Translate this page into:
Identification, molecular characterization, and plant growth promoting activities of endophytic fungi of Jasminum sambac, Camellia sinensis, and Ocimum basilicum
⁎Corresponding authors. bilal22000858@yu.ac.kr (Bilal Ahmed), assyed@ksu.edu.sa (Asad Syed)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Objectives
Endophytic fungi are widely known to reside in plant tissues causing no harm to host plants or even no noticeable change. They may colonize host plants for a part of their life cycle or may complete the entire life cycle since host provide a variety of novel metabolites. Despite being in a close relationship with medicinal host plants, the diversity and metabolic spectrum of endophytic fungi have not been researched well. Their diversity and ecological niche as plant symbionts make them attractive targets in the search for novel biochemicals.
Methods
We aimed to isolate, purify, and characterize the endophytic fungal population of medicinal host plants cultivated in Riyadh, Saudi Arabia. The goal was achieved through morphological identification and internal transcribed spacer (ITS) regions in rRNA analyses to determine phylogeny and diversity. Three aromatic and medicinal plants Jasminum sambac, Camellia sinensis, and Ocimum basilicum collected from the Riyadh region, Saudi Arabia were investigated. In total, 84 isolates were purified and they were grouped into 20 operational taxonomic units (OTUs) as per their sequences of ITS regions in rRNA.
Results
Twenty species of endophytic fungi were grouped in 12 genera i.e. Neopestalotiopsis, Trichoderma, Fusarium, Colletotrichum, Myrothecium, Chaetomium, Alternaria, Phoma, Curvularia, Cladosporium, Neodidymelliopsis, and Aspergillus and all isolates belonged to Ascomycota phylum. J. sambac was found dominant among other and had a relative frequency of 27%. C. sinensis was next with 18.7% relative frequency. The diversity was prominently recorded in leaf organs over stem and roots while roots exhibited the lowest diversity. Isolates also produced indole-3-acetic acid (IAA), 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase enzyme, and siderophores with variable magnitudes that could be assigned to their wide range of metabolic variations from species to species.
Conclusion
Conclusively, J. sambac, C. sinensis, and O. basilicum are a good source of endophytic fungi with certain plant growth-promoting traits. Overall, Alternaria was found as the most predominant genus in terms of colonization rate. Further deternminations are required to screen the beneficial compounds released by these endophytic fungi.
Keywords
Endophytic fungi
Isolation
Identification
ITS region
Medicinal and aromatic plants
1 Introduction
Fungi are profoundly important organisms in all ecosystems playing crucial roles in ecological functions such as recycling, transportation of nutrients, and organic matter decomposition (Mattoon et al., 2021). In general, medicinal plants are found loaded with endophytic fungi, and due to the presence of rich composition of metabolites in fungal population, they are considered for potent use in pharmaceutical industry (Bihari et al., 2011). Moreover, the type of association between fungal endophytes and plants have some influence on the phytochemical composition generally secondary metabolites (Faeth and Fagan, 2002). Therefore, the understanding of such associations will facilitate greater production of medicinal or aromatic compounds by utilizing the fungal relationship. Plants generally contains biomolecules of medicinal uses in all parts which have shown therapeutic potential against different infections. Besides the general range of useful compounds some medicinal plants also produce some secondary metabolites or marker compounds that are potentially useful for treating and/or preventing various ailments (Marques et al., 2020). Recently, specialized compounds from plants such as Vitex negundo, Mentha sp., Cymbopogon flexuosus, C. winterianus, Ocimum gratissimum, and Callistemon lanceolatus showed variable but potential inhibitory effects against clinical bacterial species due to variable nature of their medicinal or antioxidant composition (Sharma et al., 2020). The compounds derived from C. sinensis such as caffeine, catechins, and other compounds have shown antioxidant, antimicrobial, and anticancer activities (Aboulwafa et al., 2019). Likewise, flower extracts of J. sambac showed to have some useful bio-active compounds that were antioxidant, free radical scavengers, and non-toxic to human cells (Wu et al., 2021). Ocimum basilicum is yet another plant that produces various secondary metabolites like polyphenolic compounds (Pirbalouti et al., 2017) and provides several key benefits like antibacterial, antioxidant, and antiviral activities, and serves as hypoglycaemic, anticonvulsant, cytoprotective, and hypolipidemic agent (Rubab et al., 2017).
Due to these, it becomes imperative exploring the fungal endophytes of medicinal plants that could help finding and/or developing new alternative ways of treatments. When fungal endophytes colonize their host plants, the interaction also bring modulation of host’s physiology that may be associated with host’s response to abiotic and biotic stresses which usually benefits host plants, and therefore, fungal endophytic partners provide a native solution for crop protection and sustainable agriculture under challenging climatic conditions (Xie et al., 2020). This kind of distribution is certainly important since it determines secondary metabolites composition that may enhance the disease resistance of host plants to stresses while promoting its growth (Jia et al., 2016).
In this regard, the Arabian Peninsula particularly Kingdom of Saudi Arabia sustains a rich flora representing a significant source of medicinal plants in the Middle East (Rahman et al., 2002; Selim et al., 2012). Folk medicines based on this flora have been known since ancient times. Furthermore, medicinal plants from this flora can be exploited to produce essential drugs and other chemicals. These natural resources should be sustainably preserved and protected. Medicinal plants harbor endophytic fungi that play a vital role in the synthesis of natural pharmaceuticals (Rahman et al., 2002).
Therefore, we comprehensively designed the current study to achieve our goals: (i) collection of J. sambac, C. sinensis, and O. basilicum plant samples from the Riyadh region of Saudi Arabia, (ii) isolation of endophytic fungi, (iii) phenotypic characterization, (iv) molecular identification of fungal endophytes by ITS sequencing, and (v) plant growth promoting activities assessment of identified fungal isolates.
