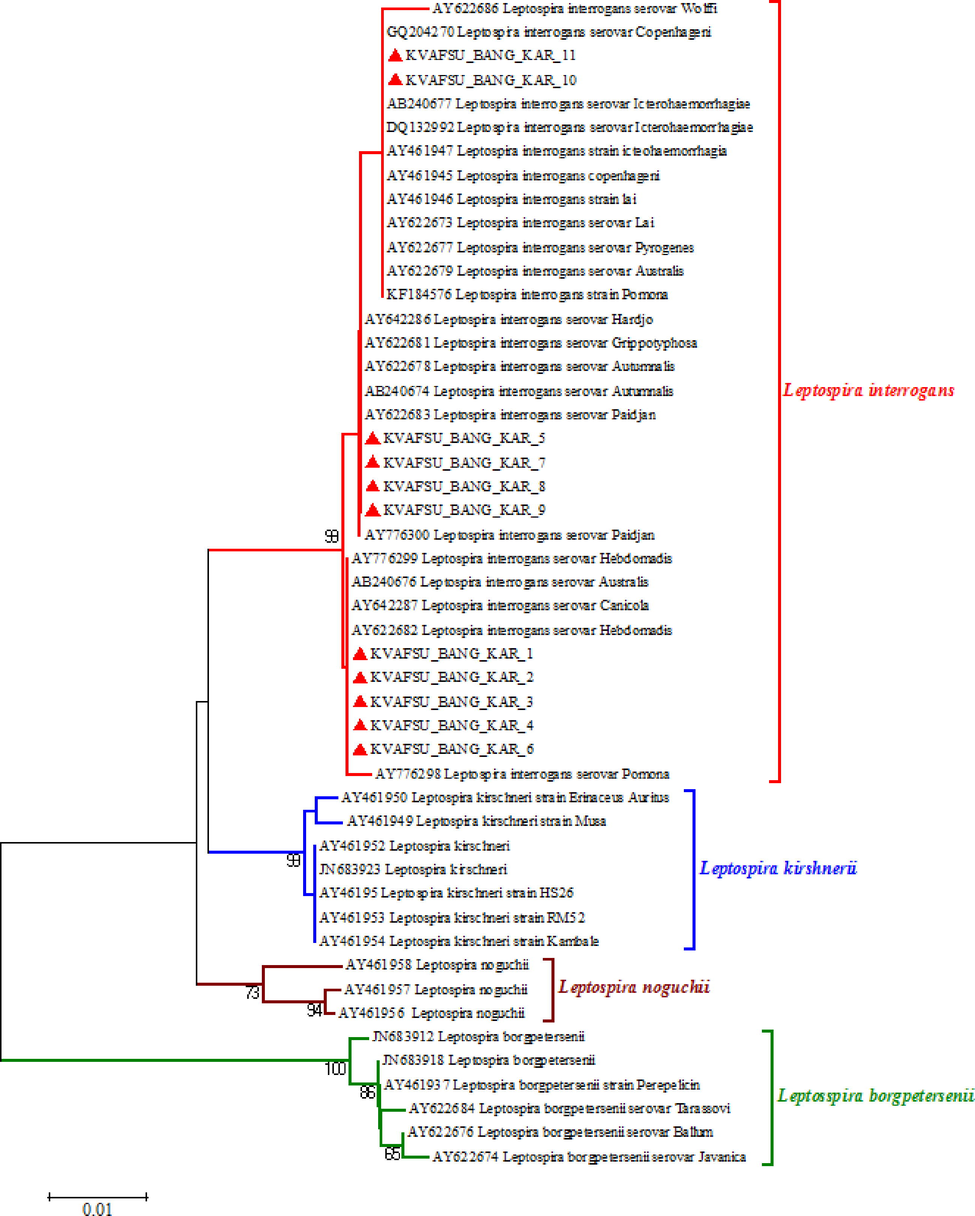
Phylogenetic tree of partial rpoB gene nucleotide sequences using neighbour-joining method.
