Translate this page into:
Intraspecific molecular variation among Androctonus crassicauda (Olivier, 1807) populations collected from different regions in saudi arabia
⁎Corresponding author. ahmedbadry@azhar.edu.eg (Ahmed Badry)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Androctonus crassicauda is one of the medically important scorpion species occurring in the Middle East. In this study, molecular variation in mtDNA cytochrome oxidase subunit I (COI) gene within seven populations of A. crassicauda from four main ecological regions of Saudi Arabia was investigated. Scorpion specimens were collected from different eco-geographical regions. DNA was extracted, subsequently 700 bp fragments of COI gene were amplified using specific scorpion primers and sequenced. The obtained sequences were analyzed, and phylogenetic trees were constructed. The phylogenetic analysis showed high levels of genetic variation among A. crassicauda populations with the existence of three distinct lineages. Also, it supports the existence of two distinct populations of A. crassicauda in Saudi Arabia, which perhaps suggestive of a putative distinct species. Further morphological studies with additional specimens from the Arabian Peninsula may reveal possible undiscovered and cryptic species in the region.
Keywords
Molecular phylogeny
Scorpiones
Phylogeography
COI
mtDNA
Ecogeographical regions
- COI
-
cytochrome oxidase subunit I
- mtDNA
-
mitochondrial DNA
- MP
-
Maximum-parsimony
- NJ
-
neighbor-joining
- BI
-
Bayesian inference
Abbreviations
1 Introduction
Androctonus Ehrenberg, 1828 is a genus of the Family Buthidae, with 30 described species (Rein, 2021; Yağmur, 2021). This genus was introduced by Ehrenberg in Ehrenberg and Hemprich (1828) with two sub-genera Prionurus Ehrenberg (=Buthus) and Leiurus Ehrenberg. Several studies dealt with species of this genus since Vachon (1948, 1952) standardized a definition of the genus Androctonus, contributing significantly to our growing knowledge on the systematics of this genus (Fet et al., 2000; Lourenço, 2005, 2008; Lourenço and Qi, 2006, 2007; Teruel et al., 2013; Kovařík and Ahmed, 2013; Lourenço et al., 2009, 2012, 2015; Rossi, 2015). Of these, Androctonus crassicauda (Olivier, 1807) is the most widely distributed species and known for its medical importance (Chippaux and Goyffon, 2008; Alqahtani and Badry 2021). The distribution of this species extended from Sinai, Egypt across Arabia, and the Middle East (Crucitti, 1999; Amr et al., 2021). It s venom contains toxins of which block potassium channels (Miller 1995), chloride channels (DeBin et al. 1993) and toxins which increase the flow of sodium ions into neurons to increase the excitability of neurons (Zlotkin et al. 1994; Benkhalifa et al. 1997). Given its medical importance and widespread distribution understanding the distribution of the species diversity in the region is important, because the correct identification of scorpion species is essential to the treatment of envenimation. Recently, molecular phylogeny of scorpions is considered very useful to provide evidence of genetic heterogeneity within and between populations of different taxa (Gantenbein et al. 1999; Ben Ali et al. 2000; Fet et al. 2003; Gantenbein and Largiadèr 2003; Ben Othmen et al. 2009; Ozkan et al., 2010). We, therefore, employed molecular analysis to assess the presence of intraspecific molecular variations within A. crassicauda populations from four main ecological regions of Saudi Arabia, by sequencing the intraspecific hypervariable region of the mtDNA cytochrome oxidase subunit I (COI) gene.
