Translate this page into:
Impact of a benzimidazole salt on gene expression, cytotoxicity, and apoptotic processes in HepG2 cell line
*Corresponding author: E-mail address: esra.bilici@usak.edu.tr (E. Bilici)
-
Received: ,
Accepted: ,
Abstract
HepG2 is crucial in liver research because of its distinctive characteristics and significant applicability in drug metabolism, hepatotoxicity, and liver diseases. Benzimidazole derivatives are potential medicinal agents for the treatment of liver cancer. A new benzimidazole-based compound was created and verified using 1H NMR, 13C NMR, and IR. We assessed the effects of a novel benzimidazole salt compound 3 on HepG2, DLD-1, MCF-7, and HEK-293T cells in culture. Compound 3 was administered to each of the three cancer cell lines for 72 hours, and the IC50 values were recorded. The IR, ADMET, real-time PCR, and the MTT assay were used to examine the effects of 3. The viabilities of MCF-7, HepG2, and DLD-1 cells after 72 hours were 22.41 µM, 25.14 µM, and 41.97 µM, respectively. In HepG2 cells, the expression levels of many molecules relevant to pathways were measured in terms of mRNA and protein. Compound 3 exhibited significant cytotoxicity, with IC50 values of 25.14 µM for HepG2, 22.41 µM for MCF-7, and 41.97 µM for DLD-1 cells, demonstrating selectivity toward cancer cells. Real-time PCR revealed elevated expression levels of pro-apoptotic markers (BAX, CASPASE-3, and CASPASE-8) and anti-apoptotic marker BCL-2, suggesting the induction of both intrinsic and extrinsic apoptotic pathways. The ADMET analysis highlighted favorable pharmacokinetic properties, including blood-brain barrier permeability and high gastrointestinal absorption. The novel benzimidazole compound 3 demonstrated significant anticancer activity and apoptotic potential in vitro, particularly against HepG2 cells. Its favorable ADMET profile and mechanism of action suggest its potential as a therapeutic agent for liver cancer. Future studies should focus on in vivo validation and further optimization of its pharmacological properties.
Keywords
Apoptosis
Bcl-2
Benzimidazole
Cytotoxicity
Gene expression
HepG2
RT-qPCR
1. Introduction
Due to rising rates of drug resistance, there is an immediate need for innovative antibiotics and anticancer medicines. A multifaceted approach, including innovative discovery methods, enhanced stakeholder collaboration, and strategic investments in research and development is required (Pham et al., 2022) to address such challenges. Novel discovery approaches, more stakeholder participation, and focused research and development expenditures are essential to combat the increasing resistance rates to antibiotics and cancer treatments. This comprehensive strategy aims to revive drug development and address the primary challenges in both industries. It is imperative to discover novel, more secure, and more efficient anticancer agents and antibiotics to address the accelerating issues of drug resistance and the efficacy of cancer treatments. Recent indicates that various compounds derived from microbial metabolites and advanced synthetic methods may enhance treatment outcomes (Dokla et al., 2020). The potent complexes that benzimidazoles may generate with nucleic acids are what could give them their anticancer properties. In addition to exhibiting additional effects like telomerase suppression, topoisomerase poisoning, and inhibition of gene transcription, these complexes have the potential to cause damage to DNA (Hedge et al., 2015; Ibrahim et al., 2018). Furthermore, it has been discovered that a few of these substances have high binding affinities with DNA and positive anticancer activity when tested against cell lines (Mamedov et al., 2022). After actively entering cells, cisplatin’s anticancer action damages DNA (Wei et al., 2015). According to Davis et al. (2014), DNA damage causes DNA replication, cell cycle arrest, and the start of cellular death. Due to chemotherapy resistance being a significant challenge in cancer treatment, much research is directed towards new bioactive chemicals (Oiso et al., 2012).
The biological process of apoptosis, also referred to as programmed cell death, is essential for preserving cellular homeostasis and getting rid of unhealthy or malignant cells. It is an important target for the creation of novel cancer therapies. Changing apoptotic pathways has the potential to overcome resistance mechanisms that often impede the efficacy of cancer treatments and increase their efficiency. The apoptotic process is often triggered by DNA damage or unchecked growth (Chen et al., 2018). The intrinsic and extrinsic paths, linked to intracellular and extracellular signals, are the two processes that initiate apoptosis (Carneiro and El-Deiry, 2020). While the extrinsic route is mostly activated by signals from the immune system, particularly via interactions with death receptors on the cell membrane, the intrinsic path is primarily triggered by DNA damage and cytokine deficiency (Cao et al., 2021). Executioner caspases, a class of cysteine caspases, break target proteins necessary for regular cell function and are the merging point of both pathways (Chen et al., 2018). When caspases are activated, apoptotic cells shrink, and the plasma membrane is altered, which eventually results in cell death (McArthur and Kile, 2018). The intrinsic process is regulated by the B-cell lymphoma-2 (BCL-2) protein family (Wang et al., 2021). BCL-2 proteins prevent apoptosis by lowering the amounts of pro-apoptotic BCL-2, Bcl-2-associated X protein (BAX), and Bcl-2 homologous antagonist killer (Green, 2022). BH3-only proteins suppress anti-apoptotic BCL-2 proteins (Hagar et al., 2023). An elevation in the BAX/BCL-2 ratio is often used as an indicator of apoptotic induction in cells (Yip and Reed, 2008).
Apoptosis is essential to cancer therapy. Numerous illnesses, including cancer, are brought on by any interference in the apoptotic process (Safwat et al., 2021). Consequently, the development of an innovative anticancer agent capable of reinstating normal apoptotic pathways is a feasible approach for cancer therapy (Cordeu et al., 2007). Tumor cells can undergo apoptosis due to a variety of natural and artificial factors (Sirion et al., 2012). The inhibition of apoptosis is a pivotal element in cancer genesis and progression, enabling cancer cells to escape programmed cell death, resulting in unchecked proliferation and therapeutic resistance (Hanahan and Weinberg, 2011). Despite having all the tools necessary for apoptosis, malignant cells frequently devise ways to thwart or interfere with systems that cause cell death (Hafezi and Rahmani, 2021). The knockout effect occurs when the apoptosis process, which suppresses cancer cell proliferation and viability, is impaired or dysfunctional (Alam et al., 2021).
