Translate this page into:
Interferon-stimulated gene (ISG12a) suppresses hepatitis B virus replication in Huh 7 cells line
⁎Corresponding authors. muhdkhurshid79@gmail.com (Khurshid Alam)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Hepatitis B virus (HBV) and its associated chronic liver disease pose a significant health hazard. Chronic HBV infection is a major contributor to the development of liver cirrhosis, liver fibrosis, and liver cancer. This study focused on ISG12a and its inhibitory effect in Huh7 cells on HBV gene expression and replication. The mammalian hepatoma cells Huh7 were transfected with the pHBV1.3 vector (pCAGGS) or HA-tagged ISG12a to determine the overexpression, to test this hypothesis further, we did knockdown or silencing by transducing Huh7 cells with lentiviruses containing shRNAs targeting ISG12a or scramble control shRNA (shControl). The expression of ISG12a was evaluated through western blot analysis. The HBsAg and HBeAg secretion in the Huh7 cells’ culture media was examined using an ELISA test. The qRT-PCR confirmed the mRNA and 3.5 mRNA Kb of ISG12a. The HBV total RNA was extracted and evaluated through a Northern blot. The isolated DNA was detected through qPCR. Additionally, a mechanistic study of enhancer II (EnhII/Cp) was examined through luciferase reporter testing. Our research findings indicate that ISG12a, an interferon-stimulated gene (ISG), plays a crucial role in suppressing the replication and gene expression of HBV. According to our study using the Huh7 cell system, overexpression of ISG12a resulted in a notable reduction in HBV protein levels as well as intracellular core-associated DNA and RNA levels. On the other hand, silencing ISG12a in Huh7 cells resulted in increased HBV RNA transcripts, DNA, and secreted proteins, indicating that ISG12a plays a role in suppressing HBV replication. Additionally, the research revealed that ISG12a inhibits the function of the EnhII/Cp promoter, which results in reduced HBV gene expression. The EnhII/Cp promoter is involved in regulating HBV gene expression, and the study showed that ISG12a restricts its activity. ISG12a may have a regulatory role in controlling the expression of HBV genes. These findings highlight the importance of ISG12a in HBV gene expression and provide valuable insights for understanding its antiviral function.
Keywords
Hepatitis B virus
ISG12a
Huh 7 cells line
DNA replication
Gene silencing
1 Introduction
The hepatitis B virus (HBV) poses a significant health challenge as it leads to chronic hepatitis B and contributes to a substantial global burden of mortality and morbidity. Despite the nearly 30-year HBV vaccination program, there are still 2 billion HBV-infected individuals worldwide, and these individuals continue to have a high risk of developing cirrhosis of the liver and hepatocellular carcinoma (HCC) (El–Serag et al.,2007; Levrero and Zucman-Rossi, 2016). Interferons and nucleos(t)ide analogues are two of the main therapy modalities. Regarding the complete elimination of viruses, these agents have thus far yielded poor results. Because of their potential negative effects, interferons cannot be utilized as a long-term therapy. In addition to the virus developing greater resistance, prolonged treatment with nucleos(t)ide analogues has also been shown to have substantial adverse effects. Global research institutions and the pharmaceutical sector have recognized the need for novel treatments for HBV (Manzoor et al., 2015).
HBV is a member of the Hepadnaviridae family and is a 3.4 Kb double-stranded, enveloped DNA virus (Bouchard and Navas-Martin, 2011). HBV targets and invades hepatocytes after infection, whereupon it attaches to the sodium taurocholate co-transporting polypeptide (NTCP) surface receptor (Yan et al., 2012). The HBV relaxed circular DNA (rcDNA) is repaired and transformed into covalently closed circular DNA (cccDNA) upon entry of the HBV genome into the cell nucleus. The cccDNA acts as a template for the transcription of various mRNAs of a virus, including the pregenomic (pg), precore RNA, X (HBx protein), PreS1, and PreS2/S. These mRNAs encoded precore, core, large (L), middle (M), and small (S) surface and polymerase (pol) proteins. Reverse transcription of viral DNA occurs while pgRNA and polymerase are encapsulated into the capsid, and finally, the hepatocytes release the HBV virions. (Kim et al., 2016). Four HBV-specific promoters (core, preS1, preS2, and X), two enhancer elements (EnhI and EnhII), and a negative regulatory cis-acting element control the transcriptional activity of HBV RNAs (Nishitsuji et al., 2018). The nuclear factors of the host and cytokines are essential for controlling the transcriptional activity of the HBV promoters, comprising signal transducer and activator of transcription (STAT1), retinoid X receptor (RXR), and hepatocyte nuclear factor 4 (HNF-4) (Waris and Siddiqui, 2002).
