Translate this page into:
Identification and molecular study of medicinal Plectranthus species (Lamiaceae) from Saudi Arabia using plastid DNA regions and ITS2 of the nrDNA gene
⁎Corresponding author. wsjuhani@uqu.edu.sa (Widad Saleem Al-Juhani),
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Plectranthus is a genus of the Lamiaceae family that includes many species of medicinal and agricultural importance. However, this genus has been the subject of taxonomic debate and contains species that are difficult to distinguish. The present study focused on six Plectranthus species commonly found in Saudi Arabia: P. arabicus, P. tenuiflorus, P. barbatus, P. pseudomarrubioides, P. asirensis, and P. hijazensis. P. hijazensis is endemic to Saudi Arabia. The capacities of five different plastid DNA barcodes (matK, rbcL, trnH-psbA, and ITS1 and ITS2 regions of the nrDNA gene) to identify and distinguish between Plectranthus species were evaluated. The following analytical methods were used to evaluate the efficiencies of the selected markers: BLAST, inter- and intraspecific distance, barcode gap, secondary structure of ITS2, and maximum likelihood (ML) phylogenetic trees. The results demonstrated that the nuclear ITS2 region can be successfully amplified and sequenced (100%), leading to a strong ability to discriminate between species and a clear barcode gap. Furthermore, there were significant differences in the ITS2 secondary structure among Plectranthus spp. Samples of Plectranthus formed monophyletic groups according to species in the ML tree, with high supported values. Our results establish that all Plectranthus species in Saudi Arabia can be classified into two groups within the Coleus clade. To our knowledge, this is the first time that local and endemic Plectranthus spp. have been identified and compared with Plectranthus samples of different geographical origins. Our results confirm the diversity of Plectranthus species growing naturally in southwestern Saudi Arabia. In addition, P. hijazensis, which is endemic to Saudi Arabia, was determined to be genetically distinct from other Plectranthus species and should, therefore, be the focus of future research, in addition to the preservation of the natural environment of these species.
Keywords
DNA-Barcode
Taxonomy
Plectranthus
Medicinal Herbs
1 Introduction
Plectranthus is one of the most important genera in the Lamiaceae family. The genus has been the subject of taxonomic debate for a variety of reasons, including the strong similarity between species and existence of multiple names for each species. In addition, it is difficult to identify species of genera that are closely related to Plectranthus (Musila et al., 2017). Initial classification attempts for Plectranthus were based on morphological characteristics, which resulted in many contradictions in the arrangement and number of subgenera and sections belonging to the genus. Morphological taxonomic classification of this tribe has not successfully classified the clades within the sub-tribe (Paton et al., 2004).
Paton et al., (2004) studied the molecular phylogenetics of the Ocimeae tribe based on three plastid DNA regions and argued that the Plectranthus genus is paraphyletic, supporting the hypothesis that Ocimeae is of Asian origin. Recently, (Paton et al., 2018) reviewed Plectranthus and Coleus based on the distribution and medicinal attributes of the phylogenetic plastid genes trnL-F, rps16, and trnS-G and showed that the Plectranthus genus is distributed within two major clades of Plectranthus and Coleus.
Seven species of Plectranthus have been reported in the Kingdom of Saudi Arabia (KSA) (Collenette, 1999). However, (Chaudhary, 2000) revised these species and accepted only six: P. arabicus, P. cylindraceus, P. tenuiflorus, P. lanuginosus, P. barbatus, and P. asirensis. Recently, (Abdel Khalik, 2016a) recorded a new species, Plectranthus hijazensis.
P. asirensis is a Saudi Arabian species used as an antibacterial to treat diaper rash and has antiseptic properties (Abulfatih, 1987). P. barbatus grows naturally in Saudi Arabia and is used as a remedy for stomach, intestine, and liver disorders, heart problems, respiratory diseases, and as an insect repellant (Grayer et al., 2010). P. tenuiflorus, another local Saudi Arabian species, is used to treat ear infections and as an ornamental plant (Abulfatih, 1987).
Al-Qurainy et al., (2014) compared and identified the relationships among Saudi Arabian P. asirensis and Plectranthus spp. of different geographical origins using nrDNA-internal transcribed spacer (ITS) and chloroplast barcodes. Abdel Khalik and Osman (2017) studied the genetic diversity among seven species of the genus Plectranthus in Saudi Arabia using inter simple sequence repeats (ISSR) and random amplification of polymorphic DNA (RAPD) markers.