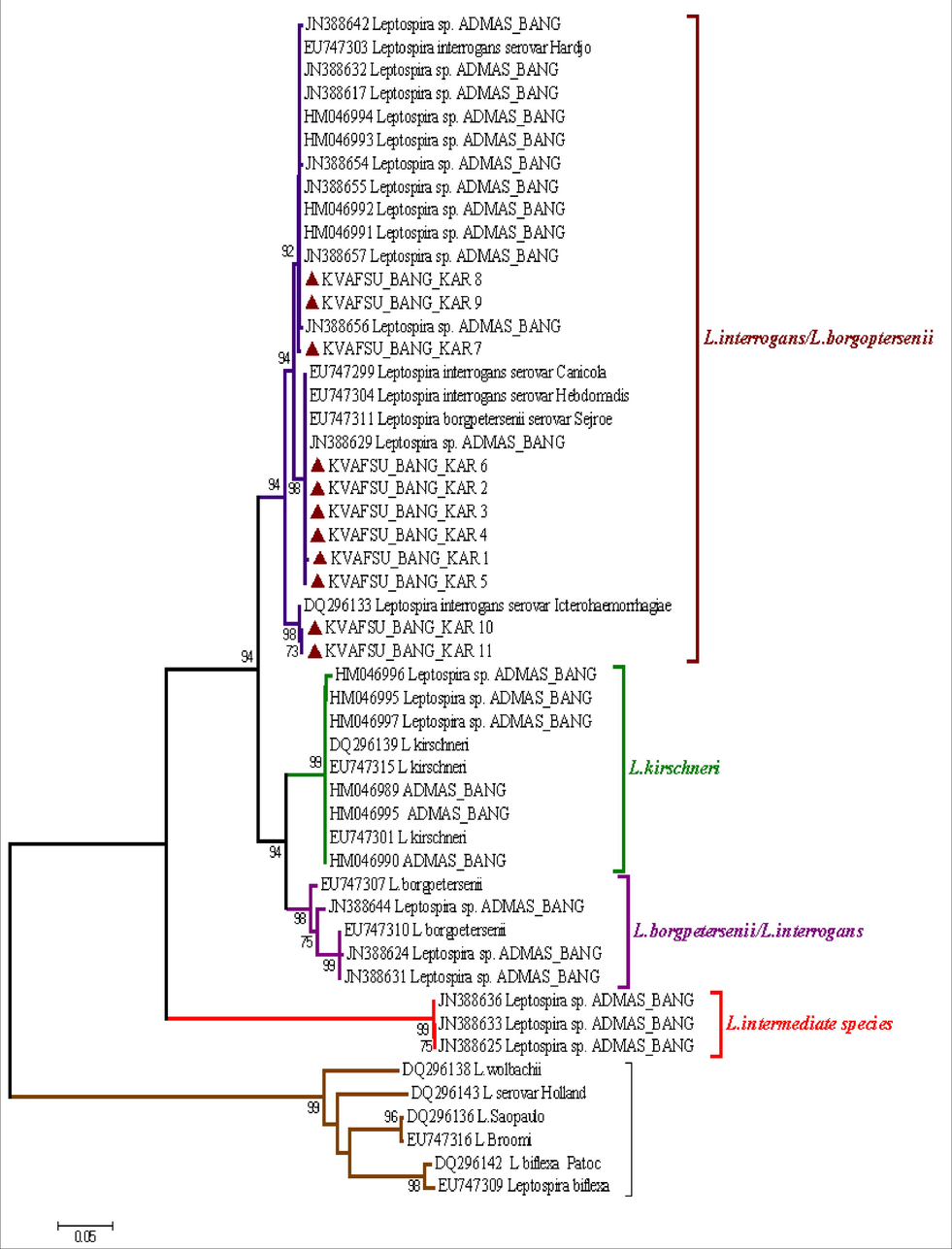
Phylogenetic tree of LipL41 gene nucleotide sequences of Leptospira constructed based on neighbour-joining method.