Type I interferon (Type I IFN) triggers a variety of proteins that prevent virus replication and restrict viral propagation from cell to cell (Boasso, 2013; Stetson and Medzhitov, 2006). It is a primordial cytokine, has an essential role in the immunity of the host, and provides a first-line innate defense against viruses and virus-related pathogens. IFNs also have the property of playing a role as anti-proliferative and immunomodulatory. IFN-α (which has 13 human subtypes), IFN-β, IFN-ω, IFN-κ, and IFN-ε all belong to type I IFNs, and among these, IFN-α and IFN-β have a significant role in treating chronic infections of hepatitis B and C. After virus infection, the type I IFN receptors, i.e., Interferon alpha/beta receptor 1 (IFNAR1) and Interferon alpha/beta Receptor Subunit 2 (IFNRA2), bind to the virus, activating the Janus kinase (JAK)-signal transducer and activator of transcription (STAT) signaling cascade, which causes 300 interferon-stimulated genes to be expressed in the host. These ISGs have antiviral properties, and they can block various viruses (Waris and Siddiqui, 2002; van Pesch et al., 2004). These ISGs have antiviral activity, including hindering different phases of viral replication and transcription (Hayes and Chayama, 2017; Ivashkiv and Donlin, 2014). ISG-based antiviral approaches signify exciting novel therapeutic targets. Indeed, current investigation efforts have extended our information considerably. Upcoming approaches are expected to demonstrate the importance of this new understanding of clinical trials and the urgent need to develop novel drugs against prevailing and emergent viruses (Raftery and Stevenson, 2017).
The interferon-stimulated gene 12a (ISG12a) is a hydrophobic protein of mitochondria that has 122 amino acids and is located in the q32 band of human chromosome 14. IFI6, ISG12a, ISG12b, and ISG12c are the four FAM14 genes found in humans, while ISG12a, ISG12b1, and ISG12b2 are found in mice. Type I IFN can significantly induce the human ISG12a gene (Cheriyath et al., 2011). Our previous studies demonstrated that ISG12a (IFI27) has an antiviral effect in human hepatoma cells (HepG2) on HBV replication and transcription (Rasmussen et al., 1993). Our other study demonstrated that IFI6, a member of the FAM14 family, limits the expression of the HBV gene both in vitro and in vivo (El-Serag and Rudolph, 2007). This proposes that ISG12a might play an important role as an antiviral in innate immunity and may offer important clues for exploring the mechanism of suppression of innate immunity against HBV infection (Parker and Porter, 2004).
The current study focused on ISG12a and its inhibitory effect in Huh7 cells on HBV replication. In Huh7 cells, overexpression inhibits and knockdown enhances HBV replication and transcription. Our research also revealed that ISG20a functions as a potential repressor by binding to the HBV EnhII/Cp region, suggesting that it has anti-HBV activity. Importantly, we further confirmed the effect of ISG12a on HBV replication in vivo. These data suggest that ISG12a could be an anti-HBV target and assess the possible therapeutic application of anti-HBV therapy.
2 Materials and methods
2.1 Cell culture and transfection
The Huh7 cells were derived from a human hepatoma cell line. Dulbecco’s modified Eagle's medium (DMEM) supplemented with 10 % fetal bovine serum was used for the growth of cells in an environment containing 5 % CO2 at 37 °C. According to the manufacturer's instructions, Lipofectamine 3000 (Invitrogen) was used to transfect the cells.
2.2 Plasmid constructs
In our previous work, we reported that ISG12a was cloned into pCAGGS at the HindIII and EcoRI sites with an HA-tag at the C terminus (Sajid et al., 2021). Our lab provided the pHBV1.3-expressing plasmid (HBV genotype D) and reporter plasmids. In pLKO-1-puro, an ISG12a short hairpin RNA (shRNA) and a non-targeting shRNA (shControl) were introduced. The shRNAs' intended targets are: ShControl: 5ʹ-GCAGAAGAACGGCATCAAG-3ʹ and ShISG12a: 5ʹ-CTCCGGATTGACCAAGTTCAT-3ʹ.
2.3 Western blotting
The western blot procedure was reported previously [19]. The antibodies used in this study involved rabbit anti-ISG12a (Catalog no. orb337785, Biorbyt) and β-actin (ABclonal, United States).
2.4 Detection of HBsAg and HBeAg
The HBV antigens, HBsAg and HBeAg, were extracted from the culture supernatant for analysis and determination by ELISA (Kehua Shengwu, China).
2.5 Reporter assays
The pRL-TK (50 ng) and promoter-luciferase reporter plasmids (450 ng each) were co-transfected in Huh7 cells. By using the Dual-Glo System (Promega) for luciferase activity assays to assess luciferase activity.
2.6 Quantitative PCR (RT-qPCR) for HBV RNA
Following the manufacturer's instructions, total RNA from Huh7 cells was extracted using the TRIzol reagent (Invitrogen, USA). To perform reverse transcription (RT-PCR) for cDNA from total RNA (Takara), the PrimeScript RT reagent kit was utilized. To amplify the cDNA through qRT-PCR, the SYBR Green Fast qPCR Master Mix was used (Yeasen Biotechnology, Shanghai). Each sample's threshold cycle (Ct) value was determined, and the relative mRNA level was then normalized to the GAPDH mRNA value. The primer sequences are listed in Table 1:
Primer
Sequence
Base pair
ISG12a RT-F
5′-GCCTCTGCTCTCACCTCATC-3′.