2 Materials and methods
Biological material. Specimens of A. crassicauda were collected from nine location representing four main ecological regions in Saudi Arabia, including the North Arabian Desert (NAD), Central Arabian Desert (CAD), the Arabian Sand Desert (ASD), Southwestern Arabian Escarpment and Highlands (SAEH) (Fig. 1, Table1). Scorpions were collected during the period from January 2021 to July 2021, using mainly ultraviolet lights at night and randomly searched in their hiding places during the daytime (Williams 1968; Stahnke 1972). The collected scorpions were preserved in 95%ethanol for DNA isolation as described by (Prendini et al., 2003).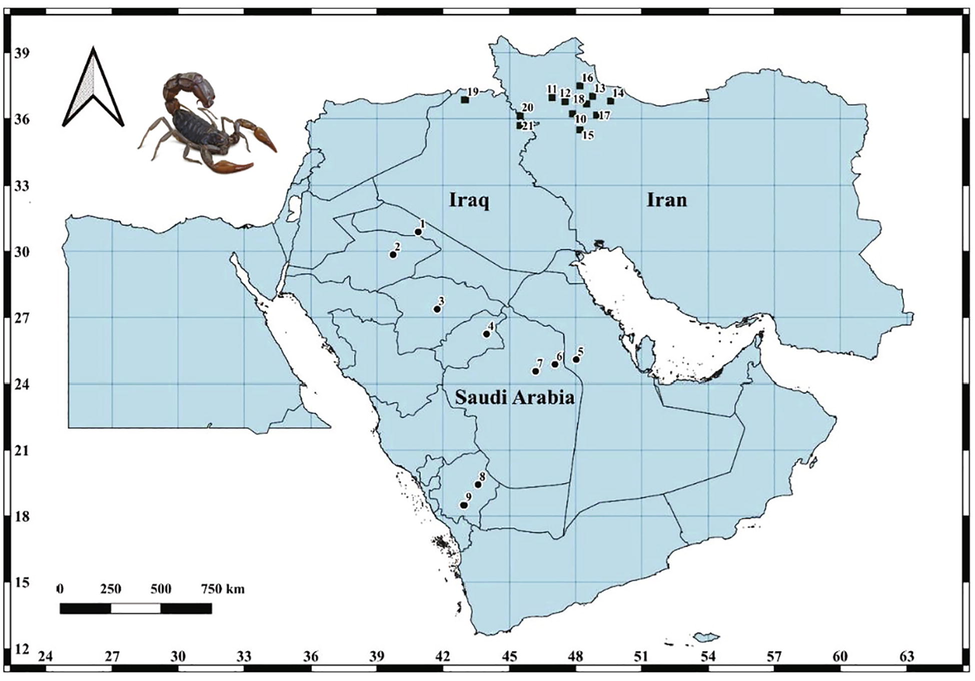
Collection localities of Androctonus crassicauda samples from Saudi Arabia and GenBank sequences from Iraq and Iran that are given in Table 1.
No
Species
Location
Region
Ecogeoraphical region
Country
N
Lat.
Long.
Accession number
Authority
1
A. crassicauda
Arar
Northern Borders Province
North Arabian Desert
Saudi Arabia
3
30.88
40.87
–
This Study
2
A. crassicauda
Dumah Al Jandal
Al Jowf
3
29.84
39.73
–
This Study
3
A. crassicauda
Hail
Hail Province
Central Arabian Desert
3
27.37
41.73
–
This Study
4
A. crassicauda
Buraydah
Al Qassim
2
26.241
43.94
–
This Study
5
A. crassicauda
Khurais
Eastern Province
Arabian Sand Desert
2
25.07
48.02
–
This Study
6
A. crassicauda
Nazeem, east of Riyadh
Riyadh
1
24.86
47.06
–
This Study
7
A. crassicauda
Dhurma
1
24.54
46.17
–
This Study
8
A. crassicauda
Tathleeth
Aseer Province
Southwestern Arabian Escarpment and Highlands
2
19.42
43.56
–
This Study
9
A. crassicauda
Wadi Al Shiq
1
18.49
42.93
–
This Study
10
A. crassicauda
Mahneshan
Mahneshan
–
Iran
1
36.22
47.86
MH352603
Gene Bank
11
A. crassicauda
Sari_Aghol
–
1
36.95
46.93
MH352604
Gene Bank
12
A. crassicauda
Sahand-e Olya
–
1
36.77
47.52
MH352605
Gene Bank
13
A. crassicauda
Darram,
Zanjan Province
–
1
37.02
48.77
MH352606
Gene Bank
14
A. crassicauda
Chavarzagh
Zanjan Province
–
1
36.80
49.66
MH352607
Gene Bank
15
A. crassicauda
Taroom,
Sansooz
–
1
35.49
48.20
MH352608
Gene Bank
16
A. crassicauda
Zanjan
Zanjan Province
–
1
36.69
48.50
MH352609
Gene Bank
17
A. crassicauda
Doasb,
Zanjan Province
–
1
36.16
48.95
MH352610
Gene Bank
18
A. crassicauda
Daneshgah
Zanjan Province
–
1
36.67
48.50
MH352611
Gene Bank
19
A. crassicauda
Sardasht,
West Azerbaijan Province
–
1
36.14
45.47
MK814934
Soltan et al., 2021
20
A. crassicauda
Sardasht,
West Azerbaijan Province
–
1
35.68
45.19
MK814933
Soltan et al., 2021
21
A. crassicauda
–
–
–
Iraq
1
36.86
42.98
MT229840
Gene Bank
DNA extraction, COI-PCR amplification, and sequencing. Genomic DNA was extracted from fresh or preserved (in 95% ethanol) scorpions either from the pedipalps or from the muscle tissue of the legs using a DNeasy extraction kit (Qiagen). The 5́ hypervariable region of the mtDNA cytochrome oxidase subunit I (COI) gene was amplified by polymerase chain reaction (PCR) in all samples, using the universal COI primers according to Folmer et al. (1994) and Coelho et al. (2014). Products of the PCR were electrophoresed in a 1 % agarose gel stained with 0.5 µg mL−1 of ethidium bromide, observed by using UV transilluminator, purified, and then sequenced on an ABI 3500 automated sequencer (Applied Biosystems Inc., USA).