2 Materials and methods
2.1 Chemicals and materials
Potato dextrose broth (PDB; M403), agar-agar (GRM666), chloramphenicol (TC204), and tryptophan (M1339) were procured from Hi-Media Laboratories Pvt. Ltd., Mumbai, India. Agarose (137049), α-ketobutyrate (K401), and ethidium bromide (E7637) were purchased from Sigma-Aldrich, USA. All other chemicals such as ethanol and sodium hypochlorite were of analytical grade and purchased from Thermo Fisher Scientific, USA.
2.2 Collection of samples
Sampling was done from different locations of Riyadh regions namely Botanical Nurseries of Al-Muzahmiya and Al-Kharj Governorates in December (2018). There were samples of various parts including roots, leaves, and stems of aromatic and medicinal plants. Samples were brought to the laboratory with utmost care that were later used for the selective isolation and purification of fungal endophytes. Samples were thoroughly washed first with running tap water followed by sterile distilled water to remove soil and dust particles, and tightly wrapped in ziplock bags under humid conditions and stored at 8 °C. The plant samples were taxonomically identified and validated by the plant herbarium unit of the Department of Botany and Microbiology, King Saud University, Riyadh, Saudi Arabia as shown in Table 1.
S. No.
Scientific name
Plant family
Plant parts
Common name
Voucher specimen
1.
J. sambac
Oleaceae
Leaves, stems, and roots
Arabic yassmin
KSU-1
2.
C. sinensis
Theaceae
Leaves, stems, and roots
Green tea
KSU-2
3.
O. basilicum
Lamiaceae
Leaves, stems, and roots
Raihan
KSU-3
2.3 Isolation of fungal endophytes
Microbes and epiphytes were removed by immersion of J. sambac, C. sinensis, O. basilicum parts sequentially for one minute in each of the following: 75% ethanol, 1–13% NaOCl, and again in 75% ethanol. Small segments measuring sizes of approximately 0.5–1.0 cm were cut and and three times rinsed with autoclaved water. These small sections were dried using towels of blotting papers. Four pieces were kept in potato dextrose agar (PDA) medium with 50 mg/L chloramphenicol to allow fungal growth at 28 ± 2 °C for two weeks. From these master plates, fungi were relocated to new PDA plates not amended with antibiotics (Suryanarayanan and Kumaresan, 2001).
2.4 Phenotypic determination of isolates
Colony morphology, secretion of pigments, spore structures, and a few characteristics of hyphae including spores were examined using standard mycological manuals (Domsch et al., 1980). Staining of mycelium was done with cotton blue dye for microscopic observation. Furthermore, extra-taxonomic papers on fungal endophytes pertaining to specific genus and species were referred (Barnett and Hunter, 1998). For the color distinction of cultures, the ‘Methuen Handbook of Color' was used as a reference (Wanscher, 1978).
2.5 Polyphasic identification of fungal endophytes
Genus-level determination of fungal endophytes was conducted based on phenotypic and microscopic investigations. For species-level characterization, a widely accepted molecular method was employed. Genomic DNAs of fungal isolates were sequenced by using the service of Macrogen Inc., Seoul, South Korea. The ITS region of the rRNA gene was analyzed using primers ITS1: 5′ (TCC GTA GGT GAA CCT GCG G) 3′ and ITS4: 5′ (TCC TCC GCT TAT TGA TAT GC) 3′.
Sequences amplified by PCR were also analyzed by the Basic Local Alignment Search Tool (BLAST) of NCBI (https://www.ncbi.nlm.nih.gov). Maximum score, total score, query cover, and percentage identity from the NCBI website were considered. Identified sequences were submitted to NCBI and accession numbers were obtained. Maximum likelihood method was followed for phylogentic analysis and tree preparation using MEGA 7.0 to establish relationships among isolates while the Maximum Composite Likelihood method to calculateevolutionary distances. DNA samples were amplified using the following PCR (reaction volume 25 µl) conditions: 95 °C for 2 min, 40 cycles at 95 °C for 30 s (denaturation), 55 °C for 30 s (annealing), and extension at 72 °C for 1 min (elongation), and final elongation at 72 °C for 10 min. PCR amplification was confirmed by running 1.2% agarose gel electrophoresis and analyzed by a gel documentation system.
2.6 Determination of IAA production by fungal isolates
Fungi were grown in PDB with added tryptophan (100 µg/ml). The cultures were incubated for four days at 26 °C for 4 days and spun for 10 min at 10,000 rpm. Supernatant served as a source of IAA using the method of Bric et al. (Bric et al., 1991). Supernatant was added to Salkowski’s reagent (1:2 ratio) and a few drops of H3PO4 were added to it. Mixtures were kept under a dark environment for at least 30 min. The absorbance of pink color solution was recorded at 530 nm. Amount of IAA in the supernatant was calculated using data from the standard curve of IAA.
2.7 Determination of ACC deaminase activity
Activity of the ACC deaminase enzyme was quantified using the method of Yedidia et al. (Yedidia et al., 1999). Briefly, 20 ml synthetic medium was inoculated with the spore suspensions (20 µl) of endophytic fungal isolates. The medium was allowed to incubate for two days at 28 °C. Fungal mycelia were washed and inoculated to a synthetic medium (5 ml) amended with 3 mM ACC but without adding ammonium. After the induction period, samples were processed as described earlier (Yedidia et al., 1999). The absorbance at λmax = 540 nm was recorded and ACC deaminase activity was measured due to the generation of α-ketobutyrate and expressed as mmol of α-ketobutyrate mg−1 protein h−1. A standard curve of α-ketobutyrate was used and values were calculated.
