Translate this page into:
In silico CD4 + T-cell multiepitope prediction and HLA distribution analysis for Marburg Virus—A strategy for vaccine designing
⁎Corresponding author. subhash.chauhan@utrgv.edu (Subhash C. Chauhan)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Marburg, a RNA virus (MRV), is responsible for causing hemorrhagic fever that affects humans and non-human primates. World Health Organization (WHO), National Institutes of Health (NIH) and Centre of Disease Control and Prevention (CDC) considered this as an extremely dangerous virus, thus categorised as risk group 4, category A priority pathogen and category “A” bioterrorism agent, respectively. Despite of all these alarming concerns, no prophylaxis arrangements are available against this virus till date. In fact, the construction of immunogenic vaccine candidates by traditional molecular immunology methods is time consuming and very expensive. Considering these concerns, herein, we have designed CD4 + T Cell multiepitopes against MRV using in silico approach. The pin-point criteria of the screening and selection of potential epitopes are, non-mutagenic, antigenic, large HLAs coverage, non-toxic and high world population coverage. This kind of methodology and investigations can precisely reduce the expenditure and valuable time for experimental planning in development of vaccines in laboratories. In current scenario, researchers are frequently using in silico approaches to speed up their vaccine-based lab studies. The computational studies are highly valuable for the screening of large epitope dataset into smaller one prior to in vitro and in vivo confirmatory analyses.
Keywords
Marburg Virus
Peptide based vaccine
CD4+ T Cell
Non-mutagenic
Antigenic
Non-toxic and High world population coverage
- MRV
-
Marburg virus
- MHF
-
Marburg Hemorrhagic Fever
- WHO
-
World Health Organization
- NIH
-
National Institutes of Health
- CDC
-
Centre of Disease Control and Prevention
- MHC
-
Major Histocompatibility Complex
- Env GPs
-
Envelope Glycoproteins
- HLA
-
Human Leukocyte Antigen
Abbreviations
1 Introduction
Marburg virus (MRV) is genus of Filoviridae family. MRV is negative stranded & non-segmented RNA virus, that is responsible for severe hemorrhagic fever, known as marburg hemorrhagic fever (MHF) in both humans and non-human primates. The MRV infection has approximately 23 to 100% fatality and lethality rates in humans and non-human primates (Mehedi et al., 2011). The systemic viral replication of MRV interferes with immune and inflammatory activities, the consequence of which are serious pathological features in patients, like hemorrhages, edema, coagulation imbalance, multiple-organ failure and shock, often resulting in death (Bente et al., 2009). Before the discovery of Ebola in 1967, the first MRV infection was observed in Germany and Serbia (Mehedi et al., 2011)followed by Zimbabwe/South Africa in 1975 (Conrad et al., 1978)Angola in 2004 (Towner et al., 2006). According to the report of CDC 2014, the recent outbreak was in Uganda from 2007 to 2014. Few studies demonstrated that MRV is highly infectious and very stable in experimental aerosol exposure (Alves et al., 2010), which raises the concern that MRV may be very suitable to be used as biological weapon (US Centres for Disease Control and Prevention/“Bioterrorism Agents/Diseases” report). Despite of all, in current scenario no treatment is available against MRV infection (Cross et al., 2018). The main enviable feature for any vaccine candidate is that the molecule should activate cell-mediated (T-Cell) and humoral (B-Cell) immune response followed by memory cell formation. CD4 + T-cells activation is mandatory for a competent humoral immune response for the induction of Immunoglobulin-G and memory B cells. CD4 + T-cells primarily recognize antigen peptides by CD4 co-receptor and only recognize the major histocompatibility complex (MHC) II protein on antigen-presenting cells; then memory B cells make a repository of infected virus for the farther prophylaxis arrangement (Clem, 2011). Filoviridae family virus consists of 7 structural proteins, among all, highly glycosylated (N- and O-linked glycans) envelope glycoproteins (Env GPs) are present over the cell surface. Host proteases like furin is responsible for the proteolysis of GP, resulting in two subunits, GP1 and GP2, linked by a disulfide bond (Volchkov et al., 2000). These GPs mediate and lead the viral entry into host cells (Takada et al., 1997),(Wool-Lewis and Bates, 1998), thus GPs are considered to be the ideal target for neutralizing antibodies against filoviruses. In current era as the electronic support increases in the life sciences, computational based approach provides access to researchers to deal with huge number of genome and proteome data of virus. Immunogenic, non-toxic and peptide-based vaccines would prove to be a good alternative treatment option for the management of MRV infection. In this article, we have focused on the special epitopes candidates those have non mutagenic tendency, which were thoroughly screened via protein variability server. We have identified various small fragments of Env GP those don’t have the mutation hot spot, which will lead the less chance of viral resistance. In current COVID scenario scientists have encountered with the toxicity issue of vaccines so here we have seriously focused on the non-toxic and highly immunogenic multi-epitopes from Env GP proteins of MRV virus. The recognition of world-wide HLA coverage analysis of CD4 + T-cell epitopes in Env GP protein was carried out by using artificial neural network algorithm (ANN) implemented in IEDB and NetMHCIIPan Server. This article, is clearly depicted that the focus of this article is to provide a rapid, cost effective and efficient vaccine candidate and process by using epitopes of viral proteins. This strategy was designed to keep in the mind of current COVID situation where we were looking safe and effective vaccine. Since a very recent statement of WHO, also claimed the inflammable problem of marburg virus as a “scary & deadly disease”. (https://www.express.co.uk/news/science/1474916/marburg-virus-news-disease-scary-deadly-world-health-organisation-africa-spt)
2 Materials and methods
2.1 Sequence retrieval and multiple sequence alignment (MSA) of retrieved proteins
681 residue long amino acid sequences of viral Env GP from 37 different strains of MRV, that are involved in the host cell binding and fusion activity, were retrieved from UniProtKB Database (www.uniprot.org). The retrieved sequences were further subjected to multiple sequence alignment using CLUSTAL Omega, to spot the non-mutated and highly immunogenic amino acid sequences for the assessment and predictions of effective epitope.