20
ISG12a RT-R
5′-ATCTTGGCTGCTATGGAGGA-3′.
20
3.5 kb mRNA RT-F
5′-GCCTTAGAGTCTCCTGAGCA-3
20
3.5 kb mRNA RT-R
5′-GAGGGAGTTCTTCTTCTAGG-3′.
20
GAPDH RT-F
5′-CCACTCCTCCACCTTTGAC-3
19
GAPDH RT-R
5′-ACCCTGTTGCTGTAGCCA-3′.
18
2.7 Northern blotting
The RNA sample used in the northern blotting procedure was resolved on 1.5 % MOPS agarose gels containing 2.2 M formaldehyde and then shifted to nylon membranes. The RNA was then hybridized with a DNA fragment that was DIG-labeled and included the entire HBV sequence. Using a PCR DIG probe synthesis kit, the probe was created (Roche, Germany). To determine the amount of total RNA, the 28 s and 18 s (rRNAs) are employed as internal controls.
2.8 HBV DNA core-associated analysis
A previously published technique was modified for the extraction of core-related HBV DNA (Jing et al., 2016). The real-time PCR was utilized to further perceive HBV DNA by using the primers; RCCCS: 5′-CTCGTGGTGGACTTCTCTC-3′ and RCCCAS: 5′- CTGCAGGATGAAGAGGAA −3′.
3 Results
3.1 ISG12a repress HBV proteins in Huh7 cells
The antiviral activity of ISG12a in HepG2 cells in terms of HBV gene expression was reported in our previous work (Sajid et al., 2021). In the current research, we explored the restriction effect of ISG12a in Huh 7 cells in regulating HBV replication. Here, we first examined the role of ISG12a on HBV protein suppression. We transfected Huh7 cells with the pHBV1.3 vector (pCAGGS) or HA-tagged ISG12a. Western blot results confirmed the expression of the ISG12a protein (Fig. 1A). As demonstrated by western blot, we found that core proteins, or HBcAg, were significantly reduced in transfected Huh7 cells (Fig. 1B). The HBeAg and HBsAg secretion in the Huh7 cells' culture media was examined using an ELISA test (Fig. 1C, D). The presence of HBsAg and HBeAg was significantly decreased in the cell culture medium, as was predicted, demonstrating the inhibitory impact of ISG12a. According to these findings, ISG12a specifically inhibits HBV proteins in Huh7 cells.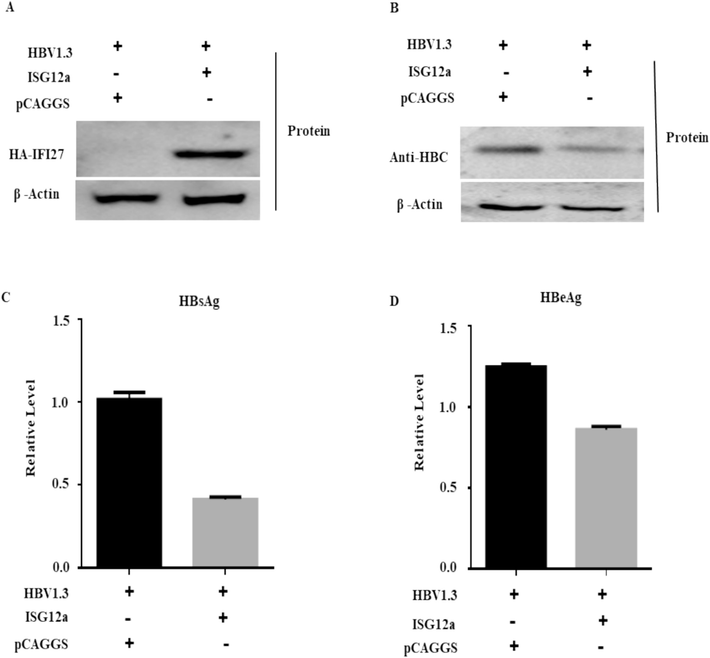
ISG12a represses HBV proteins in Huh7 cells. (A-D) Huh7 cells were co-transfected with 0.4 μg of pHBV1.3 plasmid and 0.4 μg pCAGGS-HA-Anti-ISG12a and/or pCAGGS (empty vector). The ISG12a (A) and HBcAg (B) protein expression levels were examined through western blotting. The expression of β-actin or pSV-β-Galactosidase acts as a loading control. Anti-HA and anti-HBcAg antibodies were used to detect the proteins in these experiments. (C, D) The secreted HBsAg (C) and HBeAg (D) expressions were observed through ELISA.