3.4.1 Analysis of partial rpoB gene sequences of the Leptospira
Several molecular techniques have been evaluated for the identification and characterization of Leptospira species and DNA-DNA hybridization identified 20 Leptospira species to date with nine pathogenic Leptospira species (Cerqueira and Picardeau, 2009). Analysis of a segment of rpoB may be useful as an initial screening test for the identification of a new isolate of Leptospira using a system of similarity cut-off to define species (Lascola et al., 2006).
Out of the 12 PCR positive samples, 11 samples gave a reasonably good sequence read sufficient enough for further analysis when subjected to nucleotide sequencing for partial rpoB gene (Fig. 6). By sequence analysis, the positive samples showed identity with the many published sequences of various leptospires of NIVEDI, Bangalore (formerly PD_ADMAS), India and other global isolates. In general, all the samples have shown identity with L.interrogans/L.borgpetersenii species by forming various clusters in the same species group. None of the samples have formed cluster with Leptospira krischneri and Leptospira intermediate species as well as Leptospira biflexa and few other serovars / species which were used as out group to compare the sequences.
When the above 11 amplified products were compared within the L.interrogans/L.borgpetersenii species, it was clearly evident that they formed three different clusters i.e., KVAFSU_BANG_KAR 1–6 have formed one cluster and KVAFSU_BANG_KAR 7–9 have formed another cluster. KVAFSU_BANG_KAR 10 and 11 have formed a different cluster. The cluster formation was almost matching with the findings of multiplex PCR (mPCR) where the samples KVAFSU_BANG_KAR 1–4 have shown amplication with Leptospira interrogans serovar canicola and in the cluster formation also the above four samples have shown clear identity with the L.interrogans serovar canicola. Similarly, KVAFSU_BANG_KAR 7–9 has shown amplification with Leptospira interrogans serovar sejroe and in the cluster formation also the above three samples have shown identity with the L.interrogans serovar Hardjo. KVAFSU_BANG_KAR 10 and11 has shown amplification with L.interrogans serovar Icterohaemorrhagiae and in the cluster formation also above two samples have shown identity with the L.interrogans serovar Icterohaemorrhagiae. For samples 5 and 6 even though they have shown identity with serovars Canicola, Hebdomadis and few other isolates but based on mPCR and nucleotide sequencing of partial rpoB findings it can be concluded that they belong to other serovar of L.interrogans/L.borgpetersenii species.
The present findings were in agreement with the findings of (Lascola et al., 2006) and (Bal et al., 1994) they also stated that partial rpoB gene can be effectively used for phylogenetic analysis, as well as identification of leptospires from clinical samples (Bal et al., 1994) in their findings clearly stated that 30.3% of Leptospira isolates isolated from different parts of India belong to L.interrogans/L.borgpetersenii species and in the present study also all the 11 PCR amplified products were belonging to the above species group and none of them were belonging to L.intermediate group.
3.4.2 Analysis of partial LipL41 gene sequences of the Leptospira
Out of the 12 PCR positive samples, 11 samples gave a reasonably good sequence read sufficient enough for further analysis when subjected to nucleotide sequencing for LipL41 gene (Fig. 7). The LipL41 gene includes the polymorphic sequence size of 518 coating for outer membrane lipoprotein with a PCR product size of 520 bp (Ahmed et al., 2006). In obligate intracellular bacteria, the outer membrane proteins (OMPs) play a crucial role in the process of adaptation by facilitating interactions between bacterial cells and its host (Nguyen et al., 2006).
Though more than 258 serovars in the genus Leptospira have been identified, they are antigenically distinct due to the lipoploysaccharide (LPS) and spatial arrangement of outer membrane lipoproteins (OmpL). Generally, the 16S rRNA genes are most conserved but sequence variability is noticed in outer membrane proteins (Vedhagiri et al., 2009). Keeping these points in view to identify the speciation of the leptospires and evolutionary changes LipL41 was targeted in the present study using the published nucleotide sequences.