2.2 Protein variability analysis of retrieved sequences
Protein Variability Server/PVS (http://imed.med.ucm.es/PVS/) was used to identify the variable or high mutational rate amino acid in the particular protein sequences, because one virus has several strains worldwide and they differ with each other’s on the basis of highly mutated amino acid, which is the natural tendency of viruses to show high mutational rates. This tendency is the reason of failure of most of the vaccines. (Garcia-Boronat et al., 2008)
2.3 Immunogenicity-antigenicity prediction of the viral protein
The VaxiJen V2.0 server (http://www.ddg-pharmfac.net/vaxijen/Vaxi- Jen/VaxiJen.html) was used for the assessment of immunogenicity-antigenicity of the selected protein sequences from PVS. This server runs on Auto Cross Covariance (ACC) algorithm that predicts protective and tumor antigens and subunit vaccines with the accuracy level of up to 89 %. (Doytchinova and Flower, 2007), (Janahi et al., 2017)
2.4 CD4 + epitope prediction
The Env GP protein sequence was investigated for the screening of the probable leading T-cell CD4 + epitopes using bioinformatics tool NetMHCIIpan server (Nielsen et al., 2008), which is one of the most accurate prediction servers currently available based on ANN. NetMHCIIpan server have huge pool of more than 5000 HLAs (DQ: 2912, DP: 2247, DRB4: 06, DRB1: 15, DRB3: 29, DRB5:15). The predictions output, showing binding affinity of each epitopic from core sequences with every known HLA allele. The window of peptide length was set to be 15 for HLA-II, respectively as mentioned in earlier publications (Janahi et al., 2017),(Bano et al., 2018). The epitopes were predicted on the basis of lowest percentile rank and high binding affinity.
2.5 Identification of non-toxic region of selected epitopes
The final selected epitopes were checked for the conserved regions and further subjected to ToxinPred severe (Gupta et al., 2013) for the segregation of toxic or nontoxic peptides. Support Vector Machine (SVM) and Quantitative Matrix based algorithm were used to generate quantitative matrix on the basis of probability or frequency of amino acid at a particular location.
2.6 Population coverage analysis
The population coverage rate of the final selected epitopes was calculated by using the IEDB population coverage tool (http://tools.immuneepitope.org/tools/population/iedb_input) (Bui et al., 2006). The predicted epitopes with their all-binding HLA alleles for the worldwide distribution were tabulated. IEDB server extract all allele genotypic frequencies related data from Allele Frequency database, which comprises with allele frequencies form huge population set of 115 countries and 21 different ethnicities grouped into 16 different geographical areas. The schematic representation of the entire methodology of in silico CD4 + T-cell epitope prediction and HLA distribution of MRV is mentioned in Fig. 1.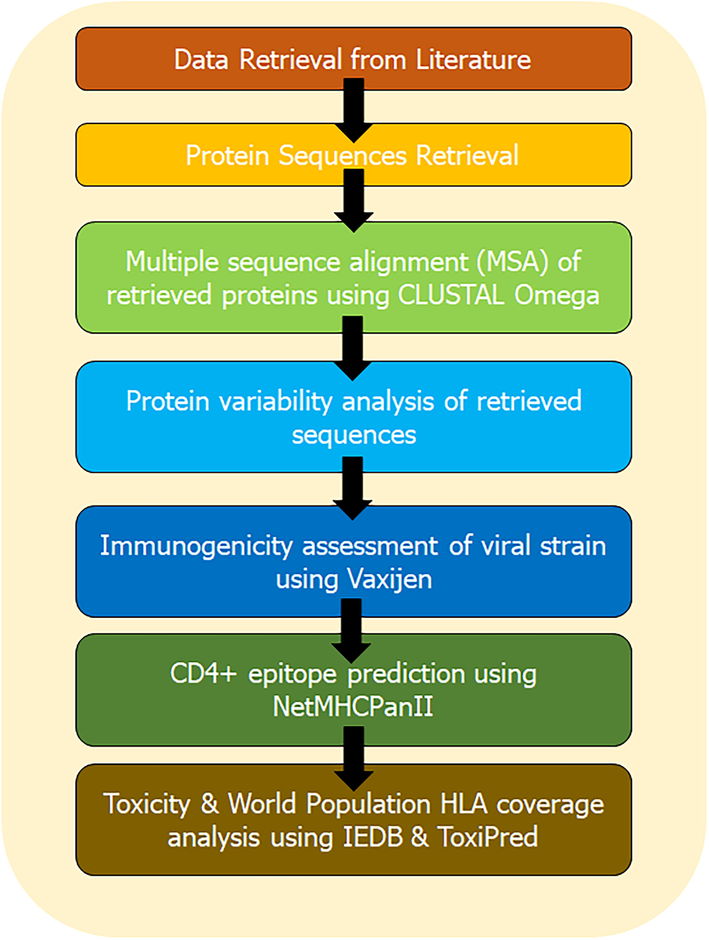
The schematic representation of the entire methodology.
2.7 IFN-gamma inducing capacity and physiochemical prediction
IFN-gamma inducing capacity predictions of all 11 epitopes were predicted by IFNepitope server (http://crdd.osdd.net/raghava/ifnepitope/index.php). In this segment all final 11 epitopes were assessed by using two categories INF-G vs non-INF-G epitopes and INF-G vs other cytokines. Followed by physiochemical properties of finally selected epitopes by using various servers
like https://pepcalc.com/; https://web.expasy.org/protparam/ & https://www.biosyn.com/peptidepropertycalculator/peptidepropertycalculator.aspx.