Variations in the macro- and micro-morphological features (including seed and pollen shape, size, coat sculpture, and trichome structure, as well as anatomical features of the leaves and stems) of the seven species of Plectranthus native to Saudi Arabia have been examined previously (Abdel Khalik, 2016b: Abdel Khalik and Karakish, 2016).
In recent years, there has been worldwide interest in using medicinal plants as a safe alternative to chemical medicines. However, difficulties in accurate identification and unethical practices in the use of these medicinal herbs pose a threat to this potential shift. Thus, the need for reliable verification tools to confirm the identity of these herbs is critical (Zahra et al., 2017). For example, P. barbatus and P. grandis are closely related species that can be mistaken for one another and used for the same medicinal purposes (Nani et al., 2015).
DNA barcoding as a molecular technique has been recommended as a useful method for species identification (CBOL Plant Working Group, 2009). DNA barcoding is not influenced by external factors or development stage, and DNA can be easily isolated from all tissues (Sucher and Carles, 2008; Seethapathy et al., 2015; Wu et al., 2015; Mishra et al., 2016), thus, providing a basis for species identification at the genetic level (Yu et al., 2017). DNA barcoding uses a short sequence of a standard part of the genome instead of the whole genome to verify the identity of samples. Portions of the rbcL and matK plastid coding genes have been suggested as barcodes for plant species (CBOL Plant Working Group, 2009) and have become the most-used loci in land plants as the rbcL region is highly suitable for amplification and sequencing (Michel et al., 2016; Mohamed, 2016). The nuclear ITS, with relatively strong discrimination power, is complementary to matK and rbcL in plants (CPBG China Plant BOL Group, 2011). The ITS2 region, a sub-region of ITS, has been described as a valuable sequence tag for identifying medicinal plants (Chen et al., 2010; Yao et al., 2010; Han et al., 2013). The ITS2 region is short in length and requires few primers for amplification. In addition, the region contains an abundance of genetic information and is located in the nuclear region, meaning that the ITS2 region can be used to overcome the issue of failure to amplify the ITS in some species and is suitable for identifying high and low level taxa (Petit and Excoffier, 2009; Naciri et al., 2012; Braukmann et al., 2017).
Overall, Saudi Arabian Plectranthus species have not received adequate attention in terms of molecular studies. In the present study, five nuclear regions (ITS1 and ITS2 of the nrDNA gene and matK, rbcL, and trnH-psbA of the plastid gene) were chosen as barcodes, and a systematic comparison of different Plectranthus specimens local to Saudi Arabia and of different geographical origins was performed. The objectives of the study were a) to determine the performance of DNA barcoding in Plectranthus specimens and b) to evaluate the species discrimination powers of different barcodes.
2 Methods
2.1 Study area
The study area included different locations in the Jazan-Fifa Mountains, Baha, and Taif. These areas are located in the Tihama mountain range, which stretches across the southwest of the KSA, with height ranging from approximately 2000 to 12,000 m. The Tihama range is located in the Afro-Alpine vegetation zone, featuring a high plant diversity. Details of sample names, dates, and collection areas are presented in Table 1.
Voucher Specimen
Accepted Scientific Name
Place of Collection
Coordinates and Altitude
GenBank Accession Numbers
ITS2
rbcL
UQU PA1
Plectranthus arabicus E.A. Bruce
Jazan, Jabal Fefa, Gardada
17°14′45.1″N 43°05′27.2″E
MN382135
MN381814
UQU PA2
MN382136
MN381815
UQU PA3
MN382137
MN381816
UQU PP4
Plectranthus pseudomarrubioides R.H. Willemse
Jazan, Jabal Fefa
17°14′45.1″N 43°05′27.2″E
MN382138
MN381817
UQU PP5
MN382139
MN381818
UQU PP6
MN382140
MN381819
UQU pH 7
Plectranthus hijazensis Abdel Khalik
Baha, Dam of Medhas
20°01′19.8″N 41°26′00.1″E
MN382141
MN381820
UQU PH8
Fail
MN381821
UQU pH 9
MN382142
MN381822
UQU PB10
Plectranthus barbatus Andrews
Jazan-Jabal Fefa
17°14′45.1″N 43°05′27.2″E
MN382143
MN381823
UQU PB11
MN382144
MN381824
UQU PB12
MN382145
MN381825
UQU PS13
Plectranthus asirensis J.R.I.Wood
Messan, Bani Malek
20°49′56.9″N 41°00′14.1″E
MN382146
MN381826
UQU PS14
MN382147
MN381826
UQU PS 15
MN382148
MN381828
UQU PT16
Plectranthus tenuiflorus (Vatke) Agnew.