3.2 ISG12a suppress HBV RNA and DNA in Huh7 cells
The impact of ISG12a on intracellular HBV RNA and DNA levels was then examined in Huh7 cells. In Huh7 cells, the control vector (pCAGGS) or HA-tagged ISG12a was transiently transfected with pHBV1.3. To verify whether ISG12a was specifically upregulated by HBV replication, the mRNA of ISG12a mRNAs was confirmed by qRT-PCR (Fig. 2A).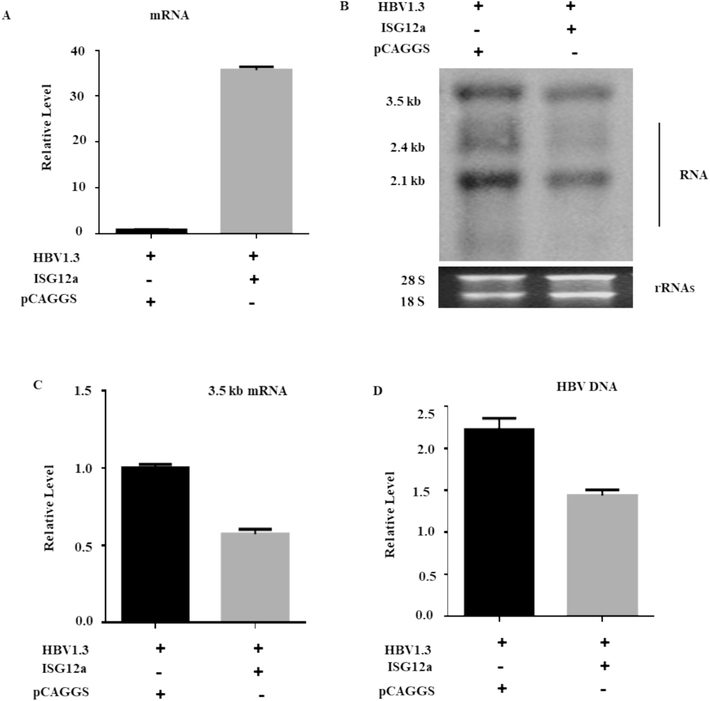
ISG12a suppresses HBV RNA and DNA in Huh7 cells. (A, B, C) Huh7 cells were co-transfected with 0.4 μg of pHBV1.3 plasmid and 0.4 μg pCAGGS-HA-Anti-ISG12a and/or pCAGGS (empty vector). The HBV total RNA was separated from Huh7 cells, which were transfected with pHBV1.3 plasmids, pSV-β-gal, along with pCAGGS-HA-ISG12a or pCAGGS (empty vector). (A) qRT-PCR was used to assess ISG12a mRNA expression. (B) A Northern blot was used to determine the total RNAs, with 28S and 18S (rRNAs) serving as loading controls. (C) The qRT-PCR was utilized to assess the intracellular 3.2 kb mRNA of HBV, where the GAPDH mRNA level of expression acted as a reference control. (D) The pCAGGS-ISG12a or pCAGGS (empty vector) were transfected into Huh 7 cells together with the pHBV1.3 plasmid vector. The intracellular core-associated HBV DNA was extracted and then measured by qPCR.
A northern blot was used to find a reduction in the three long HBV RNA transcripts, i.e., 3.5 kb, 2.4 kb, and 2.1 kb, which showed that ISG12a overexpression significantly reduced HBV RNA levels (Fig. 2B). 3.5-kb mRNA consists of two precore (pcRNA) and pregenomic (pgRNA) mRNAs. To confirm the effect of ISG12a on HBV RNA, we further examined the 3.2 kb mRNA expression. The qRT-PCR results demonstrated that ISG12a strongly reduced the amounts of 3.5 kb mRNA in Huh 7 cells (Fig. 2C). We also checked the intracellular core-associated HBV DNA in transfected cells. The isolated DNA was detected through qPCR (Fig. 2D). The HBV intracellular core-associated activity was significantly inhibited upon overexpression of ISG12a. Therefore, our data recommended that ISG12a could have a role in inhibiting HBV gene expression.