We synthesized a new benzimidazole derivative for these purposes. The chemical structure was elucidated using several spectroscopic experiments. Furthermore, the in vitro anticancer activity of this drug was assessed against human cancer cell lines, including human embryonic kidney cells (HEK293-T), hepatocellular carcinoma (HepG2), breast cancer (MCF-7), and colorectal adenocarcinoma (DLD-1), with respect to cytotoxicity.
2. Materials and methods
2.1 1-(2-Methylbenzyl)-3-(4-methylbenzyl)-1H-benzo[d]imidazol-3-ium chloride (3)
Benzimidazole and potassium hydroxide (KOH) were solubilized in 10 mL of ethyl alcohol. After a 30-minute agitation of the mixture at room temperature, 4-methylbenzyl chloride was added and allowed to reflux for 6 hours. The reaction mixture was thereafter adjusted to the ambient temperature. The potassium chloride (KCl) was filtered, and the 1-(4-methylbenzyl) benzimidazole (2) was refined by crystallization in ethanol. The synthesized compound 2 (0.363 g) was taken in a clean Schlenk tube, and 5 mL of N,N-dimethylformamide (DMF) was added. 2-Methylbenzyl chloride (216 µL) was added to the resulting reaction mixture, and the reaction was continued at 80°C for 48 hours. The DMF was extracted in vacuo after the reaction ceased. The synthesized product 3 was purified by washing several times with diethyl ether and then with hot benzene (Scheme 1). The 1H NMR, 13C NMR, and IR spectra of compound 3 have been given in Figs. 1-3.
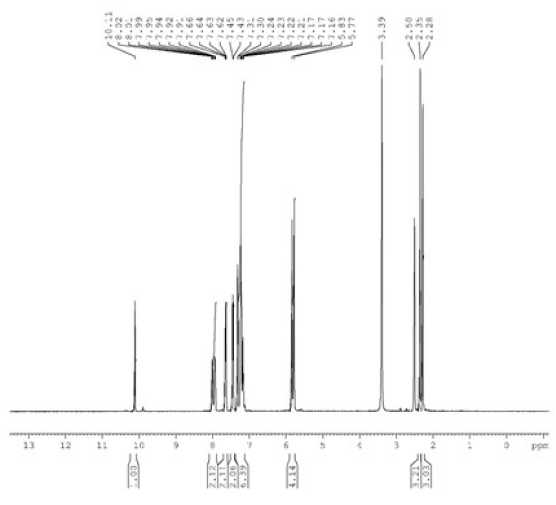
- The 1H NMR spectra of compound 3a. NMR: Nuclear magnetic resonance.
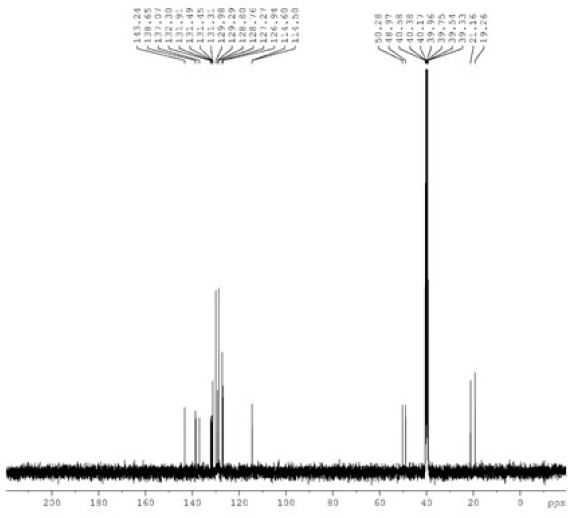
- The 13C NMR spectra of compound 3a. NMR: Nuclear magnetic resonance.
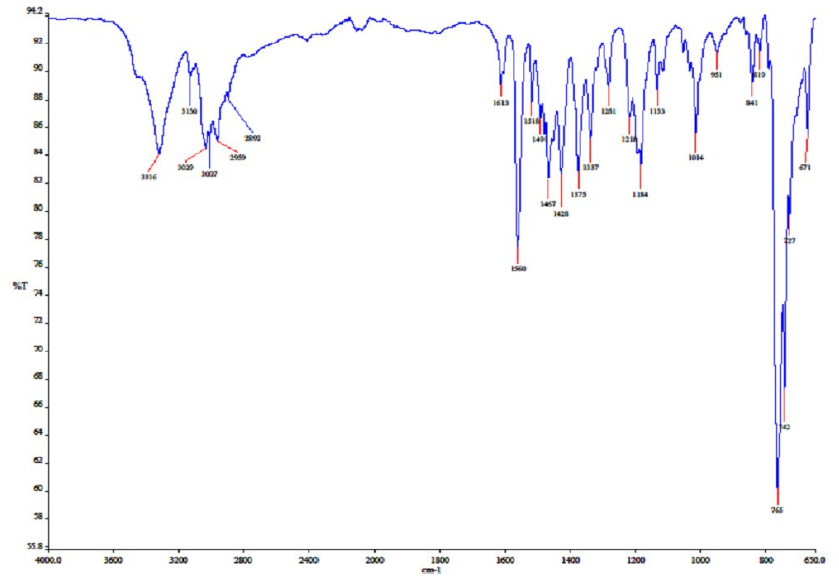
- The IR spectra of compound 3a. IR: Infrared.