Phylogenetic analysis. Sequences were screened and analyzed using Finch TV 1.4.0 (Geospiza, Inc., USA; https://www. geospiza.com). Additional sequence data were downloaded as ingroup from GenBank for A. crassicauda from Iraq and Iran (Table 1). Additional sequences data of COI region of congruent Androctonus species were also obtained from the Genbank and used as in-groups, including A. amoreuxi, A. australis, A. bicolor, A. gonneti, A. liouvillei and A. mauritanicus (Accession Numbers: KJ538436.1, KJ538184.1, KJ538333.1, KJ538381.1, KJ538220.1, and JF820097.1) respectively. Sequences data of Scorpio palmatus was downloaded as an outgroup (Accession Numbers AY156585.1). Obtained sequences were aligned with the ClustalW extension in MEGA 6 (Kumar et al. 1994), using the default settings. Nucleotide composition was calculated from the in-group sequences only. MEGA 6 was also used to estimate the genetic distances (for the entire data set). For the phylogenetic analysis, the mitochondrial COI gene data set (n = 38) was examined. The phylogenetic analysis (Maximum-parsimony (MP), Neighbor joining (NJ), and Bayesian inference (BI) were performed as described by Alqahtani and Badry (2020a). The Maximum-parsimony (MP) and neighbor-joining (NJ) analyses were performed with Paup v4 (Swofford 2001) with heuristic searches using stepwise addition followed by tree bisection reconnection (TBR) branch swapping (Swofford et al. 1996). In all alignments, gaps were treated as missing characters. Confidence within the nodes was evaluated using 1000 bootstrap replicates (Felsenstein 2002) with random addition of taxa. MrModeltest 2.3 (Nylander 2004) was used to select the best-fit models of nucleotide evolution supported by Akaike information criterion (AIC) (Akaike 1973). The geographic structure was inferred using Bayesian inference (BI) implemented with MrBayes 3.1.2 (Ronquist et al. 2012). Analyses were run for one million generations and the output parameters were visualized to determine stationarity and convergence using Tracer 1.4 (Rambaut & Drummond, 2007).
3 Results
This study comprised 38 sequences with 556 nucleotide sites of Androctonus species and the applied outgroup. In total 384 (69.0%) nucleotide sites were polymorphic and 104 (18.7%) were parsimony informative. Within the in-group, 58 (10.4%) sites were polymorphic and 45 (8.09%) were parsimony informative. Tamura and Nei (1993) genetic distances between the Arabian A. crassicauda populations ranged from d = 0.01 to 0.08, and from d = 0.10 to 0.11 between Androctonus species in-group sequences (Table 2).