3 Results
3.1 Retrieval of Env glycoprotein sequences of MRV and multiple sequences alignment (MSA)
All 37 Env glycoprotein sequences of different strains of MRV (mentioned in Supplementary Data Set part 1) with more than 89% of similarity, were retrieved from uniprot database. CLUSTAL Omega was used for the identification of evolutionary relationship (as shown in Fig. 2) and percent of similarity between all 37 protein sequences of Env protein of MRV (mentioned in Supplementary Data Set part 2). Q6UY66|VGP_MABVO was considered as reference sequence and its comparative sequence similarity coverage with other proteins more than 89%. On the basis of similarity coverage, authors obtained the information about the variation in the sequences that may lead to hot points of the virus mutations (Fig. 3). Considering this, Protein Variability Server (PVS) was used to select the non-variable fragments of the viral Env proteins, which were used for the selection and identification of the most effective and immunogenic epitopes. Total ten fragments were obtained (Table 1) by PVS study. Often, only eight fragments were selected for the identification of epitopes because fragment no. 03 (PEIKPTSTPTDAT, 13 amino acids from 240 to 252) and 06 (NLSTLS, 06 amino acids from 350 to 354) have very lesser numbers of amino acids than the selection windows length for HLA-II peptide which was 15.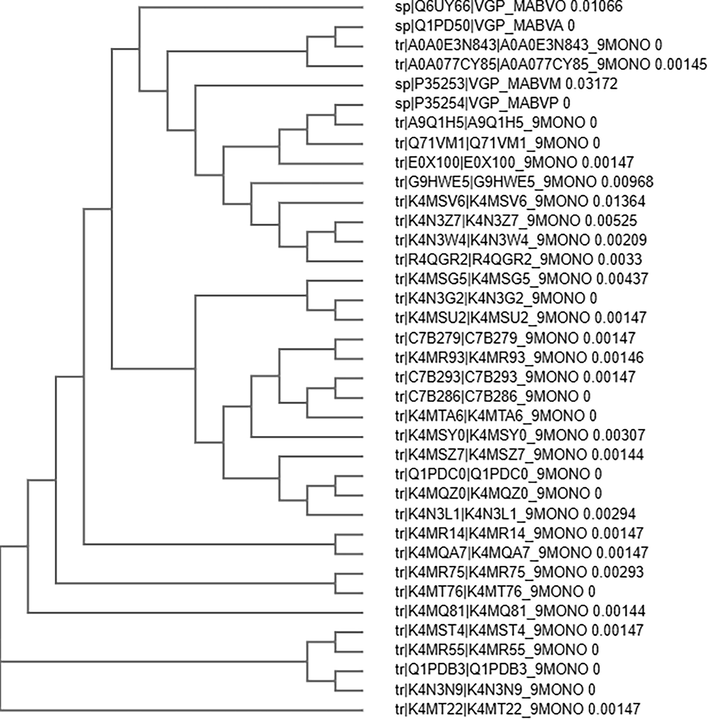
Phylogenetic relationship between all 37 Env proteins of all strains of Marburg virus.
Protein Variability Plot of Env proteins of all 37 Sequences of Marburg Virus (MRV).
Set No.
Start
End
Sequence
Vaxijen Score (0.4 Threshold)
1
3
27
TTCFFISLILIQGIKTLPILEIASN
0.6525(Probable ANTIGEN)
2
29
230
QPQNVDSVCSGTLQKTEDVHLMGFTLSGQKVADSPLEASKRWAFRTGVPPKNVEYTEGEEAKTCYNSVTDPSGKSLLLDPPTNVRDYPKCKTIHHIQGQNPHAQGIALHLWGAFFLYDRIASTTMYRGKVFTEGNIAAMIVNKTVHKMIFSRQGQGYRHMNLTSTNKYWTSSNGTQTNDTGCFGTLQEYNSTKNQTCAPSK
0.4850(Probable ANTIGEN).
3
240
252
PEIKPTSTPTDAT
1.3071(Probable ANTIGEN).
4
254
316
LNTTNPNSDDEDLTTSGSGSGEQEPYTTSDAVTKQGLSSTMPPTPSPQPGTPQQGGNNTNHSQ
0.4729(Probable ANTIGEN).
5
324
348
NTNTTAQPPMPSHNTTTISTNNTSK
0.4109(Probable ANTIGEN).
6
350
354
NLSTLS
0.8725(Probable ANTIGEN).
7
365
379
NTQSMATENEKTSAP
0.6994(Probable ANTIGEN)
8
381
405
KTTLPPTESPTTEKSTNNTKSPTTM
0.4557(Probable ANTIGEN).
9
415
496
SPSSTPNSTTQHLIYFRRKRSILWREGDMFPFLDGLINAPIDFDPVPNTKTIFDESSSSGASAEEDQHASSNISLTLSYLP
0.6654(Probable ANTIGEN).
10
498
680
SENTAYSGENENDCDAELRIWSVQEDDLAAGLSWIPFFGPGIEGLYTAGLIKNQNNLVCRLRRLANQTAKSLELLLRVTTEERTFSLINRHAIDFLLTRWGGTCKVLGPDCCIGIEDLSRNISEQIDQIKKDEQKEGTGWGLGGKWWTSDWGVLTNLGILLLLSIAVLIALSCICRIFTKYIG
0.4865(Probable ANTIGEN).