M.p.: 187.8 °C, color: white. IR: 1560 (C=N); 3316, 3130, 3029, 3007, 2959, and 2892 cm-1 (C-H). 1H NMR (400.13 MHz, DMSO-d6, 298 K), δ: 2.28 and 2.50 [s, 6 H, NCH2C6H4(CH3)-2 and NCH2C6H4(CH3)-4]; 5.77 and 5.83 [s, 4 H, NCH2C6H4(CH3)-2 and NCH2C6H4(CH3)-4]; 7.16-8.02 (m, 8 H, Ar-H); 10.11 (s, 1 H, NCHN). 13C NMR (100.13 MHz, DMSO-d6, 298 K), δ: 19.26 and 21.16 [NCH2C6H4(CH3)-2 and NCH2C6H4(CH3)-4]; 48.97, and 50.28 (NCH2C6H4(CH3)-2 and NCH2C6H4(CH3)-4); 114.50, 114.60, 126.94, 127.27, 128.76, 128.80, 129.29, 129.98, 131.31, 131.45, 131.49, 131.91, 132.30, 137.07, and 138.65 (Ar-C); 143.24 (NCHN).
2.2 Cell culture
The American Type Culture Collection (ATCC, USA) supplied the human epithelial breast adenocarcinoma cell line MCF-7 (ATCC® HTB-22TM), HepG2, and DLD-1 (ATCC® CCL-221TM). The healthy human cell line HEK-293T was used. Cytotoxicity studies were conducted in accordance with established procedures (Akkoç, 2020). HepG2, DLD-1, and HEK-293T cells were cultivated in DMEM supplemented with 10% FBS and 1% Glutamax. MCF-7 cells were cultivated in RPMI rather than DMEM.
2.3 Microculture tetrazolium (MTT) assay
In sterile 96-well plates, HepG2, DLD-1, MCF-7, and HEK-293T cells were added at a density of 5 x 103 per well. After 24 hours, cells were exposed to the chemical at four concentrations (200, 100, 50, and 25 µM) for 72 hours. The chemical was assessed in the MCF-7 cell line at concentrations of 200, 100, 50, 25, and 12.5 µM. Each well was incubated for 2 hours with a stock solution of MTT at a concentration of 5 mg/mL after the incubation period. A Promega plate reader recorded an absorbance of 590 nm. The IC50 values were determined using GraphPad Prism 5.
2.4 RNA isolation and cDNA synthesis
HepG2 cells were cultivated in DMEM supplemented with 10% FBS after being seeded at a density of 5.0 × 104 cells/cm2 in six-well plates. A.B.T.TM Blood/Tissue RNA Purification Kit for Leukemia was used to isolate total RNA after cells were collected and pelleted after a few days. Using nanodrop (Thermo ScientificTMNanoDropTM 2000/2000c Spectrophotometers), the concentration of total RNA was determined. The VitaScriptTM FirstStrand cDNA Synthesis Kit was used to synthesize cDNA by using 0.5 μg of total RNA as a template for reverse transcription. cDNA was used as a template for qPCR after being diluted 1:20 in nuclease-free water.
2.5 Real-time RT-PCR analysis
cDNA was generated from RNA by a reverse transcription process utilizing an RT-PCR kit (CFX96 Touch Real-Time PCR Detection System). Primers for CAS3, CAS8, BAX, BCL-2, and β-actin proteins were produced by Sangon Biotechnology Company. Primer sequences have been shown in Table 1. The LightCycler equipment (Roche Diagnostic, Mannheim, Germany) was used for real-time PCR in a 25 μL volume, with 12.5 μL of SYBR Green I for fluorescence detection. PCR thermal cycling was 5 minutes at 95°C, 40 cycles of 15 seconds denaturation, and 30 seconds at 60°C annealing/extension. A final 30 second 72°C extension was done. Melting curve studies confirmed amplified product specificity. A reference gene of β-actin was used to preserve cDNA loading uniformity across samples. The β-actin levels were used to standardize the differences in gene expression between the treated and untreated cells. Using separate plates without templates allowed us to avoid contaminating the original sample with reagents. The ΔΔCt approach was used to assess gene expression; this method normalized the ΔCt of every gene to the average of untreated Hep-G2 cells. The equation (fold change = 2−Δ(ΔCt), where ΔCt = target gene - β-actin and Δ(ΔCt) = treated – untreated) was applied to determine the fold change in gene expression between the control and treated cells, with respect to β-actin as the standard. The Wilcoxon rank-sum test was used to assess the statistical importance by comparing ΔCt values (cycle count at logarithmic fluorescence threshold) among groups, with a two-sided significance level of p < 0.05. Results are presented as mean ± SD. Gene expression fold changes were estimated using average ΔΔCt values and the formula fold = 2−ΔΔCt (Hassan & Ibrahim, 2014; Korashy et al., 2017).
| Aim | Primer | Oligonucleotide array (5’-3’) | |
|---|---|---|---|
| cas3 | F | TGC TAT TGT GAG GCG GTT GT | |
| R | TCC AGA GTC CAT TGA TTC GCT | ||
| cas8 | F | AAG CTC TCC CCA AAC TTG CTT | |
| R | TGC ATA AAA AGA CCC CAG AGC A | ||
| bax | F | GCC GAA ATG TTT GCT GAC G | |
| R | CGC CAC GGT GCT CTC C | ||
| bcl-2 | F | ACT TTG CCG AGA TGT CCA GC | |
| R | ATC CCA GCC TCC GTT ATC CT | ||
| Β-actin | F | GCC AAC TTG TCC TTA CCC AGA | |
| R | AGG AAG AGA GAC TTG ACC CC | ||
PCR: Polymerase chain reaction, cDNA: Complementary DNA, F: Forward primer, R: Reverse primer, PCR: Polymerase chain reaction, cDNA: Complementary DNA, cas3: Caspase 3, cas8: Caspase 8, bax: Bcl-2-Associated X Protein, bcl-2: B-cell lymphoma 2, β-actin: Beta-actin.