2.8 Siderophore production assay
A disc inoculation method was used for the detection of the siderophore activity of endophytic fungal isolates. Seven to eight days old cultures of fungi were used. Mycelial discs measuring 4 mm in size were placed on Chrome Azurol S (CAS) agar media. CAS plates with isolated strains were allowed to incubate at 30 °C for a week. The orange-colored halo zone around the disc was monitored and isolates were designated positive or negative based on the halo zone surrounding fungal inoculation on a blue medium.
2.9 Data analysis
The colonization frequency (CF) of isolated strains was calculated by using the following formula:
Relative frequency percentage for every fungus was presented as:
The isolation rate was calculated by the total number of isolated fungi divided by the total number of segments.
3 Results and discussion
Our study probably for the first time reports the isolation and describe the endophytic fungi colonizing three medicinal plants J. sambac, C. sinensis, and O. basilicum cultivated in the city of Riyadh, Saudi Arabia. Great diversity was found in species of endophytic fungi. Investigations showed that most plants had fungal partners within their tissues (Rodriguez et al., 2009). A total of 144 samples of leaves, stems, and roots were collected and screened for the isolation and characterization of fungal endophytes (Tables 2 and 3).
Aromatic and medicinal plants
No. of samples analyzed
No. of isolates
Isolate rate
CF (%)
Relative frequency (%)
J. sambac
48
39
0.81
27
46.4
C. sinensis
48
27
0.56
18.7
32.1
O. basilicum
48
18
0.37
2.5
21.5
Total
144
84
Plant parts
No. of isolates
Isolation
rateCF (%)
Relative frequency %
Leaves
40
0.83
27.7
47.62
Stems
28
0.58
19.4
33.33
Roots
13
0.27
9.02
15.48
Total
84
3.1 Isolation and morphological identification
Eighty-four isolates were collected from three aromatic and medicinal plants J. sambac, C. sinensis, and O. basilicum. Fungal isolates recovered from plants were separated into twenty morpho-types based on the similarity of their cultural characteristics that include both macro- and microscopic features including colony morphology, plate color (above and reverse), and mycelium (Fróhlich et al., 2000) (Table 4).
S. No.
Endophytic Fungi
Family
Class
Molecular identity (%)
Accession numbers
1.
Neopestalotiopsis clavispora
Sporocadaceae
Sordariomycetes
99.18
MN096884
2.
Trichoderma virens
Hypocreaceae
Sordariomycetes
99.66
MN096886
3.
Fusarium sp.
Nectriaceae
Sordariomycetes
99
MN096888
4.
Fusarium equiseti
Nectriaceae
Sordariomycetes
100
MN096904
5.
Fusarium anthophilum
Nectriaceae
Sordariomycetes
100
MN096906
6.
Colletotrichum trifolii
Glomerellaceae
Sordariomycetes
99
MN096912
7.
Chaetomium sp.
Chaetomiaceae
Sordariomycetes
98
MN096926
8.
Myrothecium inundatum
Stachybotryaceae
Sordariomycetes
99
MN096930
9.
Alternaria eichhorniae
Pleosporaceae
Dothideomycetes
99.26
MN096890
10.
Alternaria alstromeriae
Pleosporaceae
Dothideomycetes
99
MT982648
11.
Curvularia subpapendorfii
Pleosporaceae
Dothideomycetes
99
MN096895
12.
Alternaria alternata
Pleosporaceae
Dothideomycetes
100
MN096898
13.
Penicillium glabrum
Trichocomaceae
Eurotiomycetes
100
MN096900
14.
Alternaria tenuissima
Pleosporaceae
Dothideomycetes
99
MN096902
15.
Phoma multirostrata
Didymellaceae
Dothideomycetes
99
MN096914
16.
Curvularia subpapendorfii
Pleosporaceae
Dothideomycetes
99
MN096895
17.
Cladosporium tenuissimum
Davidiellaceae
Dothideomycetes
99
MN096922
18.
Cladosporium perangustum
Davidiellaceae
Dothideomycetes
98
MN096924
19.
Neodidymelliopsis sp.
Didymellaceae
Dothideomycetes
98
MN096934
20.
Aspergillus niger
Trichocomaceae
Eurotiomycetes
99.12
MN096920
Based on macro and microscopic assays done for culture characteristics and morphology, and following molecular identification, the isolates were identified as- one species of Neopestalotiopsis from four isolates, one species of Trichoderma from three isolates, four species of Alternaria from thirty-five isolates, three species of Fusarium from eight isolates, one species of Curvularia from eight isolates, one species of Colletotrichum from two isolates, one species of Phoma from three isolates, one species of Aspergillus from three isolates, two species of Cladosporium from four isolates, one species of Chaetomium from two isolates, one species of Myrothecium from three isolates, one species of Neodidymelliopsis from two isolates (Fig. 1A). Among the various isolates, the genus Alternaria was most dominant with an overall colonizing frequency (CF) of 41.5%, and this genus was also the dominant endophyte group in previous studies (Mohammad Golam Dastogeer et al., 2020) (Fig. 1 A-B).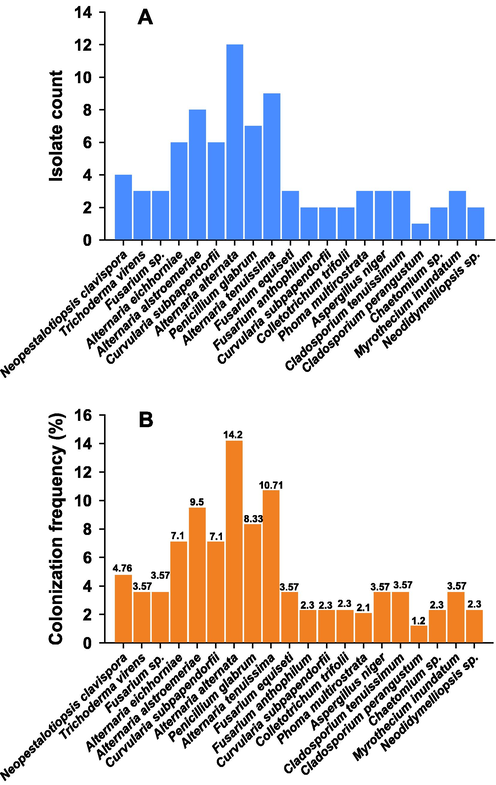
Bar diagram showing counts of fungal isolates (A) and colonization frequency (%) (B).