Populations/ Species
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
1. Northern_Borders_Province, SA
0.00
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
2. Al_Jowf, SA
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
3. Hail_Province, SA
0.07
0.07
0.00
0.00
0.00
0.00
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
4. Al_Qassim, SA
0.07
0.07
0.02
0.00
0.00
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
5. Eastern_Province, SA
0.07
0.07
0.02
0.02
0.00
0.00
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
6. Riyadh_Province, SA
0.07
0.07
0.01
0.02
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
7. Aseer_Province, SA
0.08
0.07
0.01
0.02
0.02
0.02
0.01
0.01
0.01
0.01
0.01
0.01
0.01
0.01
8. Iran
0.05
0.06
0.08
0.08
0.08
0.08
0.08
0.01
0.01
0.01
0.01
0.01
0.01
0.01
9. Iraq
0.05
0.05
0.08
0.08
0.08
0.07
0.08
0.03
0.01
0.01
0.01
0.01
0.01
0.01
10. A. liouvillei
0.10
0.10
0.11
0.11
0.11
0.11
0.11
0.09
0.09
0.01
0.01
0.01
0.01
0.01
11. A. gonneti
0.11
0.11
0.11
0.11
0.11
0.11
0.11
0.09
0.09
0.08
0.01
0.01
0.01
0.01
12. A. australis
0.10
0.10
0.10
0.11
0.10
0.10
0.10
0.10
0.09
0.11
0.10
0.01
0.01
0.01
13. A. mauritanicus
0.10
0.10
0.09
0.10
0.10
0.09
0.10
0.10
0.10
0.09
0.11
0.08
0.01
0.01
14. A._amoreuxi
0.10
0.10
0.11
0.12
0.11
0.11
0.11
0.11
0.10
0.07
0.09
0.10
0.10
0.01
15. A. bicolor
0.11
0.11
0.11
0.11
0.11
0.10
0.11
0.12
0.11
0.08
0.10
0.10
0.09
0.10
The phylogenetic tree topologies recovered by the analysis of the COI data set, from the MP, NJ, and BI analysis, were the same in outlining two major clades of A. crassicauda (Figs. 2, 3, 4). The first clade includes the population of the North Arabian Desert (NAD), and those of Iraq and Iran. This first clade is split further into two well-supported subclades. The first includes the population of the Northern part of Saudi Arabia, represented by six specimens from the Northern Border Province and Al Jowf populations. The second subclade consists of one specimen from Iraq and eleven from northern Iran. The second clade represents the remaining populations of the Central Arabian Desert (CAD), Arabian Sand Desert (ASD) and the Southwestern Arabian Escarpment and Highlands (SAEH) grouped as a sister phylogroup.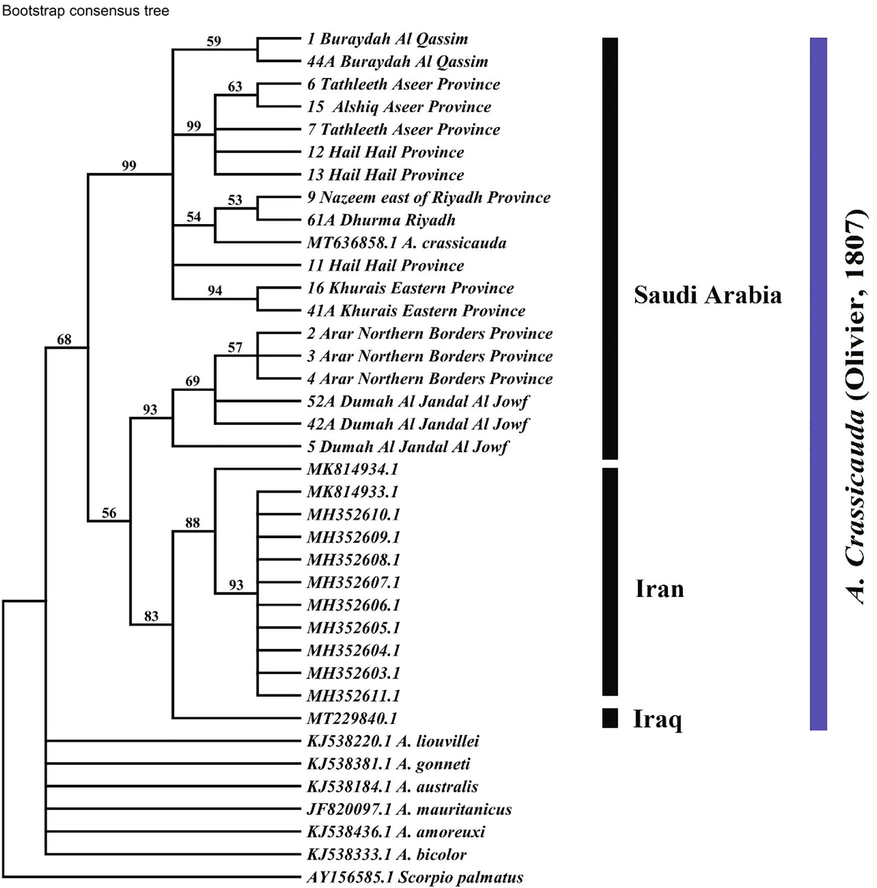
Maximum-parsimony phylogenies of Androctonus crassicauda sequences fragment of the COI gene. Numbers above the branches represent bootstrap values calculated with 1000 replicates.
