Translate this page into:
In silico prediction and experimental evaluation of potential siRNAs against SARS-CoV-2 inhibition in Vero E6 cells
⁎Corresponding author at: King Fahd Medical Research Center, King Abdulaziz University, Post Box No-80216, Jeddah 21589, Saudi Arabia. ssohrab@kau.edu.sa (Sayed Sartaj Sohrab)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Objective
The acute cases of pneumonia (COVID-19) were first reported from China in December 2019, and the pathogen was identified as SARS-CoV-2. Currently, many vaccines have been developed against this virus by using multiple genes, applying different platforms, and used for the vaccinations of the human population. Spike protein genes play an important role in host cell attachment and viral entry and have been extensively used for the development of vaccine and antiviral therapeutics. Short interfering RNA is also known as silencing RNA and contribute a significant role to regulate the expression of a specific gene. By using this technology, virus inhibition has been demonstrated against many viral diseases.
Methods
In this work, we have reported the Insilico prediction, designing, and experimental validation of siRNAs antiviral potency against SARS-CoV-2-S-RBD. The siDirect 2.0 was selected for siRNAs prediction, and secondary structure was predicted by RNAfold while the HNADOCK was used for molecular docking analysis and specific binding of siRNAs to the selected target. We have used and evaluated four siRNAs for cellular toxicity and determination of antiviral efficiency based on the Ct value of q-real-time PCR in Vero E6 cells.
Results
Based on the experimental evaluation and analysis of results from generated data, we observed that there is no cytotoxicity for any tested siRNAs in Vero E6 cells. Total four siRNA were filtered out from twenty-one siRNAs following the strict selection and scoring criteria. The better antiviral efficiency was observed in 3rd siRNAs based on the Ct value of q-real-time PCR. The results that emerged from this study encouraged us to validate the efficiency of these siRNAs in multiple cells by using alone and in a combination of two or more siRNAs to inhibit the SARS-CoV-2 proliferation.
Conclusion
The Insilico prediction, molecular docking analysis provided the selection of better siRNAs. Based on the experimental evaluation only 3rd siRNA was found to be more effective than others and showed better antiviral efficiency. These siRNAs should also be evaluated in other cell lines either separately or in combination against SARS-CoV-2 to determine their antiviral efficiency.
Keywords
siRNAs
Insilico prediction
Molecular docking
SARS-CoV-2
Vero E6 cells
Saudi Arabia
- ARDS
-
Acute respiratory distress syndrome
- COVID-19
-
The new Coronavirus Disease 2019
- MFE
-
Minimum free energy
- PCR
-
Polymerase Chain Reaction
- RBD
-
receptor binding domain
- RNAi
-
RNA interference
- SARS-CoV-2
-
Severe Acute Respiratory Syndrome Coronavirus-2
- siRNA
-
Short interfering RNA
- USFDA
-
United States Food and Drug Authority
- WHO
-
World Health Organization
Abbreviations
1 Introduction
An outbreak of unusual pneumonia was observed in hospitalized patients from Wuhan, China caused by a novel coronavirus in late December 2019. The clinical symptoms varied from asymptomatic infection to severe acute respiratory syndrome (SARS). Currently, this virus has spread to 222 countries and territories with 270,440,970 cases, 5,322,447 and 241,182,700 recoveries ( https://www.worldometers.info/coronavirus/last accessed on 13.12.2021, 06:51 GMT) and represented a great threat to global human health with approximately 2–5% mortality rate (WHO,2020a; Zhou et al., 2020). While the original SARS-CoV epidemic caused in 2002–2004 affected ∼8400 individuals with an 11% fatality rate. The causative agent was identified and based on the genomic sequences, phylogenetic relationship, and high sequences similarity with bat coronavirus (SARS-bat virus), it was tentatively designated as SARS-CoV-2 and the disease was finally designated as COVID-19 in February 2020, and pandemic was declared on March 11, 2020 (WHO,2020b). This virus falls into the family Coronaviridae, with a + ssRNA virus and the genome size is 26 to 32 kb. Acute respiratory distress syndrome (ARDS) induced by viral pneumonia is attributed to the mortality caused by COVID-19 infections. ARDS represents the most serious complication of SARS-CoV-2 infection with a mortality rate (Azoulay et al., 2020). Till now, multiple genetic variants have been emerged from different geographic locations and spread globally to infect the human population including recently emerged and known as Omicron. The management and treatment guidelines mainly focused on vaccination, prevention, control, and supportive intensive care involving oxygen supplementation and mechanical ventilation as required (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/).