In the current study, J. sambac showed the highest number of fungi as 39 isolates with a colonization rate of 27 % and isolation rate (IR) of 0.81, followed by 27 isolates from C. sinensis, CF of 18.7 % and IR 0.56, and O. basilicum has recorded the lowest CF, 12.5 %, by 18 isolates. In a previous study, (Sharmila et al., 2017) reported the isolation of endophytic fungi and the colonization rate was recorded in leaf tissues as well as few reports conducted on J. sambac, therefore, our study confirmed this endophytic biodiversity (Table 4). Xie et al. recently reported the isolation of endophytic fungi from C. sinensis (L.), the fungal diversity, and biological functions (Xie et al., 2020). They have also reported the plant growth-promoting activities of those fungi such as siderophore production, secretion of phytohormones, ACC deaminase enzyme, etc. Moreover, the variations in the composition of fungal isolates were assigned to seasonal or cultivar changes were shown (Xie et al., 2020). Endophytic fungal isolates have also been obtained from various habitats, environments, and climates. Similar to these conditions, Riyadh's vicinity conditions are characterized by extreme temperatures, the prevalence of drought, and floral diversity in natural habitats. Fungi generally show the ability to survive in such extreme conditions and to adapt to most habitats. Moreover, heterogeneous conditions may encourage a high diversity of fungal communities in desert ecosystems (Verma et al., 2012).
Among the isolated endophytic fungi, 13 species Trichoderma virens, Alternaria eichhorniae, Alternaria alstroemeriae, Alternaria alternata, Alternaria tenuissima, Fusarium sp., Fusarium equiseti, Fusarium anthophilum, Colletotrichum trifolii, Aspergillus niger, Cladosporium tenuissimum, Penicillium glabrum, and Chaetomium sp. were distributed through the four aromatic and medical plants, whereas other species have shown a specific tendency for their hosts were: Neopestalotiopsis sp., Curvularia subpapendorfii, Phoma multirostrata, Cladosporium perangustum, Myrothecium inundatum, and Neodidymelliopsis sp. (Figs. 2 and 3). In this investigation, endophytic fungi have displayed host or organ specificity, and there was little intersection between the leaf, stem, and root assemblages, which have been consistent with previous studies (Rajulu et al., 2016).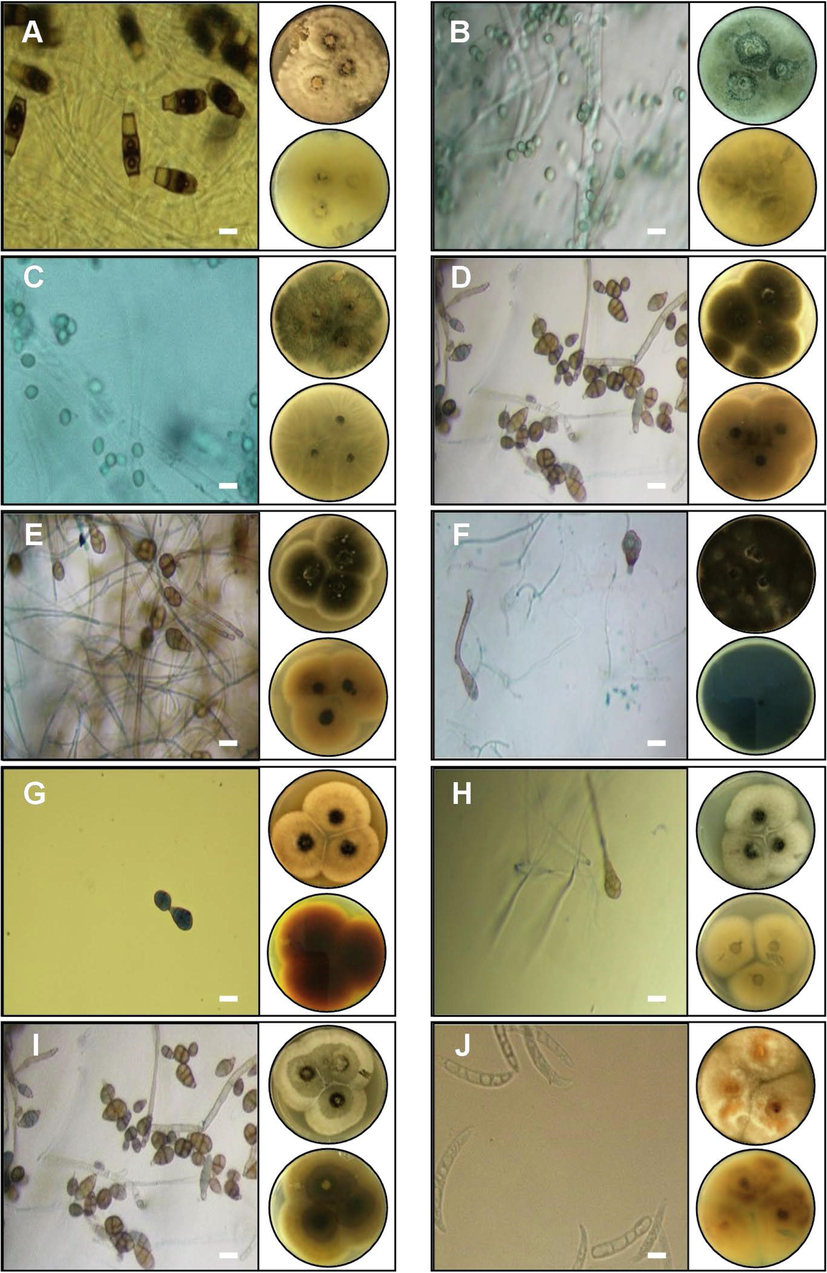
Macroscopic and microscopic features of five genera of fungal endophytes: A- Neopestalotiopsis clavispor, B- Trichoderma virens, C- Fusarium sp., D- Alternaria eichhorniae, E- Alternaria alternata, F- Alternaria alstroemeriae, G- Curvularia subpapendorfii, H- Penicillium glabrum, I- Alternaria tenuissima, and J- Fusarium equiseti. Images of Petri dishes in each panel represent pictures from the upper and lower side. The length of the scale bar is 20 µm.