3.2 Antigenicity prediction of the viral protein fragments
The VaxiJen V2.0 online server was used for identification of antigenicity of the viral envelope (Env) protein fragments of different strains of MRV, by keeping the threshold at 0.4 (Table 1). The results obtained suggest that the viral protein fragments were probable antigens with a score of 0.6525 (Set 1, TTCFFISLILIQGIKTLPILEIASN from 3 to 27 position), 0.4850 (Set 2, QPQNVDSVCSGTLQKTEDVHLMGFTLSGQKVADSPLEASKRWAFRTGVPPKNVEYTEGEEAKTCYNSVTDPSGKSLLLDPPTNVRDYPKCKTIHHIQGQNPHAQGIALHLWGAFFLYDRIASTTMYRGKVFTEGNIAAMIVNKTVHKMIFSRQGQGYRHMNLTSTNKYWTSSNGTQTNDTGCFGTLQEYNSTKNQTCAPSK, from 29 to 230), 0.4729 (Set 4, LNTTNPNSDDEDLTTSGSGSGEQEPYTTSDAVTKQGLSSTMPPTPSPQPGTPQQGGNNTNHSQ, from 254 to 316), 0.4109 (Set 5, NTNTTAQPPMPSHNTTTISTNNTSK from 324 to 348), 0.6994 (Set 7, NTQSMATENEKTSAP, from 365 to 379), 0.4557 (Set 8, KTTLPPTESPTTEKSTNNTKSPTTM, from 381 to 405), 0.6654 (Set9, SPSSTPNSTTQHLIYFRRKRSILWREGDMFPFLDGLINAPIDFDPVPNTKTIFDESSSSGASAEEDQHASSNISLTLSYLP, from 415 to 496) and 0.4865 (Set 10, SENTAYSGENENDCDAELRIWSVQEDDLAAGLSWIPFFGPGIEGLYTAGLIKNQNNLVCRLRRLANQTAKSLELLLRVTTEERTFSLINRHAIDFLLTRWGGTCKVLGPDCCIGIEDLSRNISEQIDQIKKDEQKEGTGWGLGGKWWTSDWGVLTNLGILLLLSIAVLIALSCICRIFTKYIG, from 498 to 680).
3.3 HLAs distribution analysis, antigenicity and toxicity profiling
NetMHCIIpan server was used for the identification and screening of putative CD4 + T-cell core epitope sequences among the 08 protein fragments of Env protein. Total 29 putative T-cell epitopes were extracted from Env protein, as shown in Table 2. ILIQGIKTLPILEIA was the best epitope which has highest HLA coverage (1034 no. of HLA covers), extracted, and screened from Env protein fragments. Of all 29 epitopes, 14 epitopes were identified as probable antigens. ToxinPred server was used for the toxicity profiling of all peptides (whether these peptides were toxic or non-toxic). The resultant of this profiling was that all peptides were found in non-toxic category.
Sr. No.
Epitopes
Position
HLA Coverage
Score of ANTIGENICITY and TOXICITY Profiling
1
ILIQGIKTLPILEIA
Set 1 from11 to 25
1034
0.6513(Probable ANTIGEN & NON-TOXIC
2
IALHLWGAFFLYDRI
Set2 from 134 to 148
458
−0.0643(Probable NON-ANTIGEN & NON-TOXIC).
3
ALHLWGAFFLYDRI
Set2 from 135 to 149
646
−0.1142(Probable NON-ANTIGEN & NON-TOXIC).
4
LIQGIKTLPILEIAS
Set 2 from 12 to 26
127
0.5381(Probable ANTIGEN & NON-TOXIC)
5
WGAFFLYDRIASTTM
Set 2 from 139 to 153
84
0.5262(Probable ANTIGEN & NON-TOXIC).
6
LHLWGAFFLYDRIAS
Set 2 from 136 to 150
147
−0.0445(Probable NON-ANTIGEN & NON-TOXIC).
7
AFFLYDRIASTTMYR
Set 2 from 141 to 155
242
0.4031(Probable ANTIGEN & NON-TOXIC)
8
GIALHLWGAFFLYDR
Set 2 from 133 to 147
11
0.1535(Probable NON-ANTIGEN & NON-TOXIC).
9
HLWGAFFLYDRIAST
Set 2 from 137 to 151
282
0.1553(Probable NON-ANTIGEN & NON-TOXIC)
10
GAFFLYDRIASTTMY
Set 2 from 140 to 154
113
0.3971(Probable NON-ANTIGEN & NON-TOXIC).
11
LILIQGIKTLPILEI
Set 1 from10 to 24
336
0.6611(Probable ANTIGEN & NON-TOXIC).
12
QHLIYFRRKRSILWR
Set 9 from 425 to 439
154
1.2274(Probable ANTIGEN & NON-TOXIC)
13
IAAMIVNKTVHKMIF
Set 2 from 164 to 178
14
0.0057(Probable NON-ANTIGEN & NON-TOXIC)
14
GKSLLLDPPTNVRDY
Set 2 from 101 to 115
58
0.0791(Probable NON-ANTIGEN & NON-TOXIC).
15
SLILIQGIKTLPILE
Set 1 from 9 to 23
5
0.6252(Probable ANTIGEN & NON-TOXIC).
16
TAGLIKNQNNLVCRL
Set 10 from 545 to 559
16
0.7029(Probable ANTIGEN & NON-TOXIC).
17
ERTFSLINRHAIDFL
Set 10 from 580 to 594
1
0.9379(Probable ANTIGEN & NON-TOXIC).
18
SKRWAFRTGVPPKNV
Set 2 from 67 to 81
85
0.6970(Probable ANTIGEN & NON-TOXIC).
19
GNIAAMIVNKTVHKM
Set 2 from 163 to 177
1
0.2493(Probable NON-ANTIGEN & NON-TOXIC).
20
ISLILIQGIKTLPIL
Set 1 from 8 to 22
2
0.7761(Probable ANTIGEN & NON-TOXIC).
21
GKVFTEGNIAAMIVN
Set 2 from 156 to 170
142
0.0750(Probable NON-ANTIGEN & NON-TOXIC).
22
HLWGAFFLYDRIAST
Set 2 from 137 to 151
116
0.3992(Probable NON-ANTIGEN & NON-TOXIC)
23
KVFTEGNIAAMIVNK
Set 2 from 157 to 171
62
0.2910(Probable NON-ANTIGEN & NON-TOXIC).
24
VFTEGNIAAMIVNKT
Set 2 from 158 to 172
311
0.4674(Probable ANTIGEN & NON-TOXIC).