2.6 ADME and toxicity (ADMET) properties
SMILE (CC1=CC=CC=C1CN(C=[N+]2CC3=CC=C(C)C=C3)C4=C2C=CC=C4.[Cl-]) of synthesized compound 3 was generated by using ChemBioDraw Ultra 14.0 software. This code was used to calculate ADME and Toxicity (ADMET) properties. To calculate ADME properties, SwissADME (Daina et al., 2017) webserver was used, while the Protox 3.0 web server was used to calculate toxicity properties (Banerjee, et al., 2018).
2.7 Statistical analysis
The SPSS software was used to conduct statistical analyses. The mean and standard deviation (SD) reflect the results of the student’s t-test and one-way analysis of variance (ANOVA). Three distinct experiments in triplicate were conducted. Results were regarded as significant if the p-value was <0.05.
3. Results
3.1 Synthesis of a new compound, 3
The structure of compound 3 was verified by 1H NMR, 13C NMR, and IR. In 1H NMR, it was seen that the protons belonging to the methyl groups (CH3) resonated at δ 2.28 and 2.50 ppm as singlet signals. It was determined that the methylene protons (CH2) resonated at δ 5.77 and 5.83 ppm in the lower field as a singlet. One of the most important signals belonging to NCHN, which shows the accuracy of the structure, gave a singlet signal at δ 10.11 ppm in 1H NMR spectra. The protons of the aromatic groups were found to resonate as multiplets between 7.16 ppm and 8.02 ppm, confirming the structure. In 13C NMR spectra, the signals at δ 19.26 and 21.16 ppm represent CH3 carbons and the signals at δ 48.97 and 50.28 ppm represent NCH2 carbons. The signal at δ 143.24 indicates the presence of an NCHN carbon in the benzimidazole ring. In the IR spectra, the signal at 1560 cm-1 indicates the presence of a C=N double bond in the benzimidazole ring.
3.2 Cytotoxicity of compound 3 on various cell lines
MTT assay was performed to determine the viability of MCF-7, HepG2, DLD-1, and HEK-293T cells after treatment with 1-(2-methylbenzyl)-3-(4-methylbenzyl)-1H-benzo[d]imidazol-3-ium chloride. The cytotoxicity activity of compound 3 for 72 hours has been expressed as IC50 values as shown in Table 2.
| Compounds | IC50 (µM) | |||
|---|---|---|---|---|
| MCF-7 | HepG2 | DLD-1 | HEK-293T | |
| 3 | 22.41 | 25.14 | 41.97 | 38.46 |
| Cisplatin | 83.06* | 37.32 | 53.35 | 14.88 |
*for paclitaxel, IC50: Half Maximal Inhibitory Concentration, MCF-7: Michigan Cancer Foundation-7, HepG2: Hepatocellular carcinoma G2, HEK-293T: Human Embryonic Kidney 293T
The MTT test results revealed that compound 3 exhibited strong cytotoxicity at a concentration of 200 μM against cancer cell lines MCF-7, HepG2, DLD-1, and normal cell line HEK-293T. The MTT test results of paclitaxel, tested as a control group, did not exhibit significant anti-cancer activity in the MCF-7 cell line at the determined concentrations compared to cisplatin used in other cell lines (Fig. 4).
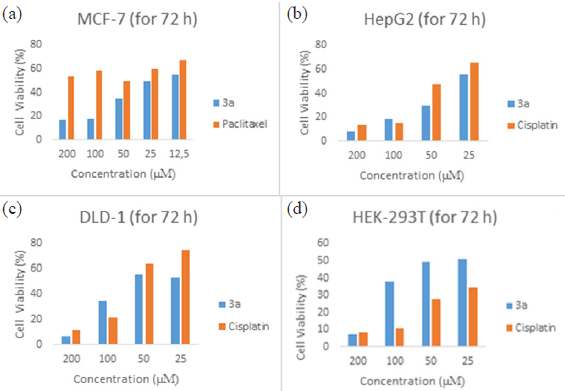
- Effects of cisplatin and compound 3a on human cancer and normal cell survival. The following drug concentrations were applied to the cells for the specified amounts of time: (a) 3a and paclitaxel (25–200 µM) were given to MCF-7 cells for 72 hours; (b) 3a and cisplatin (25–200 µM) were given to HepG2 cells for 72 hours; (c) 3a and cisplatin (25–200 µM) were given to DLD-1 cells for 72 hours; and (d) 3a and cisplatin (25–200 µM) were administered to HEK-293T cells. The MTT test was used to determine cell viability
The hepatocyte cell model showed higher sensitivity to compound 3 exposures compared to DLD-1 cells, presenting lower IC50 values for 72-hour exposure. When compared between cell lines, the IC50 value of compound 3 for DLD-1 cells was found to be 1.5 times higher than the IC50 values of HepG2 cells at 72 hours of exposure, indicating that DLD-1 cells have lower sensitivity to compound 3. Therefore, the HepG2 cell line was selected to determine different mechanisms of 3-induced cytotoxicity.
3.3 Expression analysis of Bax, Bcl-2, Caspase-3 and Caspase-8 genes using real-time PCR
The activation of essential effector caspase-3 and initiator caspase-8 was evaluated in HepG2 cells during a real-time PCR experiment to verify the anti-apoptotic properties of compound 3. The levels of the pro-apoptotic proteins BAX, CASPASE-8, CASPASE-3, and the anti-apoptotic protein BCL-2 were measured using real-time quantitative polymerase chain reaction (RT-PCR) to clarify the mechanism by which the tested medications caused cell death (Fig. 5).