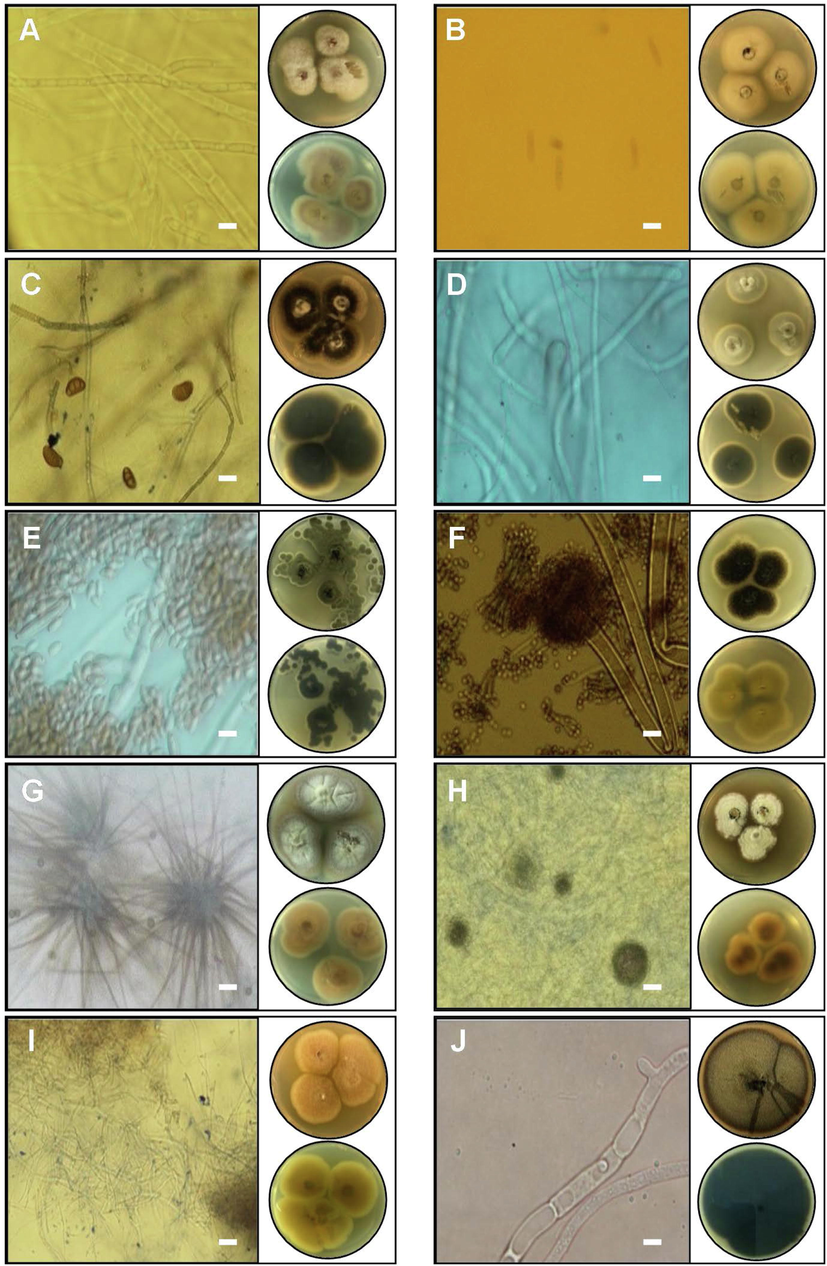
Macroscopic and microscopic characteristics of nine genera endophytes fungi isolated from aromatic and medical plants: A- Fusarium anthophilum, B- Colletotrichum trifolii, C- Curvularia subpapendorfii, D- Cladosporium tenuissimum, E- Cladosporium perangustum, F- Aspergillus niger, G- Chaetomium sp., H- Myrothecium inundatum, I- Curvularia subpapendorfii, and J- Neodidymelliopsis sp. Images of Petri dishes in each panel represent pictures from the upper and lower side. The length of the scale bar is 20 µm.