Most of the published sequences belong to human and less data was available pertaining to animals. This was probably the first study to compare the animal isolates or samples to compare with the published human isolates for the LipL41 gene region in India. Interestingly, unlike with rpoB gene region, all the sequenced amplified products shown the identity with the L.interrogans species and there was no overlapping of L.interrogans/L.borgpetersenii species observed. When the above 11 amplified products were compared within the L.interrogans species it was clearly evident that they formed three different clusters i.e., KVAFSU_BANG_KAR_ 1–4 and 5 have formed one cluster and KVAFSU_BANG_KAR_6 and 7–9 have formed another cluster. KVAFSU_BANG_KAR_ 10 and 11 have formed a different cluster. The cluster formation was almost matching with the findings of mPCR where the samples KVAFSU_BANG_KAR_ 1–4 have shown amplication with L. interrogans serovar Canicola and in the cluster formation also the above four samples have shown clear identity with the L.interrogans serovar Canicola. Similarly, KVAFSU_BANG_KAR_ 7–9 has shown amplification with L.interrogans serovar Hardjo and in the cluster formation also the above three samples have shown clear identity with the L.interrogans serovar Hardjo. KVAFSU_BANG_KAR_ 10 and 11 has shown amplification with L.interrogans serovar Icterohaemorrhagiae and in the cluster formation also above two samples has shown clear identity with the L.interrogans serovar Icterohaemorrhagiae. For sample 5 even though it has shown more identity with serovars Autumnalis, Hardjo, Paidjan and Grippotyphosa and few other isolates but based on mPCR and nucleotide sequencing of LipL41 findings, it was concluded that it belongs to other serovars of L.interrogans species. For sample 6 even though it has shown more identity with serovarss Hebdomadis, Canicola, Australis and Pomona but based on multiplex PCR and nucleotide sequencing of LipL41 findings, it was concluded that it belongs to other serovar of L.interrogans species.
The observations were in agreement with the other’s findings (Vedhagiri et al., 2009), in their study also L.interrogans has formed a separate cluster when compared with the L.borgpetersenii species. Apart from this the present observation of forming a single cluster of L.interrogans species was in agreement with the findings of previous report, where the LipL41 gene has highly conserved sequences that are expressed both in cultivated organisms and during infection in mammals (Haake et al., 1999). Furthermore, the present finding also manifest that the PCR positive samples have indicated same result with respect to conserved sequence during the infection.
4 Conclusion
Interestingly, results analysis of both rpoB and LipL41 genes were compared with the species differentiation, which could not be done completely when rpoB gene sequences alone was targeted, as in that there was an overlapping/superimposition between the L.interrogans/L.borgpetersenii species was observed as reported earlier. It is noteworthy that these findings in our present work clearly pointed out that LipL41 can be more efficiently used for species identification of leptospires when compared to rpoB gene sequences alone, but this needs to be ascertained on a larger number of samples from different geographical areas of the endemic region.
Funding
The study was funded by IAH&VB, Hebbal, Bengaluru, India.
Acknowledgements
Authors thank the Director, IAHVB and VC, KVAFSU for providing necessary funding and guidance throughout the study. The authors extend their appreciation to the Researchers Supporting Project Number (RSP-2020/201), King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Levett, P.N., 2011. Leptospirosis. Clin. Microbiol. Rev.14, 296–326.
- Reproductive performance of dairy herds infected with Leptospira interrogans serovars hardjo relative to the year of diagnosis. Vet. Rec.. WA.1996.;138:272-276.
- [Google Scholar]
- Infertility and abortion among first lactation on dairy cows seropositive or seronegative to Leptospira interrogans serovars hardjo infection. J. Am. Vet. Med. Assoc.. 1999;215:515-518.
- [Google Scholar]
- PCR based technique for detection and differentiation of Pathogenic and Saprophytic Leptospira species. Intl. J. Microbiol. Res.. 2011;2:43-48.