Currently, many vaccines have been approved by World Health Organization (WHO) and are being used for vaccination of the human population globally, but still, there is no antiviral therapeutics approved by the USFDA available to combat this disease as well for long term protection from this viral infection. A promising approach for the development of highly specific antiviral therapy can be adopted by using an RNA interference (RNAi) mechanism to regulate the expression of desired genes by degrading the corresponding mRNAs. The use of short interfering RNAs (siRNAs) has been proven as significant oligonucleotide-based therapy against many viral diseases (Sohrab et al., 2018; Sohrab et al., 2020; Uludağ et al., 2020; Donia and Bokhari 2021). The basic criteria for designing and selecting the potential siRNAs have been described and published (Sohrab et al., 2018). Currently, more than 20 clinical trials are running to evaluate the RNAi-based biopharmaceuticals for oligonucleotide-based therapeutics (Alnylam, 2020). Recently, we have experimentally evaluated the MERS-CoV siRNAs and observed promising results to inhibit the MERS-CoV proliferation in various cell lines (Sohrab et al., 2020; Sohrab et al., 2021a; Sohrab et al., 2021b; Sherif Aly El-Kafrawy et al., 2021). This finding encouraged us to design and evaluate the siRNAs against SARS-CoV-2.
Additionally, the designing of advanced siRNAs, aptamers/siRNAs chimera, siRNA-Nanoparticle, siRNA-peptide dendrimer formulation based therapy against SARS-CoV-2 using various genes and leader sequences have recently been reported and discussed in many published reports (Bappy et al., 2021; Chen et al., 2020; Chowdhury et al., 2021; Idris et al., 2021; Khaitov et al., 2021; Khan ali, 2020; Panda, 2020; Pandey and Verma, 2021; Rohani, 2021; Shawan et al., 2021; Sohrab et al., 2021a,b; Wu and Luo, 2021). But most of the above published reports discussed only the prediction and designing of potential siRNAs, except only one report has discussed the designing and experimental evaluation of a few siRNAs against SARS-CoV-2 and found significant antiviral efficacy in Vero cells (Niktab et al., 2021).
So, based on the status and urgent requirement, we report here about the Insilico prediction, designing, molecular docking, cytotoxicity, and experimental validation of siRNAs in Vero E6 cells against SARS-CoV-2. The different concentrations of siRNAs were delivered to Vero E6 cells by Lipofectamine-2000 (Invitrogen, USA), and the Multiplicity of infection (MOI) 0.1of the virus was used to infect the Vero E6 cells. The purification of viral RNA was done from the lysed cells and subjected to quantitative real-time PCR for the determination of viral load reduction based on Ct value. The data generated and analyzed results showed that only 3rd siRNAs showed the significant and better antiviral activity in tested cells.
2 Materials and methods
2.1 In-silico prediction, and selection of siRNAs
The Spike protein gene sequence of an isolate from Saudi Arabia, SARS-CoV-2/human/SAU/85791C/2020 genome (GenBank: MT630432.1) was retrieved from NCBI-PubMed. The receptor-binding domain (RBD-S) sequences were used to predict and design the siRNA by using the online tool siDirect version 2.0 (http://siDirect2.RNAi.jp/) (Naito et al., 2009). To reduce any off-target effect, the maximum Tm value was fixed to 21.5 °C for seed target duplex stability. The predicted siRNAs were further screened by using the National Centre for Biotechnology Information (NCBI-PubMed) human genomic and transcript database against any similarities with Homo sapiens sequences, and siRNAs with any off-target sequences were eliminated. Generally, to predict and screen the highly potential siRNAs, strict criteria should be followed such as the guide strand should contain A/U at 5′ terminus with a minimum of 4 A/U in the seven base pairs, while G/C should be in passenger strands at 5′ terminus. The minimum Tm value for both guide and passenger strands with a smaller number of off-target sequences should be considered for the best siRNAs selection. The software resulted in twenty-one siRNA, but we selected only four siRNAs following the criteria of selection with threshold and algorithm scores (Naito et al., 2009; Sohrab et al., 2018).
2.2 Secondary structure prediction of siRNAs
The RNAfold 2.4.18 server ( http://rna.tbi.univie.ac.at) was used to predict the siRNAs secondary structure. The sequences of the guide strand and passenger strand were exported to software and the program was run to generate the secondary structure. The secondary structure was visualized and downloaded in forna format. The software generated the secondary structure of each siRNAs with minimum free energy (MFE). The MFE of each siRNAs were variable. Based on the current studies, the secondary structure of siRNAs plays a significant role in the efficient binding of target sequences and silencing efficiency.