In this study, some genera have been isolated for the first time confirming that Saudi flora is rich in endophytic fungi. One hundred forty-four segments (leaves-steams-roots) of J. sambac, C. sinensis, and O. basilicum have produced 39, 27, and 18 endophytic strains with colonization rates at 27 % 18.7 % and 12.5 %, respectively (Table 2). Environmental conditions significantly affect the structure of fungal communities and populations (Gashgari et al., 2016). Further, plants that belong to the same geoenvironmental locations may show a higher degree of smilalrity between the taxa and species (D’Amico et al., 2008).
Interestingly, the section of leaves recorded the highest relative frequency (27.7 %), while the frequencies for stems (19.4 %) and roots (9.02 %) were notably less (Table 3). Therefore, it seems that endophytes favorably colonize the leaves due to greater surface area, rich nutrition, and thin walls (Figs. 2 and 3).
Eighty-four endophyte fungi have been classified into 12 genera sequentially subdivided under three classes: dothideomycetes, sordariomycetes, and eurotiomycetes. Dothideomycetes have included six genera as Alternaria, Curvularia, Phoma sp., Cladosporium, Neodidymelliopsis, six genera of isolates were classified in sordariomycetes class as Neopestalotiopsis, Trichoderma, Fusarium, Colletotrichum, Chaetomium, Myrothecium, while eurotiomycetes has contributed only two genera of endophytic fungi that were Aspergillus and Penicillium. The majority of endophytes were filamentous ascomycetes supporting the former studies belonging to the Ascomycota phylum (Clay, 1990). Similar results were also obtained with seven medicinal plants collected from the Al-Gouf governorate's salt marshes in North Saudi Arabia (Gashgari et al., 2016).
3.2 ITS sequence and phylogenetic analysis
Twenty-five isolates were subjected to morphological assays followed by molecular analysis to confirm the identification of fungi based on ITS1 and ITS4 sequences in rDNA genes. Identification of isolates was constructed on the highest similarity from BLAST results. Gene bank accession numbers of twenty endophytic fungi were obtained (Table 4).
Besides morphological, molecular charactrization by ITS region of the rRNA gene was employed to confirm initial observations and was an extremely useful and appropriate tool for estimating species diversity. The ITS region is reliable for the identification fungi (Gherbawy and Elhariry, 2016). The isolates that have been used for the sequencing analysis, their codes and GenBank numbers are provided in Table 4. Phylogenetic tree for twenty isolates of endophytic fungi (Table 5) was constructed by MEGA 7.0 program that confirms all strains belonging to Dothideomycetes, Sordariomycetes, or Eurotiomycetes classes of Ascomycota division (Fig. 4). *L = leaf, S = Stem, R = Root.
Endophytic fungi
Aromatic and medical plants
Total
J. sambac
C. sinensis
O. basilicum
L
S
R
L
S
R
L
S
R
Neopestalotiopsis clavispor
2
1
1
–
–
–
–
–
–
4
Trichoderma virens.
–
–
1
–
–
–
1
1
–
3
Fusarium sp.
1
1
–
1
–
–
–
–
–
3
Alternaria eichhorniae
2
1
–
1
1
–
1
–
6
Alternaria alstromeriae
1
2
–
1
1
1
1
–
1
8
Curvularia subpapendorfii
1
1
1
1
1
–
1
–
–
6
Alternaria alternata
2
2
1
2
1
1
–
2
1
12
Penicillium glabrum
2
1
–
1
1
1
1
–
–
7
Alternaria tenuissima
1
1
1
1
1
1
2
1
–
9
Fusarium equiseti
–
–
1
–
1
1
–
–
–
3
Fusarium anthophilum
1
–
1
–
–
–
–
–
–
2
Curvularia subpapendorfii
–
–
–
1
1
–
–
–
–
2
Colletotrichum trifolii
–
–
–
–
–
–
1
1
–
2
Phoma multirostrata
2
–
1
–
–
–
–
–
–
3
Aspergillus niger
1
1
–
–
–
1
3
Cladosporium tenuissimum
1
–
–
1
1
–
–
–
–
3
Cladosporium perangustum
–
–
–
1
–
–
–
–
–
1
Chaetomium sp.
–
–
–
1
1
–
–
–
–
2
Myrothecium inundatum
2
1
–
–
–
–
–
–
–
3
Neodidymelliopsis sp.
–
–
–
–
–
1
1
–
2
Total
19
12
8
12
10
5
9
6
3
84
39
27
18
84

Maximum composite likelihood phylogenetic tree of endophytic fungi isolated from aromatic and medical plants based on ITS sequences in rRNA genes. The Clustal W was used for sequence alignment in MEGA 7.0. Bootstrap percentage values obtained from 1000 replications of the data set are shown at nodes. The scale bar shows the average number of nucleotide substitutions per site.