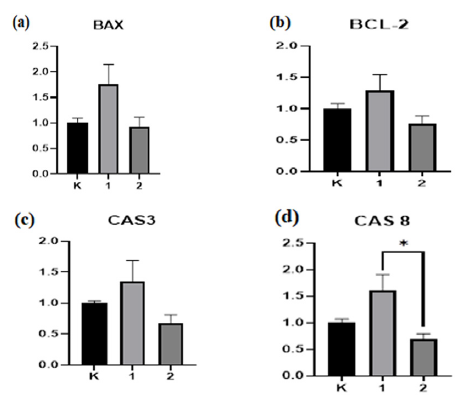
- Effect of compound 3a on the levels of CASPASE-3, CASPASE-8, BCL-2, and BAX mRNA expression in HepG2 cells. Following treatment with 3a, the mRNA expression levels for (a) BAX, (b) BCL-2, (c) CASPASE-3, and (d) CASPASE-8 were measured. The information is presented as the average ± standard error of the average obtained from three separate trials. *P < 0.05 in relation to the group under control. CASPASE-3: Cysteinyl aspartate specific protease 3, CASPASE-8: Cysteinyl aspartate specific protease 8, BCL-2: B-cell lymphoma 2, BAX: Bcl-2-Associated X Protein, HepG2 cells: Human liver cancer cell line 2, RT-qPCR: Reverse transcription-quantitative polymerase chain reaction.
The compound elevated CASPASE-3 expression levels in HepG2 cells compared to the control group. Bax expression was elevated. CASPASE-8 expression levels were significantly increased. Bcl-2 expression was significantly elevated compared to control cells not exposed to 3. Consequently, compound 3 elevated the BAX levels relative to the control, indicating its ability to trigger apoptosis in HepG2 cells.
The expression of the bax genes increased after compound 3 treatment. We observed that 3-treated cells had an elevated BAX ratio. Furthermore, the gene expression of the CASPASE-3 protein elevated following treatment and emerged as the most potent anticancer agent. CASPASE-3 facilitates apoptosis. IC50 Treatment of HepG2 cells for 72 hours modifies gene expression. HepG2 cells subjected to compound 3 for 72 hours exhibited elevated levels of BAX, BCL-2, CASPASE-3, and CASPASE-8 compared to the control group, demonstrating its effectiveness in cancer cells. The difference between Cas8 and the control group was statistically significant (*p<0.001). Each experiment was performed thrice, using β-actin as an internal control.
3.4 ADME and toxicity (ADMET) properties
The four-letter acronym ADME, which stands for absorption, distribution, metabolism, and excretion, is mostly employed in the fields of pharmacology and pharmacokinetics. The four letters represent descriptors, which quantify the way a certain medication affects the body over time. Thus, determining ADME parameters of new substances is crucial in the drug discovery field. These parameters consist of physicochemical, lipophilicity, solubility, pharmacokinetics, and drug-likeness. Here, the SwissADME web server was used to determine these parameters, and Table 3 presents the findings.
| Physicochemical properties | Druglikeness properties | |||
|---|---|---|---|---|
| Properties | Value | Requirement | Value | Compatible |
| Compound 3 | Compound 3 | Compound 3 | ||
| Lipinski’s rule | ||||
| Molecular formula | C23H23ClN2 | MW ≤ 500 | 362.90 |
No MLOGP>4.15 |
|
Molecular weight (MW, g/mol) |
362.90 |
M LOGP ≤ 4.15 |
4.90 | |
| Number of heavy atoms | 26 | HBA atoms ≤ 10 | 0 | |
| Number of aromatic heavy atoms (AHA) | 21 | HBD atoms ≤ 5 | 0 | |
| Number of rotatable bonds (RB) | 4 | Ghose’s rule | ||
| Number of H-bond acceptors (HBA) | 0 |
160 ≤ MW ≤ 480 |
362.90 | No |
| Number of H-bond donors (HBD) | 0 |
-0.4 ≤ WLOGP ≤ 5.6 |
1.65 | |
| Molar refractivity (MR) | 111.55 |
40 ≤ MR ≤ 130 |
111.55 | |
| TPSA (Å2) | 8.81 |
20 ≤ atoms ≤ 70 |
49 | |
| Lipophilicity | Veber’s rule | |||
| Log Po/w (iLOGP) | -2.42 | RB ≤ 10 | 4 | Yes |
| Log Po/w (XLOGP3) | 6.29 | TPSA ≤ 140 | 8.81 | |
| Log Po/w (WLOGP) | 1.65 | Egan’s rule | ||
| Log Po/w (MLOGP) | 4.90 | WLOGP ≤ 5.88 | 1.65 | Yes |
| Consensus Log Po/w | 3.08 | TPSA ≤ 131.6 | 8.81 | |
| Pharmacokinetics | Muegge’s rule | |||
| GI absorption | High | 200 ≤ MW ≤ 600 | 362.90 |
No XLOGP3 > 5 |
| BBB permeant | Yes | -2 ≤ XLOGP3 ≤ 5 | 6.29 | |
| P-gp substrate | Yes | TPSA ≤ 150 | 8.81 | |
| CYP1A2 inhibitor | No | Number of rings ≤ 7 | 4 | |
| CYP2C19 inhibitor | No | Number of carbon > 4 | 23 | |
| CYP2C9 inhibitor | No | Number of heteroatoms > 1 | 2 | |
| CYP2D6 inhibitor | Yes | RB ≤ 15 | 4 | |
| CYP3A4 inhibitor | No | HBA ≤ 10 | 0 | |
| Log Kp (skin permeation, cm/s) | -4.05 | HBD ≤ 5 | 0 | |
| Water solubility | Predicted toxicity properties | |||
| Log S (ESOL) Class | -6.39 Poorly soluble | LD50 (mg/kg) | 900 | |