A synonym of Plectranthus aegyptiacus (Forssk.) C. Chr.Abha, Raydah village
18°12′20.7″N 42°24′35.7″E
MN382149
MN381829
UQU PT17
MN382150
MN381830
UQU PT18
MN382151
MN381831
UQU PC22
Taif, Jabal Thaqif
21°04′32.5″N 40°18′37.4″E
MN382152
MN381832
UQU PC23
MN382153
MN381833
UQU PC24
MN382154
MN381834
2.2 Plant material
Samples (leaves, flowers, and stems) of six Plectranthus species were collected between 2014 and 2018. Voucher specimens were prepared and identified by experts in taxonomy according to Collenette (1999) and Chaudhary (2000) and were deposited at the Herbaria of King Saud University/KSA, Kew and Edinburgh herbarium/UK, and Biology Department, Faculty of Applied Science, Umm Al-Qura University, Mecca/KSA after comparison with voucher specimens. At least three accessions per species were selected for molecular analysis from these voucher specimens.
2.3 DNA extraction
Initially, the following DNA extraction techniques were tested to select the best method: DNeasy Plant Mini Kit extraction (Qiagen/USA) following commercial protocols, traditional CTAB method (Doyle and Doyle, 1987), and modified DNA extraction method of Sahu et al. (2012), as described in Manikandan et al., (2017). The quantity of the extracted DNA was estimated using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA). DNA was tested via 1% agarose gel electrophoresis and ultraviolet light.
2.4 Primer sequences
Different DNA barcodes were amplified using the following primers: a-F (5′-ATGTCACCACAAACAGAGACTAAAGC-3′) and a-R (5′-GTAAAATCAAGTCCACCRCG-3′) for rbcL; KIM3F (5′-ACCCAGTCCATCTGGAAATCTTGGTTC-3′) and KIM1R (5′-CGTACAGTACTTTTGTGTTTACGAG3′) for matK, according to CBOL Plant Working Group (2009); 2F (5′-CGTAGCTACTTCTTCGCAGC-3′) and 5R (5-CCTTATCATTTAGAGGAAGGAG-3′) for ITS2, according to Chen et al., (2010); for ITS1; 2F (5′-ATGCGATACTTGGTGTGAAT-3′) and 3R (5′-GACGCTTCTCCAGACTACAAT-3′) and F (5′-ACTGCCTTGATCCACTTGGC-3′) and R (5′-CGAAGCTCCATCTACAAATGG-3′) for trnH-psbA, according to (Kress and Erickson (2007).
2.5 DNA amplification and sequencing
DNA regions were amplified according to the method described by Maloukh et al., (2017), using 25 µL reaction volumes containing 12.5 µL Master Mix, 8.5 µL nano-pure water, 1 µL of each primer, and 2 µL DNA. The thermal cycle included an initial denaturation level of 94 °C for 5 min, followed by 94 °C for 45 s. An annealing temperature dependent on the type of primer (ITS1, 50 °C; ITS2, 56 °C; matK, rbcL, and trnH-psbA, 55 °C) was applied for 45 s, followed by 72 °C for 1.5 min, and a final extension step at 72 °C for 10 min, in 40 cycles, using a Mastercycler (Eppendorf Vapo Protect, NY, USA). The quality of the PCR products was confirmed by electrophoresis on 1.5% agarose gel, and products were purified with a 1.0% agarose gel using the QIAquick purification Kit (Qiagen, USA). Sequencing was performed using a genetic analyzer (Applied Biosystem, CA, USA).
2.6 Bioinformatic analysis
Forward and reverse sequences were assembled and edited per sample and per gene in CodonCode Aligner v.8.0.2 (Condon Code Co., USA). The ITS2 region of each specimen was detected using a hidden Markov model to remove the 5.8S and 28S regions, which may overlap with the ITS2 region (Keller et al., 2009). A multiple sequence alignment was run for each gene using the Muscle algorithm tool in MEGA 7.0.27 (Kumar et al., 2016). The sequences were manually adjusted, and ambiguous regions were removed.