3.3 Plant growth-promoting (PGP) traits of fungal endophytes
Three PGP traits of all identified fungal isolates were explored (Table 6). Biosynthesis of IAA by fungal isolates ranged from 2.1 to 56 µg/ml. Among those, Aspergillus niger and Penicillum glabrum were found as high producers of IAA (56.3 and 48.2 µg/ml, respectively) with 100 µg/ml tryptophan. Some isolates showed low production of IAA that include Myrothecium inundatum, Chaetomium sp., and Curvularia subpapendorfii while some strains like Cladosporium tenuissimum and Neodidymelliopsis sp. did not produce IAA. Similarly, variable results were observed for ACC deaminase activity and siderophore production. Here, Penicillum glabrumi and Aspergillus niger showed a positive reaction for siderophore production and also produced high amounts of α-ketobutyrate (18.2 and 15.2 μmol α-KB mg−1 Protein h−1, respectively) as a result of ACC deaminase activity. Only 12 isolates were positive for siderophore activity. Overall, aromatic, and medical plants are vital stores of endophytes fungi, wherein it is infrequently finding a fungus-free plant. The medicinal and aromatic plants used in the current study are known to be rich for their medicinally active phytochemical compositions such as the production of catechins, caffeine, and other polyphenolic compounds by C. sinensis (Aboulwafa et al., 2019), various bioactive phytochemicals by J. sambac (Wu et al., 2021), and polyphenolic secondary metabolites by O. basilicum (Pirbalouti et al., 2017) that have shown few or many clinically useful properties including radical scavenging, antioxidant, antimicrobial, antiviral, anticancer, and anticonvulsant, hypoglycaemic, and hypolipidemic properties (Rubab et al., 2017). Similar to their host plants, the endophytic microbes also produce some valuable phytochemical with a variable range of activities. For example, endophytic microbes associated with C. sinensis produced secondary metabolites that could produce antimicrobial activities against Staphylococcus aureus and Bacillus subtilis (Shan et al., 2018). Moreover, endophytic microbes produced some plant growth-regulating substances like IAA and ACC deaminase promoting plant (Shan et al., 2018). In corroboration with our results, J. sambac also showed to have various species of the genus Colletotrichum (Gioia et al., 2020). Also, J. sambac contained Phyllosticta capitalensis fungus living in endophytic mode. Similar to the antimicrobial and antioxidant activities of O. basilicum (Pirbalouti et al., 2017), the extracts from its endophytic fungi (total of eight genera) also produced antibacterial and antioxidant effects against nine clinical pathogens of humans (Atiphasaworn et al., 2017). Among them, Nigrospora sp. Exhibited the highest antibacterial and antioxidant potential. This suggests that in some cases the medicinally important host and its endophytic fungi share common bioactive properties. Moreover, the biostimulants produced by the endophytic fungus can also modulate the production of secondary metabolites of the host plant as has been shown in the case of O. basilicum L (Saia et al., 2021). IAA = indole-3-acetic acid; α-KB = α-keto butyrate; ‘+’ shows a positive reaction while ‘−’ represents a negative reaction; ND = not detected. *IAA production was checked with the addition of 100 µg/ml tryptophan.
S. No.
Genus and species
IAA production (µg/ml)*
ACC deaminase (μmol α-KB/mg Protein/h)
Siderophore production test
1.
Neopestalotiopsis clavispora
12.2 ± 2.1
4.2 ± 1.1
−
2.
Trichoderma virens
16.85 ± 3.5
17.25 ± 0.65
+
3.
Fusarium sp.
7.27 ± 1.36
23.2 ± 3.1
+
4.
Alternaria eichhorniae
5.26 ± 2.01
15.32 ± 2.1
+
5.
Alternaria alstromeriae
7.21 ± 1.41
ND
−
6.
Curvularia subpapendorfii
6.23 ± 0.86
ND
+
7.
Alternaria alternata
5.05 ± 0.47
4.2 ± 0.32
+
8.
Penicillium glabrum
48.23 ± 2.65
18.2 ± 0.78
+
9.
Alternaria tenuissima
71.56 ± 4.1
ND
−
10.
Fusarium equiseti
10 ± 2.35
ND
+
11.
Fusarium anthophilum
15.2 ± 4.65
11.05 ± 0.45
−
12.
Curvularia subpapendorfii
2.1 ± 0.53
ND
+
13.
Colletotrichum trifolii
14.23 ± 3.58
2.3 ± 0.12
+
14.
Phoma multirostrata
4.42 ± 0.36
ND
−
15.
Aspergillus niger
56.32 ± 4.78
15.26 ± 0.36
+
16.
Cladosporium tenuissimum
ND
4.21 ± 1
+
17.
Cladosporium perangustum
6.14 ± 0.58
7.14 ± 0.85
−
18.
Chaetomium sp.
5.4 ± 0.68
3.2 ± 0.2
+
19.
Myrothecium inundatum
4.23 ± 0.23
ND
−
20.
Neodidymelliopsis sp.
ND
ND
−
4 Conclusion
Aromatic and medicinal plants are colonized by fungal endophytes that can produce novel metabolites for potential pharmaceutical applications. Here, we isolated twenty endophytic fungi from three well-known aromatic and medicinal plants J. sambac, C. sinensis, and O. basilicum. The endophyte colonization frequency is significantly affected by various factors including plant age, plant tissue, and geographical site. We suggest that the diversity of endophytic fungi having different colonization frequency vary with medicinal plant species and plant organs where leaves possess the highest number. This study unveils the variety in the endophytic fungal population of J. sambac, C. sinensis, and O. basilicum collected from Riyadh, Saudi Arabia and it will yield unique records and if extended further, will probably lead to the identification of novel species producing secondary metabolites.
Funding
This work was funded by Researchers Supporting Project Number (RSP2023R15).
CRediT authorship contribution statement
Helal F. Al-Harthi: Methodology, Software, Validation, Formal analysis. Abdallah M. Elgorgan: Conceptualization, Validation, Formal analysis, Investigation, Resources, Data curation, Writing – original draft, Writing – review & editing, Visualization, Supervision, Project administration. Bilal Ahmed: Software, Validation, Writing – original draft, Writing – review & editing. Ali H. Bahkali: Conceptualization, Investigation, Data curation, Supervision, Funding acquisition. Mohamed ElSheshtawi: Validation, Formal analysis, Writing – review & editing. Jilani Purusottapatnam Shaik: . Abdullah Msaad Al-Falih: Methodology, Formal analysis. Asad Syed: Conceptualization, Software, Validation, Formal analysis, Investigation, Resources, Data curation, Writing – original draft, Writing – review & editing, Visualization, Supervision, Project administration.