2.3 Molecular docking analysis of predicted siRNAs for binding affinity
We perform the molecular docking analysis using online HNADOCK software (https://huanglab.phys.hust.edu.cn/hnadock/) to determine the efficient binding affinity of predicted siRNAs with the target region of RBD-S gene sequences (He et al., 2019). The docked images were visualized in PyMOL 2.5.2 online software (https://pymol.org/2/). The SARS-CoV-2-RBD-S gene sequence and the guide and passenger strand sequences of predicted siRNAs were used as input to evaluate the binding affinity with the target sequences. Based on the lowest Tm value for the guide strand and with the least number of passenger strand and the highest binding score of SARS-CoV-2-RBD-S gene sequences off-targets were considered during docking analysis. Based on the online software using siRNA Direct 2.0, HNADOCK for molecular docking analysis, secondary structure prediction, we have selected the four most potential siRNAs for chemical synthesis and further used them for experimental validation of SARS-CoV-2 inhibition in Vero E6 cells.
2.4 siRNAs transfection to Vero E6 cells
The delivery of siRNAs to Vero E6 cells (ATCC# CRL-1586) was performed through the reverse transfection using a standardized and published protocol (Sohrab et al., 2020; Sohrab et al., 2021b). The cells were propagated in DMEM media at 37 °C and transfected at 60–80% confluency. The different concentrations of siRNAs (0.1–50 nM) were made with a lipid complex by using the Lipofectamine-2000 (Invitrogen, USA). The lipid complex was delivered slowly to the grown Vero E6 cells. All the experiments were performed in triplicates with proper negative and positive controls. The MTT assay was used to determine the cytotoxicity after 72 h post-transfection.
2.5 Cytotoxicity assay
The cytotoxicity of siRNAs in Vero E6 cells was evaluated by Vybrant™ MTT Cell Proliferation Assay Kit as per instructions (ThermoFisher Scientific) as well as by using the published protocol (Sohrab et al., 2020; Sherif Aly El-Kafrawy et al., 2021). The cells were replenished with fresh DMEM media following the addition of 12 mM-MTT (10 μl) and incubated at 37 °C for 4 h. The OD value was measured at 570 nm using the SpectraMax i3x imaging cytometer. The mean OD value was used for the calculation and determination of cytotoxicity by using the standard formula. Corrected absorbance: % cytotoxicity = (100 × (control - sample)).
3 Analysis of viral load by quantitative real-time PCR in Vero cells
3.1 Propagation of cells, siRNAs transfection, and virus inoculation
The Vero E6 cells were propagated in DMEM media using 96-well plates and transfected at 90–95% confluency. The SARS-CoV-2 replicates very efficiently in Vero E6 cells and enables achieving the high virus titers in these cells (Ogando et al., 2020). The reverse transfection method was used for siRNA transfection and the Lipofectamine 2000 (Invitrogen, USA) was used for the delivery of siRNAs. Briefly, various dilutions (0.1 to 50 nM) of siRNAs were made from 50 mM stocks by adding the 100 l Opti-MEM medium and Lipofectamine and incubated at room temperature for 30 min. Finally, the siRNA-lipid complex (1 µl) was mixed gently to the Vero E6 cells and incubated for 24 h at 37 ◦C for further growth.
The propagation and inoculation of the virus to Vero E6 cells were performed as per published protocol (Azhar et al., 2020; Alshukairi et al., 2021). All the work with live SARS-CoV-2 has performed in biosafety laboratory level 3 (BSL3) facilities at Special Infectious Agents, King Abdulaziz University, Jeddah, Saudi Arabia. The SARS-CoV-2/human/SAU/85791C/2020 isolate was derived from a positively tested in Saudi Arabia. The virus was grown in Vero E6 cells containing DMEM media and titer was determined by plaque assay. Briefly, the Vero E6 cells were grown in DMEM media at 90–95% confluency and inoculated by the virus at MOI 0.1 and incubated for 1 h at 37 °C with gentle shaking every 15–20 min, Subsequently, the inoculum was replaced by 25 ml of viral inoculation medium (DMEM with 2% FBS, 1% penicillin/streptomycin and 10 mmol/l HEPES [pH 7.2]) and the cells were incubated in a humidified incubator at 5% CO2 and 37 °C until 80%-90% of cells showed a cytopathic effect (CPE). After full CPE, the cells were harvested by centrifugation at 500×g for 5 min and used for viral RNA isolation and purification.
3.2 Viral RNA extraction and q-real-time PCR
The purification of viral RNA was performed by using QIAampViral RNA Mini Kit (Qiagen) following the manufacturer instructions and subjected for q-real-time PCR by using PowerCheck SARS-CoV-2 real-time-PCR kit (Kogenebiotech-Korea) following the kit protocol. Briefly, the total volume of PCR mixture was made up to 20 μl containing 10 μl 2X RT-PCR master mix, 5 μl primer/probe mix, and 5 μl template viral RNA. The results based on the Ct value were interpreted as per kit instructions. The positive results should have the Ct value ≤ 37 and the negative sample should have Ct value greater than 37 or not detected.