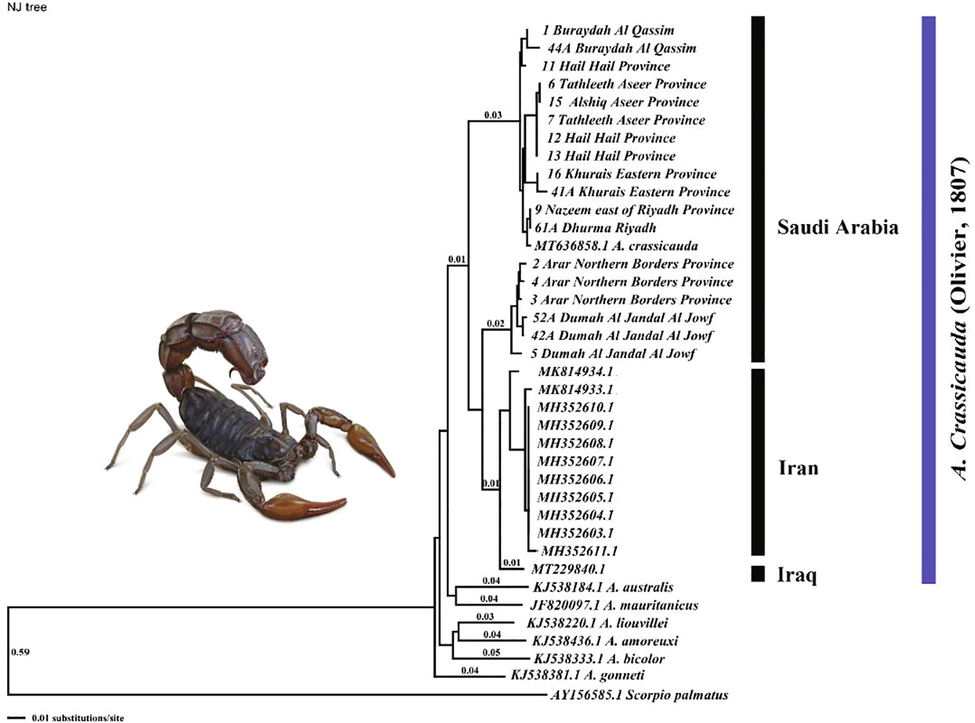
Neighbor-joining phylogenies of Androctonus crassicauda sequences fragment of the COI gene. Numbers above the branches represent distance values.
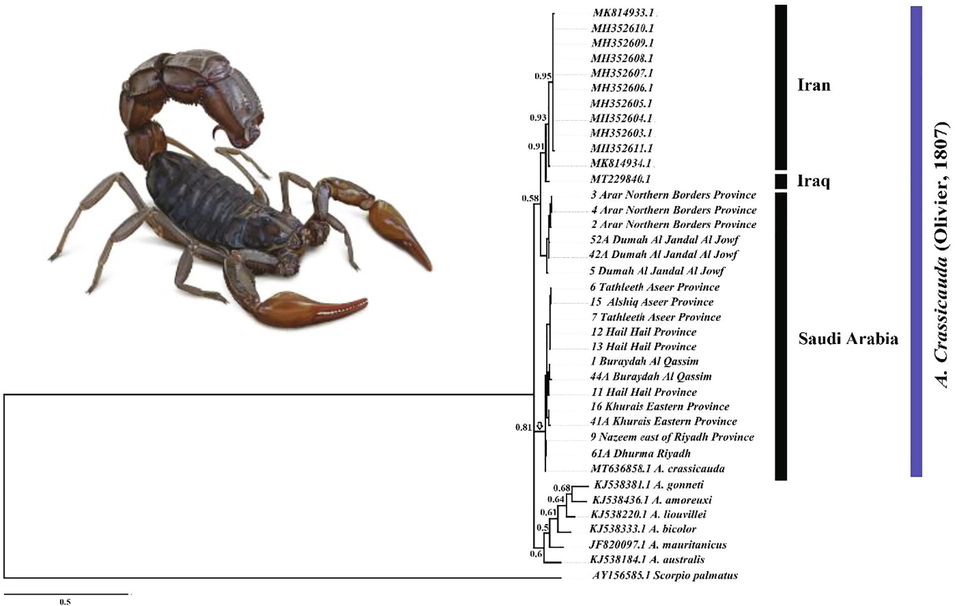
Bayesian inference tree phylogenies of genus Androctonus crassicauda sequences fragment of the COI gene. Numbers above the nodes represent the posterior probabilities.