3.3 Silencing of ISG12a enhanced HBV protein in Huh 7 cells
The results described above suggested that ISG12a might hinder the replication of HBV. We transduced Huh7 cells with lentiviruses containing shRNAs targeting ISG12a or scramble control shRNA (shControl) to test this hypothesis further. The shRNA showed an efficient knockdown of ISG12a compared to shControl in transduced cells (Fig. 3A). The effect of ISG12a silencing on HBV proteins was first examined in Huh7 cells, and the downregulation of ISG12a resulted in increased levels of HBcAg antigen, as evidenced by western blot (Fig. 3B). ELISA was used to further assess the HBsAg and HBeAg expression in the supernatant of cell culture. (Fig. 3C, D). The result showed that secreted HbcAg, HBsAg, and HBeAg were potently increased. Together, we recommended that the ISG12a silencing strongly increase HBV gene expression and replication in Huh7 cells.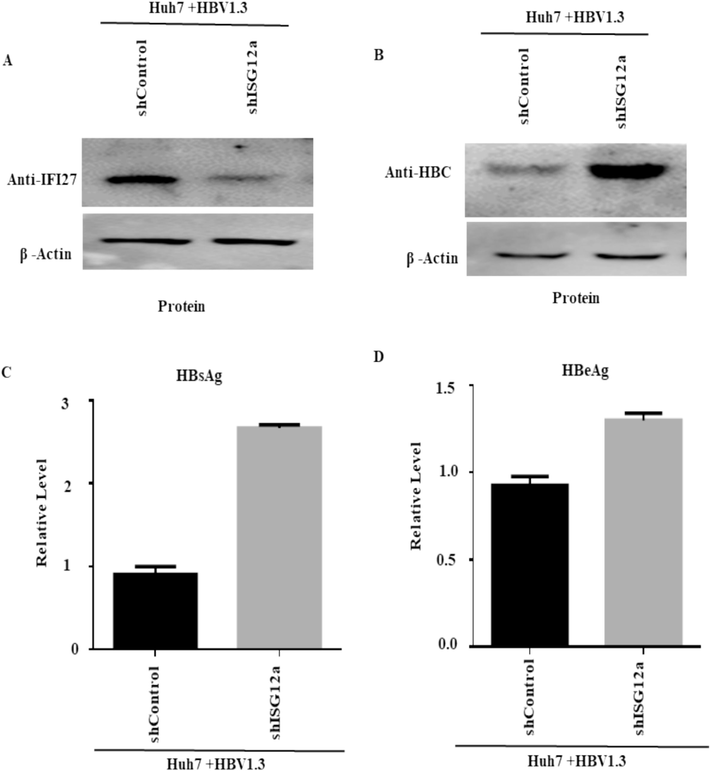
Silencing of ISG12a-enhanced HBV protein in Huh 7 cells: (A-D) Huh7 cells were transfected with plasmids containing 0.8 µg of pHBV1.3 and 0.2 µg of pSV-β-gal. After 24 h, the cells were transduced with lentiviruses expressing shRNAs to target ISG12a (shISG12a) or a control shRNA that was scrambled (shcontrol). Seven cells were collected three days after being transduced by the lentivirus. (A) ISG12a expression and (B) HBcAg levels of protein were detected using western blotting. β-actin acts as a loading control, and ISG12a-specific antibodies were applied to detect these proteins. (C, D) ELISA determined the secreted HBsAg (C) and HBeAg (D) proteins.
3.4 Silencing of ISG12a increased HBV RNA and DNA in Huh7 cells
To provide additional evidence of ISG12a's function in HBV gene expression and replication, we first check the silencing of ISG12a in HBV-transfected and shRNA-lentiviral-transduced Huh7 cells. The ISG12a mRNAs were specifically upregulated by HBV replication and were confirmed by qRT-PCR (Fig. 4A). The downregulation of ISG12a activity and HBV RNA transcripts was considerably increased at the steady-state level (Fig. 4B). Here, we also observed the inhibitory effect of ISG12a on level 3.5 kb mRNA determined by qRT-PCR (Fig. 4C).According to the qRT-PCR results, the HBV 3.5 kb mRNA level increased when ISG12a was silenced. Concordantly, gene silencing of ISG12a significantly augmented HBV intracellular core-associated DNA, as determined by qPCR (Fig. 4D). Overall, we find that knockdown or silencing of pgRNA significantly increases HBV transcription.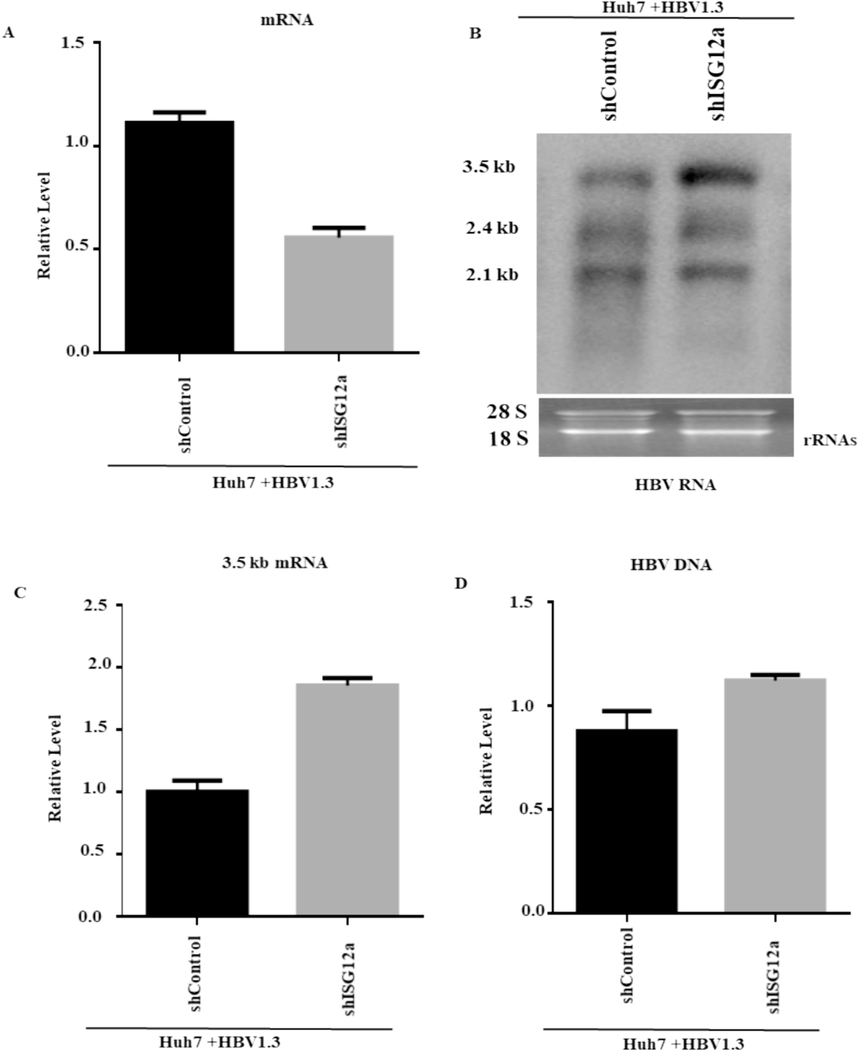