| Log S (Ali) Class | -6.26 Poorly soluble |
Toxicity class (1 to 6, worst to best) |
4 | |
| Log S (SILICOS-IT) Class | -7.86 Poorly soluble | |||
MW: Molecular weight, MLOGP: Molecular LogP (Partition Coefficient), HBA: Hydrogen bond acceptors, HBD: Hydrogen bond donors, RB: Rotatable bonds, MR: Molar refractivity, TPSA: Topological polar surface area, Log Po/w (iLOGP): Logarithm of octanol/water partition coefficient (iLOGP), Log Po/w (XLOGP3): Logarithm of octanol/water partition coefficient (XLOGP3), Log Po/w (WLOGP): Logarithm of octanol/water partition coefficient (WLOGP), Log Po/w (MLOGP): Logarithm of octanol/water partition coefficient (MLOGP), GI Absorption: Gastrointestinal absorption, BBB: Blood-brain barrier, P-gp: P-Glycoprotein, CYP: Cytochrome P450, LD50: Lethal dose for 50% of the population, Log S (ESOL): Logarithm of solubility (ESOL method), Log S (Ali): Logarithm of solubility (Ali method)
Drug-likeness properties are crucial when evaluating ADME measures for new pharmaceuticals. Companies have contemplated regulations for new medication candidates. Organizations and regulations: Pfizer (Lipinski), Amgen (Ghose), GSK (Veber), Pharmacia (Egan), Bayer (Muegge). Lipinski’s Rule of Five is notable. Lipinski’s rule of five examines physicochemical and lipophilicity characteristics, including molecular weight, MLOGP, hydrogen bond acceptors, and donor atoms (Lipinski et al. 2001). Molecular weight must range between 150 to 500. MLOGP must not exceed 4.15. Fewer than ten hydrogen bond acceptor atoms and five donor atoms are required. Synthesized chemical 3 satisfies Lipinski’s rule of five by having a molecular weight below 500 Da, hydrogen bond donors below 5, hydrogen bond acceptors below 10, and an MLOGP over 4.15. It satisfies all criteria except for MLOGP. A substance that satisfies three of Lipinski’s requirements may overlook one and still be regarded as compliant with the criterion.
The Protox-II web server assessed the toxicity of chemical 3. IC50 values for hazardous dosages are often expressed in mg/kg of body weight. The median lethal dose (IC50) is the quantity at which 50% of subjects succumb. The GHS delineates toxicity classifications. The class was graded from worst to best, from first to sixth. Table 3 indicates that the IC50 of chemical 3 was 900 mg/kg, putting it in the fourth toxicity class.
In terms of gastrointestinal absorption, the boiled-egg model is significant. Substances that have white regions in the model are easier to absorb, while outside of white regions are not. Whether the material passes through the blood-brain barrier (BBB) is determined by the yellow area. A chemical is considered to have passed the BBB and to have the potential to treat problems of the central nervous system (CNS) if located in the yellow region of the model. When compound 3 was examined in terms of the boiled egg model, it can be said that the substance has the potential to be used for CNS disorders, because the substance is in the yellow region (Fig. 6).
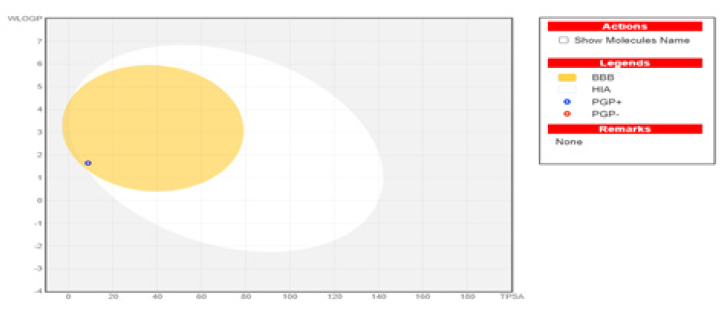
- Boiled-Egg Model. BBB: Blood-Brain Barrier, HIA: Human intestinal absorption, PGP+: P-Glycoprotein positive, PGP: P-Glycoprotein negative.
The bioavailability radar map (Fig. 7) indicated that chemical 3 was within the optimal range (pink zone) for lipophilicity, size, polarity, solubility, saturation, and flexibility. The outcome of this substance is not orally accessible as it exceeds the advised saturation limits. The medication exhibited water solubility, gastrointestinal absorption, and oral bioavailability, as anticipated. The permeability of Caco-2 for this molecule was predicted. The compound showed promise in addressing P-glycoprotein inhibitors and penetrating the BBB, presenting new opportunities for research on bioactive molecules in neurological diseases.
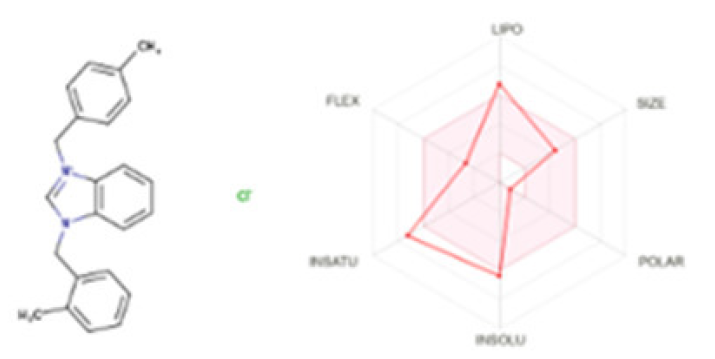
- Bioavailability radar graphic for benzimidazole salt compound 3a. The colored region represents the usual physicochemical area for oral bioavailability. The ideal value for each oral bioavailability factor is indicated in the pink area, while the predicted values for the analyzed molecule are shown in red lines.