4 Discussion
Androctonus crassicauda is one of the medically important scorpion species occurring in the Middle East. Šmíd et al., (2021) defined six different ecogeographical regions across Saudi Arabia, including the North Arabian, Central Arabian, the Arabian Sand and Tihama Deserts, as well as the Red Sea Shrublands, and Southwestern Arabian ridge and highlands. Our study of A. crassicauda scorpion populations found a relatively high population genetic diversity, retrieving three main lineages in ten investigated populations.
Previously, several studies reported similar results conducted with the same and allied genera, such as Androctonus Ehrenberg, 1828, Buthus Leach, 1815, Buthacus Birula, 1908, Hottentotta Birula, 1908, Leiurus Hemprich and Ehrenberg (1829) and Scorpio L., 1758 (Gantenbein and Largiadèr 2000; Froufe et al. 2008; Ben Othmen et al., 2009: Ozkan et al., 2010; Sousa et al. 2011; Coelhoa et al., 2014; Toprak et al., 2019; Alqahtani and Badry 2020a, b; Sarhan et al., 2020; Toprak et al., 2019). The phylogenetic analysis of the current populations of A. crassicauda has divided into three monophyletic well supported clades was unexpected. The first encompasses five Saudi Arabian populations grouped in a sister clade. These populations are from three different ecogeographical regions including the Central Arabian desert (CAD), the Arabian Sand desert (ASD), and the Southwestern Arabian Escarpment and Highlands (SAEH). The second lineage encompasses two Saudi Arabian populations from two different localities of the North Arabian Desert (NAD). The third lineage consists of twelve specimens from Iraq and Iran (Fig. 1, Table 1).
Despite that the genetic distance between Saudi Arabian clades is relatively high, this may indicate that A. crassicauda represents two cryptic species. The genetic distances between A. crassicauda obtained from the North of Arabian desert (NAD) and those obtained from other ecogeographical regions of Saudi Arabia ranged from 7 to 8% (Table 2). Also, the distance between the North Arabian Desert (NAD) populations and those from Iraq and Iran ranged from 5 to 6% (Table 2). Due to the shortage of clear morphological features used in the traditional taxonomy in many scorpions’ taxa, the occurrence of cryptic species seems to be common (Gantenbein, et al., 2000). Furthermore, analysis of genetic variations between populations of Scorpio fuliginosus (Pallary, 1928) from Morocco identified genetically distinct lineages (Froufe et al., 2008). The high intraspecific genetic variability within Androctonus scorpions may be due to prominent geographical features, which contribute to increasing scorpion inclination for diversification in association with long-term episodes of geomorphological changes and climatic changes. The divergence between the Central and Southwestern Saudi Arabia from the north and Iraqi- Iranian populations were probably associated with the Taurus-Zagros Mountains and the Saharo-Arabian developing desert belt, which may act as a biogeographic filter (Jacobs et al., 1999). Regardless of the morphological revision, our study seems to provide strong evidence for the existence of three separate lineages of A. crassicauda, which could be confirmed by further study. Generally, identification of hidden variation in scorpion species is essential, not only to revise taxonomy, but also because the many studies assessing the biochemical nature of scorpion venoms require precise species determination (Kharrat et al. 1997).
Further taxonomic and morphometric studies should be undertaken among the different populations of A. crassicauda in the Arabian Peninsula to reveal other differential characters, and perhaps describe new species.
Acknowledgements
“The authors extend their appreciation to the Deputyship for Research and Innovation, Ministry of Education in Saudi Arabia for funding this research work through the Project number (UB-46-1442). Also, many thanks to Prof. Dr. Khaled Mohammed-Geba, Department of Zoology, Faculty of Science, Menoufia University, Shebin El-Kom, Menoufia, Egypt, for reviewing this article and for providing valuable suggestions. Also, we deeply appreciate the precious aids provided Dr. Hamdy Aly, Department of Zoology, Faculty of Science, Al-Azhar University (Assiut, Egypt), for his help in the molecular study.”
Funding
“This research was funded by the Deputyship for Research and Innovation, Ministry of Education in Saudi Arabia, grant number (UB-46-1442), and the APC was funded by the same project.”
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Information theory and an extension of the maximum likelihood principle. Budapest: Akademiai Kiado; 1973. p. :267-281.
- Interspecific phylogenetic relationship among different species of the genus Buthacus (Scorpiones: Buthidae) inferred from 16S rRNA in Egypt and Saudi Arabia. Zool. Middle East. 2020;66(2):178-185.
- [Google Scholar]
- Genetic diversity among different species of the genus Leiurus (Scorpiones: Buthidae) in Saudi Arabia and the Middle East. Saudi J. Biol. Sci.. 2020;27(12):3348-3353.