4 Results
4.1 Prediction and selection of siRNAs
The RBD-S protein gene sequence of an isolate from Saudi Arabia, SARS-CoV-2/human/SAU/85791C/2020 was retrieved from NCBI-PubMed used for siRNAs prediction, designing, and selection. The online software siDirect 2.0 predicted twenty-one siRNAs, but only four siRNAs were selected for further analysis as per their specific characteristics and strict selection criteria. The details and the sequences of siRNAs (guide/passenger strand) have been provided in Table 1.
S. No
Location of siRNAs (Start-End)
Target sequence
Predicted RNA oligo sequences (5′→3′)
Seed-duplex stability (Tm/°C) Guide/Passenger strand
Minimum FreeEnergy
(MFE)(kcal/mol)
/frequency of the MFE structure (%)/
Ensemble diversityDocking score kcal/mol/ligand rmsd (Å)
1
86–108
CGCATCATTTTCCACTTTTAAGT
UUAAAAGUGGAAAAUGAUGCG
CAUCAUUUUCCACUUUUAAGU4.6/7.2
−21.71
60.36/ 0.82
−220.57/323.96
2
218–240
TGGAAAGATTGCTGATTATAATT
UUAUAAUCAGCAAUCUUUCCA
GAAAGAUUGCUGAUUAUAAUU
3.5/5.3
–22.16
47.54/ 1.06
−192.64/324.91
3
358–380
AACCTTTTGAGAGAGATATTTCA
AAAUAUCUCUCUCAAAAGGUU
CCUUUUGAGAGAGAUAUUUCA
6.6/12.2
–23.97
54.55/ 0.77
−290.50/134.02
4
227–249
TGCTGATTATAATTATAAATTAC
AAUUUAUAAUUAUAAUCAGCA
CUGAUUAUAAUUAUAAAUUAC8.0/8.7
−16.00
61.82/ 0.81
−252.62/581.31
4.2 Prediction of secondary structure
In this study, we have used the online software RNAfold 2.4.18 for secondary structure prediction for all four siRNA (Fig. 1). The results have been computed using an online program and the secondary structure of each siRNAs was visualized and downloaded into forna format with MFE structure drawing encoding base-pair probabilities. The structure is colored by base-pairing probabilities. Results of all siRNAs with the MFE are given in Table 1. The structures did not show any complexity.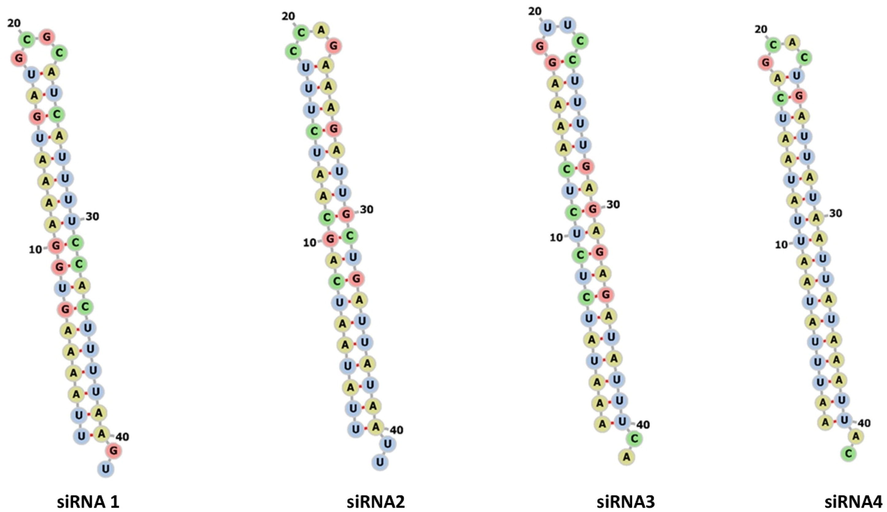
The possible folding and secondary structure prediction of Insilico predicted siRNAs molecules computed using with the online web server (1–4).