Silencing of ISG12a increased HBV RNA and DNA in Huh7 cells. (A-D) carefully transfected Huh7 cells with 0.8 µg of pHBV1.3 and 0.2 µg of pSV-β-gal. After 24 h, the cells were transduced with lentiviruses producing ISG12a-targeting shRNAs (shISG12a-4 and shISG12a-5), or shControl. The total RNA of HBV was isolated from transduced Huh7 cells. (A) ISG12a mRNA levels were analyzed by qRT-PCR, where GPADH mRNA levels of expression were used for data normalization. (B) Transcripts of HBV, i.e., 3.5 kb, 2.4 kb, and 2.1 kb, were detected through the northern blot. The 28S and 18S rRNAs were utilized as loading controls. (C) The pgRNA was validated by qRT-PCR, and the GAPDH mRNA level of expression acted as a reference control. (D) The HBV intracellular core-associated DNA were first extracted and then detected 4 days post-transduction by q-PCR.
3.5 ISG12a attenuating the activity of EnhII/Cp promoter
To elucidate the mechanism by which ISG12a affects HBV, we used a luciferase reporter experiment to analyze how ISG12a affected the activity of the SP1, SP2, EnhI/Xp, and EnhII/Cp promoters of HBV. The reporter plasmids pGL3-EnhI/Xp, pGL3-EnhII/Cp, pGL3-SP1, and pGL3-SP2 were co-transfected into Huh7 cells along with pCAGGS-ISG12a or pCAGGS. The ISG12a suppressed EnhII/Cp activity (Fig. 5A) but did not influence the activities of EnhI/Xp, SP1, or SP2 promoters, according to the results of the luciferase-based reporter study. Further, a mechanistic study of EnhII/CP will be investigated in the future. The results validate that ISG12a hinders HBV replication by suppressing the viral activity of EnhII/Cp.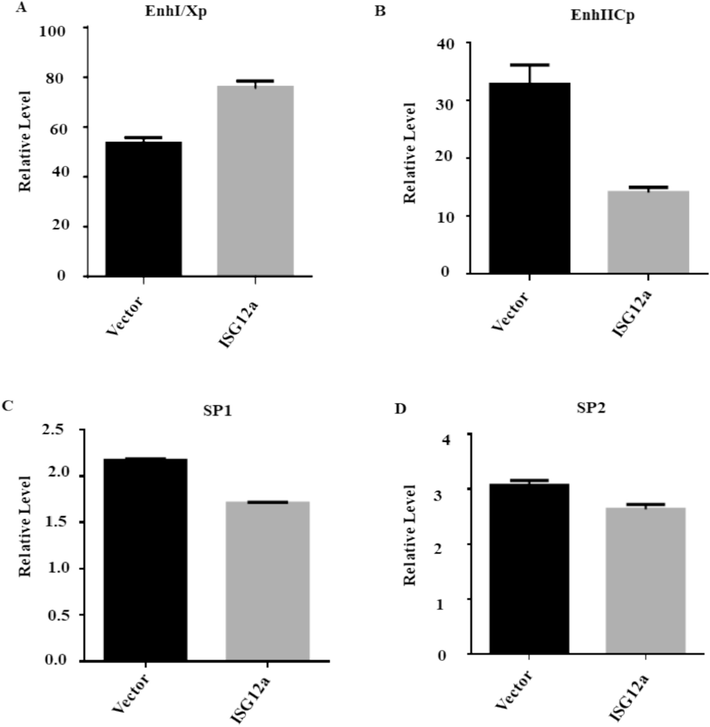
ISG12a attenuating activity of the EnhII/Cp promoter: (A-D) Huh7 cells were transfected with 200 ng of each reporter plasmid together with 250 ng of the ISG12a expression plasmid, or pCAGGS, and Luciferase activity was then measured, and the transfection efficiency was checked using 50 ng of pRL-TK.