- [Google Scholar]
- A contribution to the scorpion fauna of Saudi Arabia, with an identification key (Arachnida. Scorpiones). J. King Saud Univ. - Sci.. 2021;101396
- [Google Scholar]
- Scorpions and scorpion sting envenoming (scorpionism) in the Arab Countries of the Middle East. Toxicon. 2021;191:83-103.
- [Google Scholar]
- Comparison of Ribosomal ITS Regions Among Androctonus spp. Scorpions (Scorpionida: Buthidae) from Tunisia. J. Med. Entomol.. 2000;37(6):787-790.
- [Google Scholar]
- Phylogeography of Androctonus species (Scorpiones: Buthidae) in Tunisia: diagnostic characters for linking species to scorpionism. Acta Trop.. 2009;112(1):77-85.
- [Google Scholar]
- Refined electrophysiological analysis suggests that a depressant toxin is a sodium channel opener rather than a blocker. Life Sciences. 1997;61(8):819-830.
- [Google Scholar]
- Deep intraspecific divergences in the medically relevant fat-tailed scorpions (Androctonus, Scorpiones) Acta trop.. 2014;134:43-51.
- [Google Scholar]
- The scorpions of Anatolia: biogeographical patterns. Biogeographia-The Journal of Integrative Biogeography. 1999;20(1)
- [Google Scholar]
- Purification and characterization of chlorotoxin, a chloride channel ligand from the venom of the scorpion. American journal of physiology-cell physiology. 1993;264(2):C361-C369.
- [Google Scholar]
- The first molecular phylogeny of Buthidae (Scorpiones) Euscorpius. 2003;2003(4):1-10.
- [Google Scholar]
- Fet, V., Sissom, W. D., Lowe, G., Braunwalder, M. E., 2000. Catalog of the scorpions of the world (1758-1998). New York Entomological Society.
- DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology. 1994;3(5):294-299.
- [Google Scholar]
- Genetic diversity within Scorpio maurus (Scorpiones: Scorpionidae) from Morocco: Preliminary evidence based on CO1 mitochondrial DNA sequences. Biologia. 2008;63(6):1157-1160.
- [Google Scholar]
- The phylogeographic importance of the Strait of Gibraltar as a gene flow barrier in terrestrial arthropods: a case study with the scorpion Buthus occitanus as model organism. Mol. Phyl. Evolu.. 2003;28(1):119-130.
- [Google Scholar]
- Nuclear and mitochondrial markers reveal the existence of two parapatric scorpion species in the Alps: Euscorpius germanus (CL Koch, 1837) and E. alpha Caporiacco, 1950, stat. nov.(Euscorpiidae) Revue suisse de Zoologie. 2000;107(4):843-870.
- [Google Scholar]
- First DNA phylogeny of Euscorpius Thorell, 1876 (Scorpiones, Euscorpiidae) and its bearing on taxonomy and biogeography of this genus. Biogeographica (Paris). 1999;75(2):49-65.
- [Google Scholar]
- The origin of grass-dominated ecosystems. Annals of the Missouri Botanical Garden. 1999;86(2):590.
- [Google Scholar]
- Maurotoxin, a four disulfide bridge toxin from Scorpio maurus venom: purification, structure and action on potassium channels. FEBS letters. 1997;406(3):284-290.
- [Google Scholar]
- A review of Androctonus finitimus (Pocock, 1897), with description of two new species from Pakistan and India (Scorpiones, Buthidae) Euscorpius. 2013;2013(168):1-10.
- [Google Scholar]
- MEGA: molecular evolutionary genetics analysis software for microcomputers. Bioinformatics. 1994;10(2):189-191.
- [Google Scholar]
- Nouvelles considérations taxonomiques sur les espèces du genre Androctonus Ehrenberg, 1828 et description de deux nouvelles espèces (Scorpiones, Buthidae) Revue suisse de Zoologie. 2005;112(1):145-171.
- [Google Scholar]
- A new species of Androctonus Ehrenberg, 1828 from Togo (Scorpiones, Buthidae) Entomol Mitt Zool Mus Hamb. 2008;15(179):37-44.
- [Google Scholar]
- A new species of Androctonus Ehrenberg, 1828 from Afghanistan (Scorpiones, Buthidae) Zoology in the Middle East. 2006;38(1):93-97.
- [Google Scholar]
- A new species of Androctonus Ehrenberg, 1828 from Mauritania (Scorpiones, Buthidae) Bol SEA. 2007;40:215-219.