4.3 Molecular docking analysis
The predicted and designed siRNAs were used further for molecular docking analysis to reveal the binding efficiency to the target sequences. We have selected the best siRNAs model from the top ten docked models generated and visualized in PyMOL 2.5.2 from siRNAs and target sequence. The results showed high binding energy of siRNAs guide strand with SARS-CoV-2-RBD-S sequences which ranged from −192.64 to −290.50 kcal/mol. The RMSD score of each siRNAs ranged from 134.02 to 581.31 Angstrom (Å) (Fig. 2, Table 1). The siRNA guide strand showed high sequences complementarity with the target strand and promoted effective binding to the nucleotide bases. In docking analysis, the highest binding affinity was observed with the 3rd siRNA with the score of −290.50 kcal/mol thereby confirming the high structural affinity of this siRNA for the SARS-CoV-2-RBD-S sequence. Additionally, each off-target corresponding to the 3rd siRNA passenger strand showed only 2 mismatches at the 5′ end which reduces the possibility of binding to the undesired target and reduces the off-target effect. The present docking analysis showed that the potent siRNAs can efficiently bind with the target sequences of SARS-CoV-2 and can silence the activity of a specific target and thereby hamper the attachment of the virus to host cells.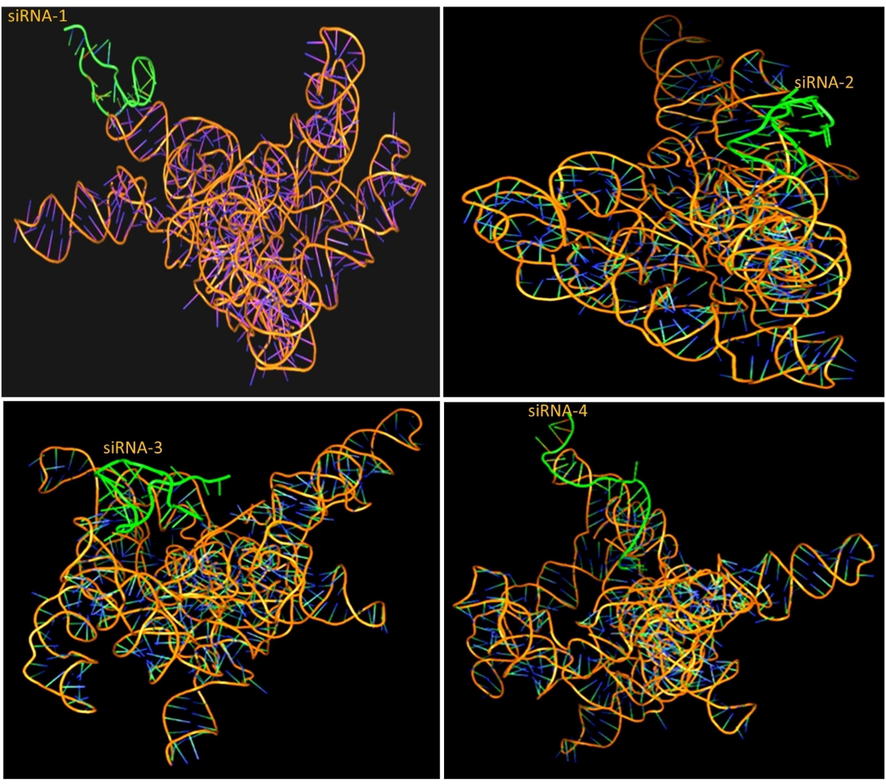
Molecular docking of predicted siRNAs (1–4-green) with SARS-CoV-2-RBD-S target gene and 3D interaction diagram of different docked complex with the target.
4.4 Cytotoxicity assay
The cytotoxicity results of all the four tested siRNAs were determined and there was no significant cytotoxicity observed in any tested siRNAs in Vero E6 cells ranged from 0.1 to 50 nM concentrations. The OD and CC-50 values of each siRNAs have been presented in Table 2 and Fig. 3.
siRNAs Concentration. (nM)
siRNA-1
siRNA-2
siRNA-3
siRNA-4
50
1.4
1.5
1.4
1.3
25
1.3
1.2
1.3
1.4
10
1.2
1.3
1.4
1.3
5.0
1.2
1.2
1.3
1.3
1.0
1.1
1.3
1.4
1.2
0.5
1.1
1.2
1.2
1.2
0.25
1.2
1.1
1.2
1.2
0.1
1.1
1.1
1.1
1.1
CC 50
49.40
41.52
48.21
45.64
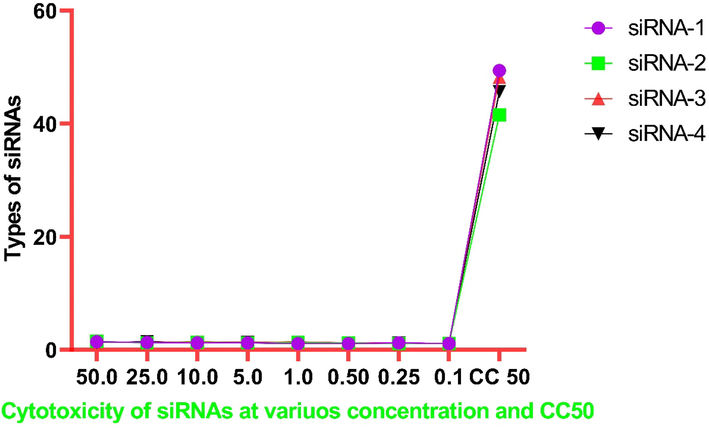
Cytotoxicity of different siRNAs (1–4) at various concentrations (0.1–50 nM) in Vero E6 cells.