4 Discussion
Interferon (IFN) comprises a set of cytokines with antiviral activity and immunoregulatory and antitumor effects that play key roles in both innate and adaptive immune responses (Jianyu and Jieliang et al., 2021). Even though various antiviral ISGs have been discovered using various technical methods (Metz et al., 2012; Schoggins et al., 2011). The production of IFN-α and ISGs is essential for cells to defend against pathogens or viruses. The expression of several effectors and ISGs eventually controls the host's response to infectious viruses. Novel studies have led us to understand the anti-viral mechanisms of innate immunity. The majority of ISGs' roles and molecular processes in inhibiting viral infection are poorly known (Lau et al., 2005). It was confirmed that the biological activity of the ISG12a protein has only been examined in a few studies. To date, pegylated IFN-α has been the main component of the sole immunotherapy for the treatment of chronic HBV. However, only 20–40 % of HBeAg seroconversion occurs when pegylated IFN-a is used for the long-term therapy of HBV [24]. Around 300 IFN-stimulated genes (ISGs) play a major role in mediating the intracellular activity of IFN-α (Dunn et al., 2009). Targeting several phases of the viral life cycle, ISGs also exhibit a strong antiviral effect. Numerous of these ISGs prevent the hepatitis C virus from replicating (Itsui et al., 2006). However, some ISGs, like USP 18 and ISG 15, increase HCV replication (Randall et al., 2006).
In previous research, the high basal ISG12a or IFI27 can prevent the Newcastle disease virus's (NDV) oncolysis and replication. ISG20 directly binds to the epsilon stem-loop region of pgRNA to stop the expression of the HBV gene. It has non-apoptotic antiviral activity through a proteasome-dependent pathway to target viral protein (NS5A) in cells infected by HCV. Murine ISG12a showed a restrictive effect against the Murine hepatitis virus (MHV) and West Nile virus (WNV) with unidentified mechanisms. Our previous studies demonstrated that ISG12a or IFI27 is greatly expressed in HepG2 upon IFN-α induction, and IFN-α-induced ISG12a is liable for IFN-α-mediated inhibition of HBV both in vitro and in vivo. With this essential viral activity of ISG12a, we provide a piece of crucial evidence for exploring the restrictive effect of ISG12a on HBV in the Huh 7 cell line. In our study, we evaluated ISG12a in the context of the HBV immune response in Huh-7 cells. In these huh7 cells, the overexpression of ISG12a decreased the transcript and protein levels of the HBV, whereas HBV gene expression and replication activity were both boosted by ISG12a depletion. The previously demonstrated article revealed that ISG20 has an antiviral effect on HBV. An ISG20, after binding to its EnhII/Cp portion of HBV, controls the HBV at the level of transcription (Parker et al., 2004). Myxovirus resistance protein 1 (MxA) interacts with the core protein of HBV, and it interferes with its capsid assembly (Li et al., 2012). ISG TRIM22 has been reported to effectively inhibit HBV and EnhII/Cp promoter activities that are associated with HBV replication (Qu et al., 2014). After validation of the previous research, we also demonstrated that ISG12a suppressed the activity of the EnhII/Cp promoter. A detailed investigation of ISG12a in Huh 7 cells will determine how ISG12a-mediated EnhII/Cp promoter inhibition is involved.
The present research determined that ISG12a has an anti-HBV effect on HBV gene expression and replication in vitro. Our research discloses that HBV replication is inhibited due to ISG12a and EnhII/Cp region interactions. Taken together, our study demonstrated an innovative mechanism of anti-HBV regulation by ISG12a, which will lead to the improvement of possible new therapeutics for the proficient treatment of HBV infection.
5 Conclusions
The study concluded that ISG12 in Huh7 has an antiviral impact on HBV gene expression and replication. It reveals that the overexpression and silencing of ISG12 augment HBV replication and transcription in Huh7 cells. In addition, by binding to the HBV EnhII/Cp region, ISG12a performs a putative repressor function, from which we assumed that it has anti-HBV activity. In the future, this knowledge could potentially contribute to the development of medications or therapeutic strategies to treat HBV infection.
CRediT authorship contribution statement
Hafiz Ullah: Methodology, Software, Writing – original draft. Amin Ullah: Conceptualization, Supervision, Writing – review & editing. Hadia Gul: Methodology, Validation. Rahat Ullah Khan: Investigation. Junaid Ahmad: Conceptualization, Formal analysis, Writing – review & editing. Rafa Almeer: Formal analysis, Funding acquisition, Software, Writing – review & editing. Khurshid Alam: Conceptualization, Methodology, Supervision, Writing – review & editing. Muhammad Ayaz: Conceptualization, Investigation, Writing – review & editing. Muhammad Ajmal Khan: Conceptualization, Investigation, Writing – review & editing. Zafar Abbass Shah: Investigation, Visualization.