25
EGNIAAMIVNKTVHK
Set 2 from 161 to 175
6
0.2464(Probable NON-ANTIGEN & NON-TOXIC).
26
FTEGNIAAMIVNKTV
Set 2 from 159 to 177
156
0.5746(Probable ANTIGEN & NON-TOXIC).
27
RGKVFTEGNIAAMIV
Set 2 from 156 to 172
71
0.2351(Probable NON-ANTIGEN & NON-TOXIC).
28
FDESSSSGASAEEDQ
Set 9 from 468 to 482
67
0.4303(Probable ANTIGEN & NON-TOXIC).
29
MFPFLDGLINAPIDF
Set 9 from 443 to 457
18
0.3241(Probable NON-ANTIGEN & NON-TOXIC).
3.4 Population coverage analysis
IEDB population coverage server was used for the identification of population coverage of our screened non-mutagenic, antigenic and non-toxic peptides. This analysis indicates that 11 peptides (ILIQGIKTLPILEIA, LILIQGIKTLPILEI, VFTEGNIAAMIVNKT, AFFLYDRIASTTMYR, FTEGNIAAMIVNKTV, QHLIYFRRKRSILWR, LIQGIKTLPILEIAS, SKRWAFRTGVPPKNV, WGAFFLYDRIASTTM, FDESSSSGASAEEDQ and TAGLIKNQNNLVCRL) show highest HLA coverage. Each region has 100% population coverage except South Africa, which makes an average of 96.79% of world population coverage, as mentioned in Fig. 4.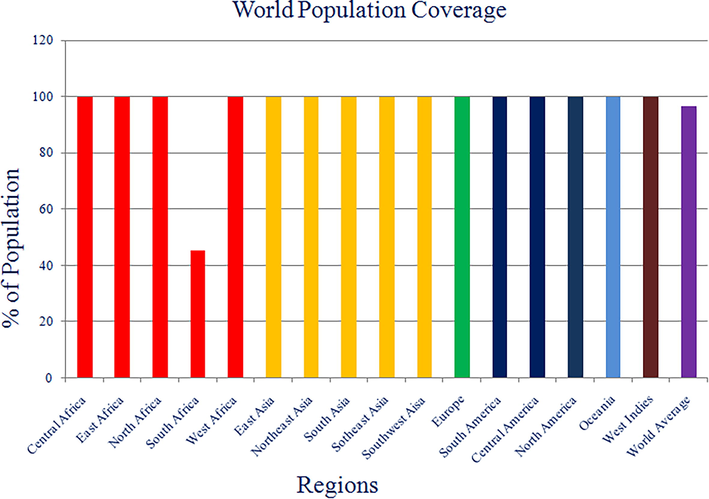
Bar-diagram representation of population coverage of final 11 epitopes against different regions of globe.
3.5 IFN-gamma inducing capacity and physiochemical prediction
IFN-gamma induction potential has been screened for all 11 epitopes by using two categories, IFN-G vs non-IFN-G and INF-G vs other cytokines. In first of category (INF-G vs non-INF-G) 05 out 11 epitopes ILIQGIKTLPILEIA, LILIQGIKTLPILEI, FFLYDRIASTTMYR, QHLIYFRRKRSILWR and LIQGIKTLPILEIAS were qualified to inducing INF-gamma potential. Meanwhile we have again cross checked all 11 epitopes in the second category (INF-G vs other cytokines), and we found that all 11 epitopes were qualified and had more potential to induce the IFN-gamma than other cytokines (Table 3). The physiochemical parameters of 3 epitopes QHLIYFRRKRSILWR, SKRWAFRTGVPPKNV and FDESSSSGASAEEDQ have hydrophilic nature; the rest of them were fall in hydrophobic in nature. AFFLYDRIASTTMYR, LIQGIKTLPILEIAS and WGAFFLYDRIASTTM epitopes were shown thermodynamically stable in biological systems, the rest of them were unstable. ILIQGIKTLPILEIA, LILIQGIKTLPILEI and LIQGIKTLPILEIAS were found to have good protein-binding potential according to Boman Index. All epitopes were showing estimated half-life less than 24hrs. expect VFTEGNIAAMIVNKT which was showing 100hrs (Table 4).
S.No.
Sequence
Method
IFN-gamma versus Non IFN-gamma
Score
Method
IFN-gamma versus other cytokine
Score
1
ILIQGIKTLPILEIA
SVM based
POSITIVE
0.19866307
MERCI
POSITIVE
1
2
LILIQGIKTLPILEI
SVM based
POSITIVE
0.26377164
SVM
POSITIVE
0.58082894
3
VFTEGNIAAMIVNKT
SVM based
NEGATIVE
−0.045341899
MERCI
POSITIVE
1
4
AFFLYDRIASTTMYR
SVM based
POSITIVE
0.18703322
SVM
POSITIVE
0.57522841
5
FTEGNIAAMIVNKTV
SVM based
NEGATIVE
−0.23191814
MERCI
POSITIVE
1
6
QHLIYFRRKRSILWR
SVM based
POSITIVE
0.28610167
MERCI
POSITIVE
17
7
LIQGIKTLPILEIAS
SVM based
POSITIVE
0.15795841
MERCI
POSITIVE
1
8
SKRWAFRTGVPPKNV
SVM based
NEGATIVE
−0.060456685
MERCI
POSITIVE
1
9
WGAFFLYDRIASTTM
SVM based
NEGATIVE
−0.01994069
MERCI
POSITIVE
1
10
FDESSSSGASAEEDQ
SVM based
NEGATIVE
−0.20096112
SVM
POSITIVE
0.40317329
11
TAGLIKNQNNLVCRL
SVM based
NEGATIVE
−0.42184921
MERCI
POSITIVE
2
S.No.
Sequence
Sequence Composition (In percentage)
Mol. Wt.
Estimated solubility & Instability index
Protein-binding Potential (Boman index)
Estimated half-life (Model: mammalian reticulocytes, in vitro).