4.5 Quantitative real-time PCR analysis for viral load
The cells were harvested for viral RNA purification from siRNAs transfected cells after full CPE in positive control using Kit. The purified RNA was subjected to quantitative real-time PCR. The Ct value of each siRNAs was used for determining the viral load in Vero E6 cells. The dose-dependent inhibition and reduction of viral load were observed. The less viral load was observed in 3rd siRNA than others at 5.0 nM and 10 nM respectively. The Ct value and analyzed results for all four siRNAs have been provided in Table 3 Fig. 4.
Concentration of siRNAs (nM)
siRNA-1
siRNA-2
siRNA-3
siRNA-4
Negative/Positive Control
Ct values of Realtime-RT-PCR
50.0
14.56
14.32
14.99
14.75
49.40/14.21
25.0
16.32
16.37
17.69
16.28
10.0
18.57
17.56
19.84
17.32
5.0
16.88
18.25
18.71
17.38
1.0
16.67
16.31
18.65
14.58
0.50
16.82
16.86
19.99
14.37
0.25
17.54
17.46
16.87
15.53
0.1
16.58
17.95
17.21
17.45
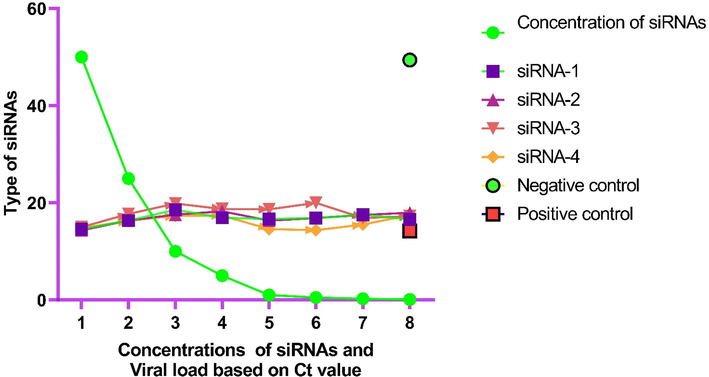
Graphical representation of Ct value of RT-PCR result for the inhibition of SARS-CoV-2 in Vero E6 cells.
5 Discussion
Coronavirus Disease 2019 (COVID-19) was caused by SARS-CoV) and based on the genomic sequence similarity with Bat-Coronaviruses, this virus was finally designated as SARS-CoV-2. As of 12.12.2021, this virus has spread to 222 countries and territories with 5,322,447 deaths (06:51 GMT). This virus particularly damages the pulmonary system and lungs tissues, resulting in the malfunctioning of respiratory functions and the death of infected individuals. At the onset of the pandemic, there were no USFDA drugs, antivirals, and approved vaccines, and there was an urgent need to develop effective antiviral therapeutics against SARS-CoV-2. Currently, there are many vaccines have been approved and is used to vaccinate the human population against SARS-CoV-2. But there are still many antiviral drugs, therapeutic molecules including oligonucleotide-based therapeutics are under the stage of an investigation. The RNAi approach is being used to regulate the silencing of a particular gene/target by using siRNA/shRNA molecules.
The potential efficiency of a siRNA is determined and evaluated by the availability and binding of target sites as well as the degree of its potentiality. So, the target accessibility of each siRNAs and its binding to the target is crucial for efficient gene silencing (Gatta et al., 2018). The RNAi technology has played a significant role and produced considerable results towards different diseases caused by various viruses (Jeang, 2012; Chakraborty et al., 2017). Currently, using RNAi technology, a significant number of siRNAs/shRNAs have been designed, developed, and experimentally evaluated and showed promising results against many viruses including MERS-CoV and SARS-CoV-2 against the multiple targets of a viral genome (Sohrab et al., 2018; Sohrab et al., 2020; Uludağ et al., 2020; Panda et al., 2020; Chen et al., 2020; Sohrab et al., 2021a; Sohrab et al., 2021b; El-Kafrawy et al., 2021; Khaitov et al., 2021; Idris et al., 2021; Pandey and Verma, 2021; Donia and Bokhari 2021; Bappy et al., 2021; Niktab et al, 2021; Khanali et al., 2021; Shawan et al., 2021; Chowdhury et al., 2021; Wu and KQ, 2021; Rohani, et al., 2021; Khaitov et al., 2021). The SARS-CoV-2 uses the S protein to attach with host cells and many vaccines have been developed by using this target, but many variants have been emerged globally by mutations in the Spike (S) gene and making it difficult to develop vaccines and antivirals for broad-spectrum resistance against SARS-CoV-2 (Harvey et al., 2021; Greaneyet al., 2021; Khateeb et al., 2021; Starr et al., 2021; (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/).