BLAST was run against known specimens in GenBank. The identification process took place at three levels: family, genus, and species. The query sequences were detected by selecting the highest maximum score and the lowest E-value. Outcomes were classified into three levels, as described in (Meier et al., 2006).
Genetic distances were calculated using the Kimura two-parameter (K2P) model in MEGA v.7.0.27 (Kumar et al., 2016). Inter- and intraspecific distances were calculated as the barcoding gaps by using TaxonDNA v.1.7.8 (Meier et al., 2006). The secondary structures of the ITS2 sequences were predicted using the ITS2 ribosomal RNA database ( http://its2.bioapps.biozentrum.uni-wuerzburg.de/) (Schultz et al., 2005).
Two phylogenetic trees were built in MEGA 7.0.27 (Kumar et al., 2016): a neighbor‐joining (NJ) tree and a maximum likelihood (ML) tree. Both trees were created by running 1,000 bootstrap replicates and K2P model of selection. The gamma distributed invariant site (G 1) nucleotide model was used for for ML tree. The number of discrete gamma categories was set as five (Liu et al., 2012; Kumar et al., 2016).
3 Results
3.1 Evaluation Efficiency of DNA barcoding
The preliminary results obtained from the DNA isolation experiments showed that the method of (Manikandan et al., 2017) allowed for the extraction of relatively more and better quality DNA than other methods, and therefore, DNA was extracted from all samples using this method.
The 21 plant specimens for each gene yielded 21 (100%) PCR products for ITS2 and rbcL (Table 2). The lowest percentages of samples yielding PCR products occurred using the matK and trnH-psbA genes. All 21 (100%) specimens were successfully sequenced for their rbcL genes and 20 (95%) specimens for their ITS2 loci. The ITS2 region had the highest guanine-cytosine content (Table 2). Thus, two rbcL and ITS2 DNA barcodes were selected for the following analyses. (GC): Guanine-cytosine content.
Variable
nrITS regions
cpDNA regions
ITS1
ITS2
matK
rbcL
trnH-psbA
Number of samples
21
21
21
21
21
Mean and range of GC content (%)
49 (46–50)
67 (71.6–64.5)
35 (34–35.1)
44 (42.1–44.4)
27 (26–28)
Efficiency of PCR amplification (%)
17 (80.9%)
21 (100%)
7 (33.3)
21 (100%)
8 (38)
Success rate of sequencing (%)
7 (33.3%)
20 (95%)
6 (28.5)
21 (100%)
8 (38)
Amplified product length (bp)
∼200
∼200
∼800
∼600
∼400
Mean and range of sequenced length (bp)
210 (200–270)
250 (210–270)
767 (763–869)
641 (610–658)
414 (310–448)
3.2 Species resolution and barcode analysis
Based on BLAST, 100%, 85.7%, and 28.5% of the ITS2 sequences were correctly identified at the family, genus, and species levels, respectively (Fig. 1). Furthermore, 100%, 71.4%, and 14.2% of the rbcL sequences were successfully identified at the family, genus, and species levels, respectively. E-values were (0) for each gene.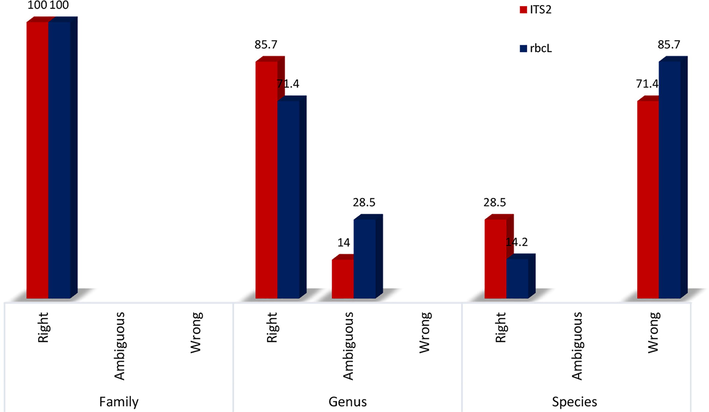
Successful identification of Plectranthus spp., local to the Kingdom of Saudi Arabia at three levels (family, genus, and species) using BLAST and matching sequences drawn from the NCBI database. Values listed in the columns are estimated in percentages (%).