1
ILIQGIKTLPILEIA
Acidic: 6.67
1635.18 g/mol
Poor water solubility, unstable
−1.44 kcal/mol
20 h
Basic: 6.67
Neutral: 26.67
Hydrophobic: 60
2
LILIQGIKTLPILEI
Acidic: 6.67
1677.27 g/mol
Poor water solubility, unstable
−1.65 kcal/mol
5.5 h
Basic: 6.67
Neutral: 26.67
Hydrophobic: 60
3
VFTEGNIAAMIVNKT
Acidic: 6.67
1607.97 g/mol
Poor water solubility, unstable
0.19 kcal/mol
100 h
Basic: 6.67
Neutral: 33.33
Hydrophobic: 53.33
4
AFFLYDRIASTTMYR
Acidic: 6.67
1855.2 g/mol
Poor water solubility, Stable
1.7 kcal/mol
4.4 h
Basic: 13.33
Neutral: 20
Hydrophobic: 60
5
FTEGNIAAMIVNKTV
Acidic: 6.67
1607.87 g/mol
Poor water solubility, unstable
0.19 kcal/mol
1.1 h
Basic: 13.33
Neutral: 20
Hydrophobic: 60
6
QHLIYFRRKRSILWR
Acidic: 0
2072.46 g/mol
Good water solubility, unstable
3.59 kcal/mol
0.8 h
Basic: 40
Neutral: 13.33
Hydrophobic: 46.67
7
LIQGIKTLPILEIAS
Acidic: 6.67
1609.09 g/mol
Poor water solubility, stable
−0.88 kcal/mol
5.5 h
Basic: 6.67
Neutral: 33.33
Hydrophobic: 53.33
8
SKRWAFRTGVPPKNV
Acidic: 0
1743.11 g/mol
Good water solubility, unstable
2.49 kcal/mol
1.9 h
Basic: 26.67
Neutral: 40
Hydrophobic: 33.33
9
WGAFFLYDRIASTTM
Acidic: 6.67
1779.11 g/mol
Poor water solubility, stable
0.48 kcal/mol
2.8 h
Basic: 6.67
Neutral: 26.67
Hydrophobic: 60
10
FDESSSSGASAEEDQ
Acidic: 33.33
1545.49 g/mol
Good water solubility, unstable
3.52 kcal/mol
1.1 h
Basic: 0
Neutral: 46.67
Hydrophobic: 20
11
TAGLIKNQNNLVCRL
Acidic: 0
1657.06 g/mol
Poor water solubility, stable
1.38 kcal/mol
7.2 h
Basic: 13
Neutral: 40
Hydrophobic: 46.67
4 Discussion
Immunization is one of the major, successful and cost-effective preventive strategy for community health to combat against the fatal infectious diseases globally (Chabot et al., 2004). Although, there is continuous development in the area of vaccines, classical vaccination like whole pathogen immunization is still popular. These types of immunizations are known to produce long lasting and strong immunity, but the major concern is that it may induce strong allergic reactions (Skwarczynski and Toth, 2016). So, peptide-based vaccines (PBVs) or multi epitope vaccines have now become a better choice for safe vaccination. PBVs are a striking alternative approach that depends on selection and usage of short peptide fragments to engineer the stimulation of extremely targeted immuno-protective responses, avoiding allergenic sequences (Li et al., 2014). In this context multi epitope-based vaccines are competent of stimulating strong immunogenic responses and safer option than whole protein-based vaccines. Earlier various studies in public domain are also encouraging the efficacy and impact of multiple epitopes based in silico vaccinology (Chaitra et al., 2005; Parida et al., 2007; Wiwanitkit, 2007; Gupta et al., 2010; Shey et al., 2019).
As we all are aware of antimicrobial resistance, which is a severe problem of healthcare at present and affecting millions of people around the globe. Antiviral resistance on the other hand, has been considered as a lesser threat than antibiotic resistance because unlikely drugs, vaccines are used for the prophylactic roles (Kennedy and Read, 2017). However, lately vaccine resistance is also becoming an important and inflammable problem.
Viruses are known for the high mutational rate in very short replication time, which is led by the nucleotide sequence context on the template molecule as well as by external environmental factors. This kind of genetic variation is the guarantee of virus survival in extreme conditions, as the significance of high rate mutation escort to formation of quasi-species or new viral strains (Lauring and Andino, 2010). Single-stranded RNA virus like influenza and marburg often carries error prone polymerases, which habitually induce at least one (range 0.1–10) incorrect base selection during every round of replication and initiate rapid materialization for vaccine resistant (Domingo and Holland, 1997). In this article authors pinpoint three nodes of vaccine that are identification of non-mutagenic, highly antigenic and non-toxic peptides from Env gylcoprotein of marburg virus (MRV), which may have potential to point at a direction in designing of a new vaccine to combat marburg virus induced infections.