Based on the status and urgent need globally, we have performed this work and discussed the antiviral potential of selected siRNAs in the Vero E6 cells. The outputs of online software resulted in twenty-one siRNAs but only four siRNAs were selected, based on their specific selection criteria as described (Naito et al., 2009; Sohrab et al., 2018). The predicted and selected siRNAs were further analyzed by molecular docking analysis and secondary structure prediction for their effective binding efficiency to the target region. Better inhibition of a target can be expected with the lower binding energy of a given siRNAs. The MFE of the selected siRNAs was varied from −16.00 to –23.97 kcal/mol. The high docking score shows better binding efficiency with the target. The molecular docking score ranged from −192.64 to −290.50 kcal/mol while the RMSD scores varied from 134.02 to 581.31 Å (Table 1). The secondary structure analysis results provided a piece of vital information and efficient binding to the specific target. The analyzed siRNAs showed highly effective binding to the viral genome with their variable MFE as well as with the free energy of the thermodynamic ensemble (TE). The molecular docking analysis results showed the effective binding to the target region. The 3rd siRNAs were found to be the best as compared to other siRNAs.
The cytotoxic effect of each siRNAs was evaluated before the antiviral evaluation and there was no significant cytotoxicity observed at any tested concentration. The tested siRNAs were found to have better potential to reduce the viral load as determined by Ct value of q-real-time PCR. The high Ct value is correlated with the lower copy number of the target gene. The copy number of viral-based mRNA was lower in our tested group, and it appears that siRNA has affected the virus proliferation and a similar Ct value has been reported for COVID-19 patients (Carter et al., 2020).
Based on the results obtained, we will evaluate these siRNAs in a combination using other cell lines as well as different methods of siRNAs delivery in our future work. Interestingly, based on the toxicity and siRNA evaluation study, it appears that Vero E6 cells were not affected by any siRNAs, and it could be used as oligonucleotide-based antiviral therapy for SARS-CoV-2 shortly to protect the human population globally.
6 Conclusions
Based on the data analysis and results obtained from Insilico prediction, selection, secondary structure prediction, molecular docking, cytotoxicity, and virus inhibitory analysis using q-real-time PCR, it is concluded that the designed siRNAs could be effectively used as oligonucleotide-based antiviral therapeutics against SARS-CoV-2.
Author contributions
SSS, SAE conceptualized, designed, and executed the experiments, SSS, SAE wrote the manuscript. EIA: Critically reviewed the manuscript. All authors provided critical feedback and analysis of the manuscript. All authors reviewed the manuscript and approved it.
Acknowledgments
The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number IFPRC-204-141-2020 and King Abdulaziz University, DSR, Jeddah, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Alnylam Pharmaceuticals., 2020. Vir and Alnylam Expand Collaboration to Advance Rnai Therapeutics for the Treatment of Coronavirus Infection, Including Covid-19. Available online at: https://investors.alnylam.com/press-release?id= 24656.
- Test-based de-isolation in COVID-19 immunocompromised patients: Cycle threshold value versus SARS-CoV-2 viral culture. Internat. J. Infect. Diseases IJID. 2021;108:112-115.
- [Google Scholar]
- Amotosalen and ultraviolet A light treatment efficiently inactivates severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in human plasma. Vox Sanguinis (December). 2020;116(6):673-681.
- [Google Scholar]
- Increased mortality in patients with severe SARS-CoV-2 infection admitted within seven days of disease onset. Intensive Care Med.. 2020;46(9):1714-1722.
- [Google Scholar]
- Designing potential siRNA molecule for the nucleocapsid(N) gene silencing of different SARS-CoV-2 strains of Bangladesh: Computational approach. Comput. Biol. Chem.. 2021;92:107486.
- [Google Scholar]
- Assay Techniques and Test Development for COVID-19 Diagnosis. ACS Cent. Sci.. 2020;6(5):591-605.
- [Google Scholar]
- Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Molecular therapy. Nucleic acids. 2017;8:132-143.
- [Google Scholar]
- Computational identification of small interfering RNA targets in SARS-CoV-2. Virol. Sin.. 2020;35(3):359-361.