4. Discussion
To progress in the medical domain, researchers are working on many projects to create novel chemicals with potent anticancer properties. Numerous anticancer medications can stop cancers from growing and stop them from spreading (Sakamaki et al, 2019). Of all the medications based on metals, platinum pharmaceuticals are the most widely used. Despite being one of the most often used medications for cancer treatment, cisplatin has drawbacks that restrict its application (Cheng, 2017). In our work, we synthesized 3 as a novel benzimidazole derivative to create potent anticancer prospects. The common anticancer chemotherapeutic drugs paclitaxel and cisplatin, which are employed in various other investigations, served as the chemotherapeutic references for our investigation.
The primary cause of cancer is a disturbance in the equilibrium between cell division and death. Numerous processes, including necrosis, autophagy, and apoptosis, lead to cell death. The physiological process by which programmed cell death takes place is called apoptosis. Cancer typically occurs when there is a disturbance in the apoptotic process. Dysregulation of the regular cell cycle is a characteristic shared by cancer patients. The activation of oncogenes results in the overexpression of growth factors that promote the expression of cyclin-dependent kinase/cyclin D protein. This could be the mechanism behind cancer. Cancer is caused by the overgrowth of cells that avoid the checkpoints of the cell cycle, which is triggered by this phase in the synthesis of proteins (Pucci et al., 2000; Malumbres and Barbacid, 2009).
In this study, a new compound was synthesized in three steps. The characterization of this compound was done in detail using IR and NMR data. Proton and carbon NMR data provide confirmation of the structure.
Compounds of 2-aryl benzimidazole, a novel class of derivatives, have been identified to trigger apoptosis in HepG2 cancer cells (Li et al., 2011). Ghani et al. (2012) investigated a platinum (II) compound in conjunction with 1H-benzimidazol-2-ylmethyl-N-(4-bromo-phenyl)-amine for its anticancer efficacy in MCF7 cells. The compound has an IC50 of 10.2 μM, whereas cisplatin has an IC50 of 9.9 μM. We used MTT, cell cycle analysis, ADMET, and molecular expressions of the antiapoptotic protein BCL-2, proapoptotic protein BAX, as well as CASPASE-3 and CASPASE-8 markers to elucidate the mechanism by which benzimidazole derivative 3 combats cancer in vitro. The MTT assay demonstrated a significant decrease in viability across MCF-7, HepG2, DLD-1, and HEK-293T cell lines, yielding IC50 values of 22.41, 25.14, 41.97, and 38.46 μM, respectively (Table 2). HepG2 cells were used to investigate the cytotoxicity pathways generated by 3. Subsequent research on many cancer cell lines and the mechanisms of action may uncover effective chemotherapeutic agents.
Numerous synthetic benzimidazole derivatives have undergone in vitro testing for breast, colon, and lung malignancies (Safwat et al., 2021). We discovered that 3, a new benzimidazole derivative, induced cytotoxicity in human cancer cell lines. Compound 3 induced concentration- and time-dependent intrinsic and extrinsic apoptotic pathways. Research into the mechanism of action is required. Our results provide the foundation for future research on the function of apoptosis in cancer therapy.
5. Conclusion
The present study successfully synthesized and analyzed a new benzimidazole-derived chemical (compound 3) exhibiting interesting anticancer attributes. Compound 3 exhibited substantial cytotoxicity against several cancer cell lines, particularly affecting HepG2 cells, and successfully regulated apoptotic pathways by enhancing pro-apoptotic markers (BAX, CASPASE-3, and CASPASE-8). Its exceptional ADMET profile highlights its potential as a therapeutic option. The results highlight the compound’s effectiveness in triggering apoptosis and overcoming chemoresistance, presenting it as a promising option for additional preclinical and in vivo investigations to assess its potential as a new therapy for hepatocellular carcinoma and other cancers.
CRediT authorship contribution statement
E. Bilici and S. Akkoc: Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Declaration of Generative AI and AI-assisted technologies in the writing process
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
References
- Design, synthesis, characterization, and in vitro cytotoxic activity evaluation of 1,2-disubstituted benzimidazole compounds. J. Phys. Org. Chem.. 2020;34:e4125..
- [Google Scholar]
- Synthesis, anticancer activity, and in silico modeling of alkylsulfonyl benzimidazole derivatives: Unveiling potent bcl-2 inhibitors for breast cancer. ACS Omega. 2024;9:9547-9563. https://doi.org/10.1021/acsomega.3c09411
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Novel palladium(II) and platinum(II) complexes with 1H-benzimidazol-2-ylmethyl-n-(4-bromo-phenyl)-amine: Structural studies and anticancer activity. Eur. J. Med. Chem.. 2012;47:399-411. https://doi.org/10.1016/j.ejmech.2011.11.008
- [CrossRef] [PubMed] [Google Scholar]
- B cell lymphoma 2: A potential therapeutic target for cancer therapy. Int. J. Mol. Sci.. 2021;22:10442. https://doi.org/10.3390/ijms221910442
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Apoptosis: A review of programmed cell death. Toxicol. Pathol.. 2007;35:495-516. https://doi.org/10.1080/01926230701320337
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Mitochondrial pathway-mediated apoptosis is associated with erlotinib-induced cytotoxicity in hepatic cells. Oncology letters. 2018;15(1):783-788.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res.. 2018;46:W257-W263. https://doi.org/10.1093/nar/gky318
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Changing profiles of cancer burden worldwide and in China: A secondary analysis of the global cancer statistics 2020. Chin Med J (Engl). 2021;134:783-791.
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol.. 2020;17:395-417. https://doi.org/10.1038/s41571-020-0341-y
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Multifunctional platinum-based nanoparticles for biomedical applications. Wiley. Interdiscip. Rev. Nanomed. Nanobiotechnol.. 2017;9(2) 9 2017 Mar https://doi.org/10.1002/wnan.1410
- [Google Scholar]
- Biological profile of new apoptotic agents based on 2,4-pyrido[2,3-d]pyrimidine derivatives. Bioorg. Med. Chem.. 2007;2007:15, 1659-1669..