Acknowledgements
The authors would like to extend their sincere appreciation to the Researchers Supporting Project Number (RSP2024R96), King Saud University, Riyadh, Saudi Arabia. This research work was also supported by State Key Laboratory of Virology, Modern Virology Research Center, College of Life Sciences, Wuhan University, Wuhan, China.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Type I interferon at the interface of antiviral immunity and immune regulation: the curious case of HIV-1. Scientifica. 2013;2013
- [CrossRef] [Google Scholar]
- Hepatitis B and C virus hepatocarcinogenesis: lessons learned and future challenges. Cancer Lett.. 2011;305
- [CrossRef] [Google Scholar]
- Emerging roles of FAM14 family members (G1P3/ISG 6–16 and ISG12/IFI27) in innate immunity and cancer. J. Interferon Cytokine Res.. 2011;31
- [CrossRef] [Google Scholar]
- Temporal analysis of early immune responses in patients with acute hepatitis B virus infection. Gastroenterol.. 2009;137(4):1289-1300.
- [CrossRef] [Google Scholar]
- Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. J. Gastroenterol. 2007;132
- [CrossRef] [Google Scholar]
- Interferon stimulated genes and innate immune activation following infection with hepatitis B and C viruses. J. Med. Virol.. 2017;89
- [CrossRef] [Google Scholar]
- Expressional screening of interferon-stimulated genes for antiviral activity against hepatitis C virus replication. J. Viral Hepat.. 2006;13(10):690-700.
- [CrossRef] [Google Scholar]
- Roles of hepatocyte nuclear factors in hepatitis B virus infection. World J. Gastroenterol.. 2016;22
- [CrossRef] [Google Scholar]
- Peginterferon Alfa-2a, lamivudine, and the combination for HBeAg-positive chronic hepatitis B. N. Engl. J. Med.. 2005;352(26):2682-2695.
- [CrossRef] [Google Scholar]
- Mechanisms of HBV-induced hepatocellular carcinoma. J. Hepatol.. 2016;64
- [CrossRef] [Google Scholar]
- MxA inhibits hepatitis B virus replication by interaction with hepatitis B core antigen. Hepatol.. 2012;56(3)
- [CrossRef] [Google Scholar]
- Hepatitis B virus therapy: What’s the future holding for us? World J. Gastroenterol.. 2015;21(44):12558.
- [Google Scholar]
- Identification of type I and type II interferon-induced effectors controlling hepatitis C virus replication. Hepatol.. 2012;56(6)
- [CrossRef] [Google Scholar]
- TIP60 complex inhibits hepatitis B virus transcription. Journal of Virology. 2018;92(6)
- [Google Scholar]
- Identification of a novel gene family that includes the interferon-inducible human genes 6–16 and ISG12. BMC Genom.. 2004;5
- [CrossRef] [Google Scholar]
- Qu, C., Chen, T., Fan, C., Zhan, Q., Wang, Y., Lu, J., Lu, L. ling, Ni, Z., Huang, F., Yao, H., Zhu, J., Fan, J., Zhu, Y., Wu, Z., Liu, G., Gao, W., Zang, M., Wang, D., Dai, M., Sun, Z., 2014. Efficacy of Neonatal HBV Vaccination on Liver Cancer and Other Liver Diseases over 30-Year Follow-up of the Qidong Hepatitis B Intervention Study: A Cluster Randomized Controlled Trial. PLoS Med., 11(12). DOI: 10.1371/journal.pmed.1001774.
- Advances in anti-viral immune defence: revealing the importance of the IFN JAK/STAT pathway. Cell. Mol. Life Sci.. 2017;74:2525-2535.
- [Google Scholar]
- Silencing of USP18 potentiates the antiviral activity of interferon against hepatitis C virus infection. Gastroenterol.. 2006;131(5):1584-1591.
- [CrossRef] [Google Scholar]
- Identification of a New Interferon-α-inducible Gene (p27) on Human Chromosome 14q32 and Its Expression in Breast Carcinoma. Cancer Res.. 1993;53(17)
- [Google Scholar]
- The Functional and Antiviral Activity of Interferon Alpha-Inducible IFI6 Against Hepatitis B Virus Replication and Gene Expression. Front. Immunol.. 2021;12
- [CrossRef] [Google Scholar]
- A diverse range of gene products are effectors of the type i interferon antiviral response. Nature. 2011;472(7344)
- [CrossRef] [Google Scholar]
- Characterization of the Murine Alpha Interferon Gene Family. J. Virol.. 2004;78(15)
- [CrossRef] [Google Scholar]
- Interaction between STAT-3 and HNF-3 leads to the activation of liver-specific hepatitis B virus enhancer 1 function. J. Virol.. 2002;76(6):2721-2729.
- [CrossRef] [Google Scholar]
- Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;2012(1)
- [CrossRef] [Google Scholar]