Env gylcoprotein of any virus is supposed to be a probable target for the vaccine construction, because only Env gylcoprotein is accountable for the docking and connection of virus with any kind of human protein or receptor which lead the entry of virus into the cell (Janahi et al., 2017). In current study, authors focused on selection of non-mutagenic, immunogenic and non-toxic epitopes of Env gylcoprotein of MRV. As we know, viruses are known for their characteristics of high genomic mutation rates which provides the protection coverage for the virus and this is one of the major causes of vaccine failure (Laughlin et al., 2015). Considering this problem, we have focused on 37 variable strains of Env glycoproteins with 89–90% homology. Selected amino acid sequences were considered as input data for Protein Variability Server (PVS) to identify the most probable hot spots of mutations in Env glycoprotein. After implementation of PVS techniques, 10 different non variable/mutagenic and antigenic fragments of Env glycoprotein were generated. Out of 10, 8 fragments were selected for further screening, those have at least equal to or more than 15 peptides length. In this study authors have only focused on the selection and identification of CD4 + T cell mediated immunity because, CD4 + T-cells activation is an initial and mandatory factor for a competent humoral immune response for the induction of immunoglobulin-G and memory B-cells. CD4 + T-cells primarily recognized by antigen peptides by CD4 co-receptor and only recognized by the major histocompatibility complex (MHC) II protein on antigen-presenting cells; then memory B cells construct a repository of infected virus for the further prophylaxis arrangement (Clem, 2011). Cytotoxic T-cells or TCD8 + have different roles in immunity, which is related to therapeutic understanding not a prophylaxis segment. That’s why for rapid and cost-effective development for prophylaxis vaccine development CD4 + T cell alone is capable and important to induce protective response. NetMHCPanII server was used for the identification of predicted putative CD4 + T-cell epitopic core sequences in each of fragments of Env proteins along with their respective binding HLAs. Only CD4 + T-cell epitopes were chosen because CD4 + T-cells are the only immune cells that initially identify antigenic proteins and forms major histocompatibility complex (MHC) II, followed by setting up the configuration of memory B cells for the further prophylaxis arrangement (Gupta et al., 2010). Total 5224 HLAs were listed in the NetMHCpan server (DQ: 2912, DP: 2247, DRB4: 06, DRB1: 15, DRB3: 29, DRB5:15). All eight fragments were screened with all 5224 HLAs. After generation of this huge data, we chose only strong peptide binder among all 8 sets of fragments with lowest affinity score. Best 29, strong binder epitopes were extracted, as mentioned in Table 2. Interferon -gamma (IFN-gamma) potential was also evaluated for all 11 epitopes, and we found in category 1 (IFN-gamma vs Non IFN-gamma) 5 out 11 epitopes were showing potential to induce the IFN-gamma response against viral infection and among 5 epitopes our best top two selected epitopes (ILIQGIKTLPILEIA: 1034 HLAs and LILIQGIKTLPILEI: 336 HLAs) were present those having highest HLA coverages. Second category (IFN-gamma vs other cytokinin) were showed all 11 epitopes were showing better potential to induce the IFN-gamma as compared to other cytokines (details as mentioned in Table 3). About the physiochemical characteristics we found that most of finally selected epitopes have broad range of HLA coverage, but they were not showing good solubility in water and thermodynamically unstable in biological system, since its very usual with all small peptides and nucleosides, they are very prone to degradation via circulating proteases and nucleases. These enzymes are abundantly found in the biological systems, that’s why in this case authors will suggest the nanoparticle (NP)-formulation coting of peptides to shield themselves from proteases enzymes. NPs have the ability to transport weak antigens or vaccines to the mature DCs within the secondary lymph organs. Nano formulation is able to protect peptides, from degradation by proteases. By using NPs as a delivery system, we can initiate stronger immune responses. Once NPs reaches DCs, then controlled release of epitopes can be achieved through chemical modification on their surface, thus activation of DCs can be achieved more efficiently (Jia et al., 2018). Based on all the previously mentioned information we strongly believe that, nano formation system can prove to be an effective and potentiating delivery system for multi-epitopes. Even recently developed COVID-19 vaccine by Pfizer–BioNTech and Moderna also encapsulating the mRNA in lipid nanoparticles (Chaudhary et al., 2021) to save them from nucleases enzymes in biological system. Biodegradable nanoparticles generally made up of poly(D,L-lactic acid-co-glycolic acid)/PLGA are approved for the use of human (Elmowafy et al., 2019). PLGA based peptide NPs are very popular and efficient option of vaccine delivery system for targeting DCs and the development of DCs based cellular vaccines (Athanasiou et al., 2017). So, in that case authors will suggest PLGA nano-formulation will be most suitable candidate for this encapsulation while it already approved by FDA in drug formulations and able to protect the non-water soluble and thermodynamically unstable epitopes. Furthermore, all 29 epitopes were screened based on antigenicity and final 11 epitopes were selected as probable multi epitopes vaccine candidates, with non-toxic properties and high world population coverage (as shown in Fig. 5). This result shows that the proposed epitopes would be significant vaccine contenders for large proportion of the human population which is around 96.78% globally. In short, this study generously focused on a strong prophylactic intervention against MRV with very low possibility of resistance, highly antigenic, non-toxic and high world population coverage. However, the T-cell stimulation potential of the predicted putative CD4 + T-cell epitopes are required to be validated by wet lab experiments for their efficient use as peptide vaccine candidates against marburg virus (MRV).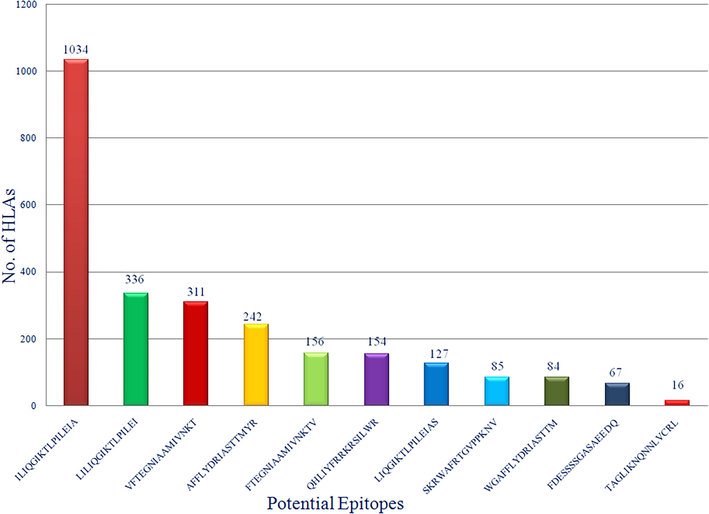
Bar-diagram representation of final 11 epitopes with their HLAs coverage analysis.