- [Google Scholar]
- SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017:7. https://doi.org/10.1038/srep42717
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- “Platinum resistant” ovarian cancer: what is it, who to treat and how to measure benefit? Gynecol Oncol.. 2014;133:624-631.
- [CrossRef] [PubMed] [Google Scholar]
- Development of benzimidazole-based derivatives as antimicrobial agents and their synergistic effect with colistin against gram-negative bacteria. Eur. J. Med. Chem.. 2020;186:111850..
- [CrossRef] [PubMed] [Google Scholar]
- The mitochondrial pathway of apoptosis part II: The BCL-2 protein family. Cold Spring Harb Perspect Biol. 2022;14:a041046. https://doi.org/10.1101/cshperspect.a041046
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Targeting BCL-2 in cancer: Advances, challenges, and perspectives. Cancers (Basel). 2021;13:1292. https://doi.org/10.3390/cancers13061292
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Chalcone/1,3,4-Oxadiazole/Benzimidazole hybrids as novel anti-proliferative agents inducing apoptosis and inhibiting EGFR & BRAFV600E. BMC Chem.. 2023;16:17, 116..
- [Google Scholar]
- Hallmarks of cancer: The next generation. Cell. 2011;144:646-674. https://doi.org/10.1016/j.cell.2011.02.013
- [CrossRef] [PubMed] [Google Scholar]
- Some genetic profiles in liver of ehrlich ascites tumor-bearing mice under the stress of irradiation. J. Radiat. Res. Appl. Sci.. 2014;7:188-197. https://doi.org/10.1016/j.jrras.2014.02.002
- [CrossRef] [Google Scholar]
- A novel benzimidazole derivative binds to the DNA minor groove and induces apoptosis in leukemic cells. RSC Adv.. 2015;5:93194-93208. https://doi.org/10.1039/c5ra16605e
- [CrossRef] [Google Scholar]
- Design, synthesis, molecular modeling and anti-proliferative evaluation of novel quinoxaline derivatives as potential DNA intercalators and topoisomerase II inhibitors. Eur. J. Med. Chem.. 2018;155:117-134. https://doi.org/10.1016/j.ejmech.2018.06.004
- [CrossRef] [PubMed] [Google Scholar]
- Sunitinib inhibits breast cancer cell proliferation by inducing apoptosis, cell-cycle arrest and DNA repair while inhibiting NF-ĸB signaling pathways. Anticancer Res.. 2017;37:4899-4909.
- [CrossRef] [PubMed] [Google Scholar]
- Discovery of benzimidazole derivatives as novel multi-target EGFR, VEGFR-2 and PDGFR kinase inhibitors. Bioorg. Med. Chem.. 2011;19:4529-4535. https://doi.org/10.1016/j.bmc.2011.06.022
- [CrossRef] [PubMed] [Google Scholar]
- Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev.. 2001;46:3-26. https://doi.org/10.1016/s0169-409x(00)00129-0
- [CrossRef] [PubMed] [Google Scholar]
- Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer. 2009;9:153-166. https://doi.org/10.1038/nrc2602
- [CrossRef] [PubMed] [Google Scholar]
- Synthesis of morpholine-, piperidine-, and n-substituted piperazine-coupled 2-(Benzimidazol-2-yl)-3-arylquinoxalines as novel potent antitumor agents. ACS Pharmacol Transl Sci. 2022;5:945-962. https://doi.org/10.1021/acsptsci.2c00118
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Apoptotic caspases: Multiple or mistaken identities? Trends Cell Biol.. 2018;28:475-493. https://doi.org/10.1016/j.tcb.2018.02.003
- [CrossRef] [PubMed] [Google Scholar]
- Involvement of NF-κB activation in the cisplatin resistance of human epidermoid carcinoma KCP-4 cells. Oncol. Rep.. 2012;28:27-32. https://doi.org/10.3892/or.2012.1801
- [CrossRef] [PubMed] [Google Scholar]
- N,2,6-trisubstituted 1 h-benzimidazole derivatives as a new scaffold of antimicrobial and anticancer agents: Design, synthesis, in vitro evaluation, and in silico studies. RSC Adv.. 2022;13:399-420. https://doi.org/10.1039/d2ra06667j
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Cell cycle and apoptosis. Neoplasia. 2000;2:291-299. https://doi.org/10.1038/sj.neo.7900101
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- Synthesis, anticancer assessment, and molecular docking of novel chalcone-thienopyrimidine derivatives in hepG2 and MCF-7 cell lines. Oxid. Med. Cell. Longev.. 2021;2021:4759821. https://doi.org/10.1155/2021/4759821
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- trans-platinum(II) thionate complexes: Synthesis, structural characterization, and in vitro biological assessment as potent anticancer agents. Chempluschem. 2019;84:1525-1535. https://doi.org/10.1002/cplu.201900394
- [CrossRef] [PubMed] [PubMed Central] [Google Scholar]
- New substituted c-19-andrographolide analogues with potent cytotoxic activities. Bioorg. Med. Chem. Lett.. 2012;22:49-52. https://doi.org/10.1016/j.bmcl.2011.11.085
- [CrossRef] [PubMed] [Google Scholar]
- Antitumor activity of dual BCL-2/BCL-xl inhibitor pelcitoclax (APG-1252) in natural killer/T-cell lymphoma (NK/TCL) Blood. 2021;138:2062. https://doi.org/10.1182/blood-2021-152207
- [CrossRef] [Google Scholar]
- Renoprotective mechanisms of morin in cisplatin-induced kidney injury. Int. Immunopharmacol.. 2015;28:500-506. https://doi.org/10.1016/j.intimp.2015.07.009
- [CrossRef] [PubMed] [Google Scholar]
- Bcl-2 family proteins and cancer. Oncogene. 2008;27:6398-6406. https://doi.org/10.1038/onc.2008.307
- [CrossRef] [PubMed] [Google Scholar]