- [Google Scholar]
- A computational approach to design potential siRNA molecules as a prospective tool for silencing nucleocapsid phosphoprotein and surface glycoprotein gene of SARS-CoV-2. Genomics. 2021;113(1):331-343.
- [Google Scholar]
- RNA interference as a promising treatment against SARS-CoV-2. Int Microbiol.. 2021;24(1):123-124.
- [Google Scholar]
- Strategies for improving the specificity of siRNAs for enhanced therapeutic potential. Expert Opin. Drug Discov.. 2018;13(8):709-725.
- [Google Scholar]
- Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat. Commun.. 2021;12(1)
- [Google Scholar]
- SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol.. 2021;19(7):409-424.
- [Google Scholar]
- HNADOCK: a nucleic acid docking server for modeling RNA/DNA-RNA/DNA 3D complex structures. Nucleic Acids Res.. 2019;47 W35-W42
- [Google Scholar]
- A SARS-CoV-2 targeted siRNA-nanoparticle therapy for COVID-19. Mol. Ther.. 2021;29(7):2219-2226.
- [Google Scholar]
- RNAi in the regulation of mammalian viral infections. BMC Biol.. 2012;10
- [CrossRef] [Google Scholar]
- Silencing of SARS-CoV-2 with modified siRNA-peptide dendrimer formulation. Allergy. 2021;76(9):2840-2854.
- [Google Scholar]
- An aptamer/ siRNA Chimera against The SARS-CoV- 2: A dual therapeutic strategy for the virus neutralizing and RNA interfering. Res. Squire. 2020
- [CrossRef] [Google Scholar]
- Emerging SARS-CoV-2 variants of concern and potential intervention approaches. Crit. Care. 2021;25(1)
- [CrossRef] [Google Scholar]
- siDirect 2.0: updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinf.. 2009;10(1)
- [Google Scholar]
- Design of advanced siRNA therapeutics for the treatment of COVID-19. Meta Gene.. 2021;29:100910
- [CrossRef] [Google Scholar]
- SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol.. 2020;101(9):925-940.
- [Google Scholar]
- Prediction of potential small interfering RNA molecules for silencing of the spike gene of SARS-CoV-2. Indian J. Med. Res. 2020
- [CrossRef] [Google Scholar]
- An in-silico analysis of effective siRNAs against COVID-19 by targeting the leader sequence of SARS-CoV-2. Adv. Cell Gene Ther.. 2021;4(2)
- [CrossRef] [Google Scholar]
- Discvering potential candidates of RNAi-based therapy for COVID-19 using computational methods. PeerJ 2021
- [CrossRef] [Google Scholar]
- Designing an effective therapeutic siRNA to silence RdRp gene of SARS-CoV-2. Infect. Genet. Evol.. 2021;93:104951.
- [Google Scholar]
- In vitro inhibitory analysis of rationally designed siRNAs against MERS-CoV replication in Huh7 Cells. Molecules. 2021;26(9):2610.
- [Google Scholar]
- Designing and evaluation of MERS-CoV siRNAs in HEK-293 Cell line. J. Infect. Public Health.. 2021;14(2):238-243.
- [Google Scholar]
- In silico prediction and designing of potential siRNAs to be used as antivirals against SARS-CoV-2. Curr. Pharm. Des.. 2021;27(32):3490-3500.
- [Google Scholar]
- Design and delivery of therapeutic siRNAs: application to MERS-Coronavirus. Curr. Pharm. Des. 2018
- [CrossRef] [Google Scholar]
- In silico prediction and experimental validation of siRNAs targeting ORF1ab of MERS-CoV in Vero cell line. Saudi J. Biol. Sci.. 2021;28(2):1348-1355.
- [Google Scholar]
- SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature. 2021;597(7874):97-102.
- [Google Scholar]
- Prospects for RNAi Therapy of COVID-19. Front. Bioeng. Biotechnol.. 2020;8
- [CrossRef] [Google Scholar]
- Statement Regarding Cluster of Pneumonia Cases in Wuhan, China 2020. 2020. Available from:
- [Google Scholar]
- WHO, 2020b. Director-General’s Remarks at the Media Briefing on 2019-nCoV on 11 February 2020. Available online: https://www.who.int/dg/speeches/detail/who-director-general-sremarks-at-the-mediabriefing-on-2019-ncov-on-11-february-2020.
- Developing effective siRNAs to reduce the expression of key viral genes of COVID-19. Int. J. Biol. Sci.. 2021;17(6):1521-1529.
- [CrossRef] [Google Scholar]
- Coronavirus disease 2019 (COVID-19): A clinical update. Front. Med.. 2020;14(2):126-135.
- [Google Scholar]