5 Conclusion
In this study, immuno-informatics tools were employed to design a putative vaccine peptide coding for multiple T-cell CD4 + epitopes. Total 11 peptides were minutely screened based on high antigenicity, non-mutagenic, non-toxic and broad HLA coverage. This computer-based study uses strong technical and logical methodologies that help this study to become more precise and reproducible in real time models. The authors are very hopeful, however, the T-cell stimulation potential of these predicted peptides containing the core amino acid sequences are to be validated by using in vitro and in vivo experiments for their competent use as multiepitope vaccine candidates against MRV infection. This study can be highly useful for designing newer vaccine strategies to prevent and/or lower the death toll attributed to MRV infection in future.
Acknowledgments
The authors are grateful to UTRGV and funding agencies. This work was partially supported by the National Institute of Health/National Cancer Institute’s funding: R01 CA210192, R01 CA206069, R01 CA204552 awarded to SCC and UTRGV Start up. Authors are also thankful to honourable Vice Chancellor of Swami Rama Himalayan University, India for providing necessary manual assistance and software supports.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Aerosol exposure to the angola strain of marburg virus causes lethal viral hemorrhagic Fever in cynomolgus macaques. Vet. Pathol.. 2010;47(5):831-851.
- [Google Scholar]
- A Poly(Lactic-co-Glycolic) Acid Nanovaccine Based on Chimeric Peptides from Different Leishmania infantum Proteins Induces Dendritic Cells Maturation and Promotes Peptide-Specific IFNγ-Producing CD8(+) T Cells Essential for the Protection against Experimental Visceral Leishmaniasis. Front. Immunol.. 2017;8:684.
- [Google Scholar]
- Bano, T., Mohammed Janahi, E., Dhasmana, A., Lohani, M., Haque, S., R, K.M., S, A.D., Jawed, A., Wahid, M., Akhter, N., and M, Y.A. (2018). In silico CD4+, CD8+ & humoral immunity associated antigenic epitope prediction and HLA distribution analysis of HTLV-I. J. Buon 23, 1514-1527.
- Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinf.. 2006;7:153.
- [Google Scholar]
- The societal value of universal childhood vaccination. Vaccine. 2004;22(15-16):1992-2005.
- [Google Scholar]
- Defining putative T cell epitopes from PE and PPE families of proteins of Mycobacterium tuberculosis with vaccine potential. Vaccine. 2005;23(10):1265-1272.
- [Google Scholar]
- mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat. Rev. Drug Discovery. 2021;20(11):817-838.
- [Google Scholar]
- Fundamentals of vaccine immunology. J. Glob. Infect. Dis.. 2011;3(1):73.
- [CrossRef] [Google Scholar]
- Epidemiologic investigation of Marburg virus disease, Southern Africa, 1975. Am. J. Trop. Med. Hyg.. 1978;27:1210-1215.
- [Google Scholar]
- Post-exposure treatments for Ebola and Marburg virus infections. Nat. Rev. Drug Discov.. 2018;17(6):413-434.
- [Google Scholar]
- RNA virus mutations and fitness for survival. Annu. Rev. Microbiol.. 1997;51(1):151-178.
- [Google Scholar]
- VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinf.. 2007;8:4.
- [Google Scholar]
- Biocompatibility, biodegradation and biomedical applications of poly(lactic acid)/poly(lactic-co-glycolic acid) micro and nanoparticles. J. Pharmaceut. Investigat.. 2019;49(4):347-380.
- [Google Scholar]
- PVS: a web server for protein sequence variability analysis tuned to facilitate conserved epitope discovery. Nucleic Acids Res.. 2008;36(Web Server):W35-W41.
- [Google Scholar]
- In silico approach for predicting toxicity of peptides and proteins. PLoS ONE. 2013;8(9):e73957.
- [Google Scholar]
- In silico CD4+ T-cell epitope prediction and HLA distribution analysis for the potential proteins of Neisseria meningitidis Serogroup B–a clue for vaccine development. Vaccine. 2010;28(43):7092-7097.
- [Google Scholar]
- In silico CD4+, CD8+ T-cell and B-cell immunity associated immunogenic epitope prediction and HLA distribution analysis of Zika virus. EXCLI J. 2017;16:63-72.
- [Google Scholar]
- Interactions Between Nanoparticles and Dendritic Cells: From the Perspective of Cancer Immunotherapy. Front. Oncol.. 2018;8:404.
- [Google Scholar]
- Why does drug resistance readily evolve but vaccine resistance does not? Proc. R. Soc. B.. 2017;284(1851):20162562.
- [CrossRef] [Google Scholar]
- Quasispecies theory and the behavior of RNA viruses. PLoS Pathog.. 2010;6(7):e1001005.
- [Google Scholar]
- Peptide Vaccine: Progress and Challenges. Peptide Vaccine: Progress and Challenges. Vaccines (Basel). 2014;2(3):515-536.
- [Google Scholar]
- Nielsen, M., Lundegaard, C., Blicher, T., Peters, B., Sette, A., Justesen, S., Buus, S., and Lund, O. (2008). Quantitative predictions of peptide binding to any HLA-DR molecule of known sequence: NetMHCIIpan. PLoS Comput Biol 4, e1000107.
- Computational analysis of proteome of H5N1 avian influenza virus to define T cell epitopes with vaccine potential. Vaccine. 2007;25(43):7530-7539.
- [Google Scholar]
- In-silico design of a multi-epitope vaccine candidate against onchocerciasis and related filarial diseases. Sci. Rep.. 2019;9:4409.
- [Google Scholar]
- A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. U.S.A.. 1997;94(26):14764-14769.
- [Google Scholar]
- Marburgvirus genomics and association with a large hemorrhagic fever outbreak in Angola. J. Virol.. 2006;80(13):6497-6516.
- [Google Scholar]
- Predicted epitopes of Lig A of Leptospira interrogans by bioinformatics method: a clue for further vaccine development. Vaccine. 2007;25(15):2768-2770.
- [Google Scholar]
- Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J. Virol.. 1998;72(4):3155-3160.
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2021.101751.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