- [Google Scholar]
- Scorpions from Ennedi, Kapka and Tibesti, the mountains of Chad, with descriptions of nine new species (Scorpiones: Buthidae, Scorpionidae) Arthropoda Selecta. 2012;21(1):307338-307340.
- [Google Scholar]
- More about the genus Androctonus Ehrenberg, 1828 (Scorpiones, Buthidae), with the description of a new species from Ethiopia. Arachnida. 2015;5:11-29.
- [Google Scholar]
- A new species of Androctonus Ehrenberg, 1828 from Morocco (Scorpiones: Buthidae) Euscorpius. 2009;2009(89):1-8.
- [Google Scholar]
- Nylander J. A. A., 2004. MrModeltest. Program distributed by the Author. Uppsala: Uppsala University, Evolutionary Biology Centre.
- A study on the genetic diversity of Androctonus crassicauda (Olivier, 1807; Scorpiones: Buthidae) from Turkey. Journal of Venomous Animals and Toxins including Tropical Diseases. 2010;16(4):599-606.
- [Google Scholar]
- Systematics and biogeography of the family Scorpionidae Latreille, with a discussion of phylogenetic methods. Invertebr. Syst.. 2003;17:185-259.
- [Google Scholar]
- Rambaut, A, Drummond, A. J., 2007. Tracer v1. 4: MCMC trace analyses tool. http://tree.bio.ed.ac.uk/software/tracer.
- J.O. Rein Scorpion files. Species List. Available from 2021.
- MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Systematic Biology. 2012;61(3):539-542.
- [Google Scholar]
- Tre nuove specie di importanza medica del genere Androctonus Ehrenberg, 1828 (Scorpiones: Buthidae) Aracnida - Rivista Arachnologica Italiana. 2015 5(Supplemento):2–20
- [Google Scholar]
- Genetic diversity within Leiurus quinquestriatus (Scorpiones: Buthidae) populations in Egypt as inferred from 16S mDNA sequence analysis. Zool. Middle East. 2020;66(3):269-276.
- [Google Scholar]
- Diversity patterns and evolutionary history of Arabian squamates. J. Biogeogr.. 2021;48(5):1183-1199.
- [Google Scholar]
- Genetic diversity of Maghrebian Hottentotta (Scorpiones: Buthidae) scorpions based on CO1: new insights on the genus phylogeny and distribution. African Invertebrates. 2011;52(1):135-143.
- [Google Scholar]
- PAUP*. Phylogenetic Analyses Using Parsimony (and Other Methods), Version 4.06b. Sunderland (Massachusetts: Sinauer Associates; 2001.
- Phylogenetic inference. In: Hillis D.M., Moritz C., Mable B.K., eds. Molecular systematics. Sunderland (Massachusetts): Sinauer Associates; 1996. p. :407-510.
- [Google Scholar]
- Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular biology and evolution. 1993;10(3):512-526.
- [Google Scholar]
- A new species of Androctonus Ehrenberg, 1828 from northwestern Egypt (Scorpiones: Buthidae). Euscorpius. 2013;177:1-12.
- [Google Scholar]
- Mitochondrial cytochrome oxidase subunit I-sequence variation in three scorpion species from southeast Turkey. Indian Journal of Animal Research. 2019;53(2):187-191.
- [Google Scholar]
- Etudes sur les scorpions. Archives de l'Institut Pasteur d'Algerie. Institut Pasteur d'Algerie. 1948;26(1):25-29.
- [Google Scholar]
- Vachon, M., 1952. Etude sur les Scorpions Alger: Institut Pasteur d’Algérie, 482 pp. Vachon, M. 1973 [1974]. Etude des caracteres utilisés pour classer les familles et les genres de scorpions (Arachnides). 1. La trichobothriotaxie en arachnologie: Sigles trichobothriaux et types de trichobothriotaxie chez les scorpions. Bulletin du Muséum National d’Histoire Naturelle Ser 3(140), 857-958.
- Scorpion preservation for taxonomic and morphological studies. Wasmann J. Biol.. 1968;26:133-136.
- [Google Scholar]
- Androctonus turkiyensis sp. n. from the Şanlıurfa Province. Turkey (Scorpiones: Buthidae). Euscorpius. 2021;No. 341
- [Google Scholar]
- Insect Specific Neurotoxins from Scorpion Venom that Affect Sodium Current Inactivation. Journal of Toxicology: Toxin Reviews. 1994;13(1):25-43.
- [Google Scholar]