- [Google Scholar]
- Developmentof species-specific PCR primer sets for the detection of Leptospira. FEMS Microbiol. Lett.. ER.2006.;264:31-39.
- [Google Scholar]
- Use of polymerase chain reaction for the detection of leptospires in clinical samples. Indian Vet. J.. 2011;78:1087-1090.
- [Google Scholar]
- Rapid diagnosis of leptospirosis using PCR and DNA hybridization techniques. Indian J. Anim. Sci.. 2001;72:1104-1106.
- [Google Scholar]
- Bevans, A.I., Fitzpatrick, D.M., Stone, D.M., Butler, B.P., Smith, M.P., Cheetam, S., 2020. Phylogenetic relationship and diversity of bat associated Leptospira and the histopathological evaluation of these infections in bats from Grenade, West Indies. PLoS. Negl. Trop. Dis. 2014,1-15.
- Use of rpoB gene analysis for detection and identification of Leptospira species by direct sequencing. Eur. J. Biotech. Biosci.. 2016;4:34-43.
- [Google Scholar]
- Partial rpoB gene sequencing for identification of Leptospira species. FEMS. Microbiol. Lett.. 2006;263:142-147.
- [Google Scholar]
- Identification of Leptospira sps from environmental sources in areas with high human leptospirosis incidence in the Philippines. Pathogen and Global Health. LR.2019.;113:109-116.
- [Google Scholar]
- Balamurugan, V., Gangadhar, NL., Mohandoss, N., Thirumalesh, SRA., Dhar, M., Shome, R., Krishnamoorthy,P., Prabhudas, K., Rahman, H.2013. Characterization of Leptospira isolates from animals and humans: phylogenetic analysis identifies the prevalence of intermediate species in India. Springer. Plus.2, 362.
- Detection of leptospires in urine by PCR for early diagnosis of leptospirosis. J. Clin. Microb.. WJ.1994.;32:1894-1898.
- [Google Scholar]
- Meenambigai, T.V., Gopalakrishnan. Ravikimar. Andy, S., Govindan, B., Chidambaram, S., Bakthavachalam, M., 2011. Simultaneous detection of LipL32 and LipL21 genes of Pathogenic Leptospira from serum samples of bovines by multiplx PCR. Vet. Sci Dev. 1, 60–62.
- Ahmed, N., Devi, S.M., Valverde, M.L., Vijayachari, P., Machangus, R.S., 2006. Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira isolates. PloS One. 5, 1533.
- Detection of seven species of pathogenic leptospires by PCR using two sets of primers. J. Gen. Microbiol.. WJ.1993.;139:1691-1700.
- [Google Scholar]
- Detection of pathogenic leptospires in animals by PCR based on LipL21 and LipL32 genes Indian. J. Expt. Biol.. 2007;45:568-573.
- [Google Scholar]
- Bansal, A.K., 2005. Bioinformatics in microbial biotechnology: A mini review. J. Micro. Cell. Fact. 4, 19.
- The neighbour-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol.. 1987;4:406-425.
- [Google Scholar]
- Felsenstein, J., 1985.Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783–791.
- Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Nat. Acad. Sci.. 2004;10:11030-11035.
- [Google Scholar]
- MEGA 5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol.. 2007;28:2731-2739.
- [Google Scholar]
- Phylogenetic analysis of general bacterial porins: a phylogenomic case study. J. Mol. Microbiol. Biotechnol.. ST.2006.;11:291-301.
- [Google Scholar]
- Evolutionary implication of outer membrane lipoprotein-encoding genes ompL1, lipL32 and lipL41 of pathogenic leptospira species. Genomics. Proteomics. Bioinf.. 2009;7:96-106.
- [Google Scholar]
- Changes in the surface of Leptospira interrogans serovar grippotyphosa during invitro cultivation. Infect. Immun.. MA.1999.;59:1131-1140.
- [Google Scholar]