Acknowledgments
The authors extend their appreciation to the Researchers Supporting Project number (RSP2023R15), King Saud University, Riyadh, Saudi Arabia
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- A Comprehensive insight on the health benefits and phytoconstituents of Camellia sinensis and recent approaches for its quality control. Antioxidants. 2019;8:455.
- [Google Scholar]
- Antibacterial and antioxidant constituents of extracts of endophytic fungi isolated from Ocimum basilicum var. thyrsiflora leaves. Curr. Microbiol.. 2017;74:1185-1193.
- [Google Scholar]
- Illustrated genera of imperfect fungi. American Phytopathological Society (APS Press); 1998.
- Pharmacognostical and phytochemical investigation of various tulsi plants available in south eastern Odisha. Int. J. Res. Pharm. Biomed. Sci.. 2011;2:605-610.
- [Google Scholar]
- Rapid in situ assay for indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl. Environ. Microbiol.. 1991;57:535-538.
- [CrossRef] [Google Scholar]
- Endophytic fungi occurring in fennel, lettuce, chicory, and celery—commercial crops in southern Italy. Mycol. Res.. 2008;112:100-107.
- [Google Scholar]
- Compendium of soil fungi. Vol Volume 1. Academic Press (London) Ltd.; 1980.
- Fungal endophytes: common host plant symbionts but uncommon mutualists. Integr. Comp. Biol.. 2002;42:360-368.
- [Google Scholar]
- Gashgari, R., Gherbawy, Y., Ameen, F., Alsharari, S., 2016. Molecular characterization and analysis of antimicrobial activity of endophytic fungi from medicinal plants in Saudi Arabia. Jundishapur J. Microbiol. 9.
- Endophytic fungi associated with high-altitude Juniperus trees and their antimicrobial activities. Plant Biosyst. Int. J. Deal. with all Asp. Plant Biol.. 2016;150:131-140.
- [Google Scholar]
- A Survey of Endophytic Fungi Associated with High-Risk Plants Imported for Ornamental Purposes. Agriculture. 2020;10:643.
- [Google Scholar]
- A friendly relationship between endophytic fungi and medicinal plants: a systematic review. Front. Microbiol.. 2016;7:906.
- [Google Scholar]
- Chemical diversity of essential oils from the Brazilian medicinal plant Lychnophora pinaster Mart from different environments. Ind. Crops Prod.. 2020;156:112856
- [Google Scholar]
- Fungal Melanins and Applications in Healthcare, Bioremediation and Industry. J. Fungi. 2021;7:488.
- [Google Scholar]
- Host Specificity of Endophytic Fungi from Stem Tissue of Nature Farming Tomato (Solanum lycopersicum Mill.) in Japan. Agronomy. 2020;10:1019.
- [Google Scholar]
- Effects of foliar of the application chitosan and reduced irrigation on essential oil yield, total phenol content and antioxidant activity of extracts from green and purple basil. Acta Sci. Pol. Hortorum Cultus. 2017;16:177-186.
- [CrossRef] [Google Scholar]
- Notes on succulent plant species of Saudi Arabia. Bangladesh J. Plant. Taxon. 2002;9:25.
- [Google Scholar]
- Endophytic fungi of orchids of Arunachal Pradesh, North Eastern India. Curr. Res. Environ. Appl. Mycol.. 2016;6:293-299.
- [Google Scholar]
- Biomedical description of Ocimum basilicum L. J. Islam. Int. Med. Coll.. 2017;12:57-69.
- [Google Scholar]
- An Endophytic Fungi-Based Biostimulant Modulates Volatile and Non-Volatile Secondary Metabolites and Yield of Greenhouse Basil (Ocimum basilicum L.) through Variable Mechanisms Dependent on Salinity Stress Level. Pathogens. 2021;10:797.
- [Google Scholar]
- Shan, W., Zhou, Y., Liu, H., Yu, X., 2018. Endophytic actinomycetes from tea plants (Camellia sinensis): isolation, abundance, antimicrobial, and plant-growth-promoting activities. Biomed. Res. Int. 2018.
- Synergistic antioxidant and antimicrobial activities of essential oils of some selected medicinal plants in combination and with synthetic compounds. Ind. Crops Prod.. 2020;154:112569
- [Google Scholar]
- Antibacterial Activities of Endophytic Fungal Strains Isolated from Aromatic Plants. Asian J. Pharm. Sci. Technol.. 2017;7:28-33.
- [Google Scholar]
- Suryanarayanan, T.S., Kumaresan, V., 2001. Fungal Endophytes: The Tropical. Trichomycetes and other fungal groups 197.
- Histological Investigation of Fungal Endophytes in Healthy Tissues of Azadirachta indica A. Juss. Agric. Nat. Resour.. 2012;46:229-237.
- [Google Scholar]
- Methuen handbook of colour. London: E. Methuen; 1978.
- Wu, L.-C., Lin, C.-L., Peng, C.-C., Huang, T.-L., Tsai, T.-H., Kuan, Y.-E., Chung, Y.-C., 2021. Development from Jasminum sambac Flower Extracts of Products with Floral Fragrance and Multiple Physiological Activities. Evidence-Based Complement. Altern. Med. 2021.
- Implications of endophytic microbiota in Camellia sinensis: A review on current understanding and future insights. Bioengineered. 2020;11:1001-1015.
- [Google Scholar]
- Induction of defense responses in cucumber plants (Cucumis sativus L.) by the biocontrol agent Trichoderma harzianum. Appl. Environ. Microbiol.. 1999;65:1061-1070.
- [Google Scholar]