3.3 Inter- and intraspecific distance and barcode gap
The interspecific distances for the rbcL and ITS2 barcode loci are shown in Table 3. The ITS2 locus showed a minimum interspecific distance of 0.139, while the maximum interspecific distance was 0.000. The rbcL loci exhibited a minimum interspecific value of 0.018 and a maximum intraspecific distance of 0.000. Fig. 2 shows that the ITS2 loci have a wider barcode gap than the rbcL loci.
Level
ITS2 locus
rbcL locus
Inter-specific distance
Intra-specific distance
Inter-specific distance
Intra-specific distance
Mean
0.326
0.00
0.038
0.00
Max
0.231
0.00
0.053
0.00
Min
0.139
0.00
0.018
0.00
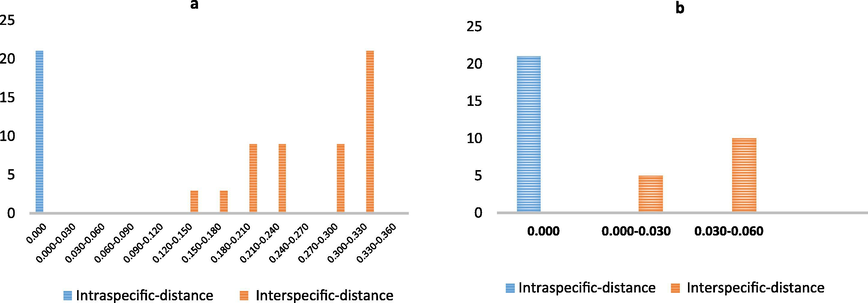
Barcode gaps for DNA barcodes, a-ITS2 and b-rbcL, based on the K2P distance and six Plectranthus spp. local in the Kingdom of Saudi Arabia. Total barcoded sequences for ITS2 = 21, rbcL = 21, three specimens per species.
3.4 ITS2 secondary structure
Fig. 3 shows the predicted secondary structures of the ITS2 sequences used to identify the Plectranthus spp. The six species shared a similar ITS2 morphological structure, with a central ring and four similar helices, namely, I, II, III, and IV. However, there were variations in the stem loop number, size, and position and degree of angle from the center of the spiral arm of the ITS2.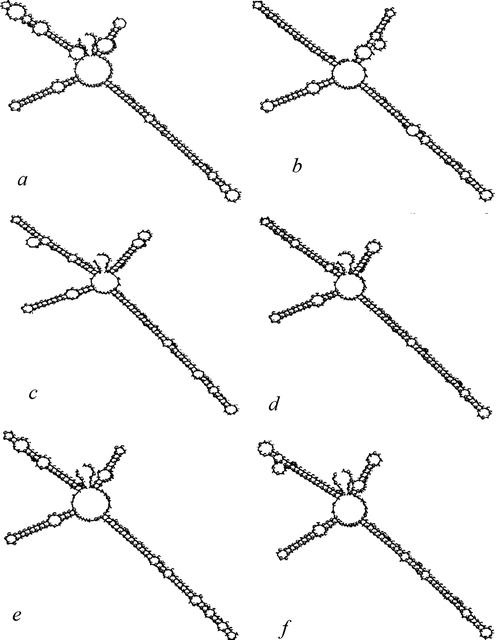
Observed variation in the secondary structure of ITS2 barcodes for the identification of Plectranthus spp. native to the Kingdom of Saudi Arabia, using an ITS2 workbench. a: Plectranthus arabicus. b: Plectranthus barbatus. c: Plectranthus hijazensis. d: Plectranthus pseudomarrubioides. e: Plectranthus asirensis. f: Plectranthus tenuiflorus.
3.5 Phylogenetic trees
The rbcL ML tree (Fig. 4) was constructed using 21 Saudi Arabian Plectranthus spp. sequences, with accessions numbers MN381814–MN381834. The ITS2 ML tree (Fig. 5) was constructed using 20 Saudi Arabian Plectranthus spp. sequences, with accessions numbers MN82135–MN382154. In addition, 36 Plectranthus spp. sequences were downloaded from GenBank representing 8 different countries in Asia, Africa, Australia, and USA.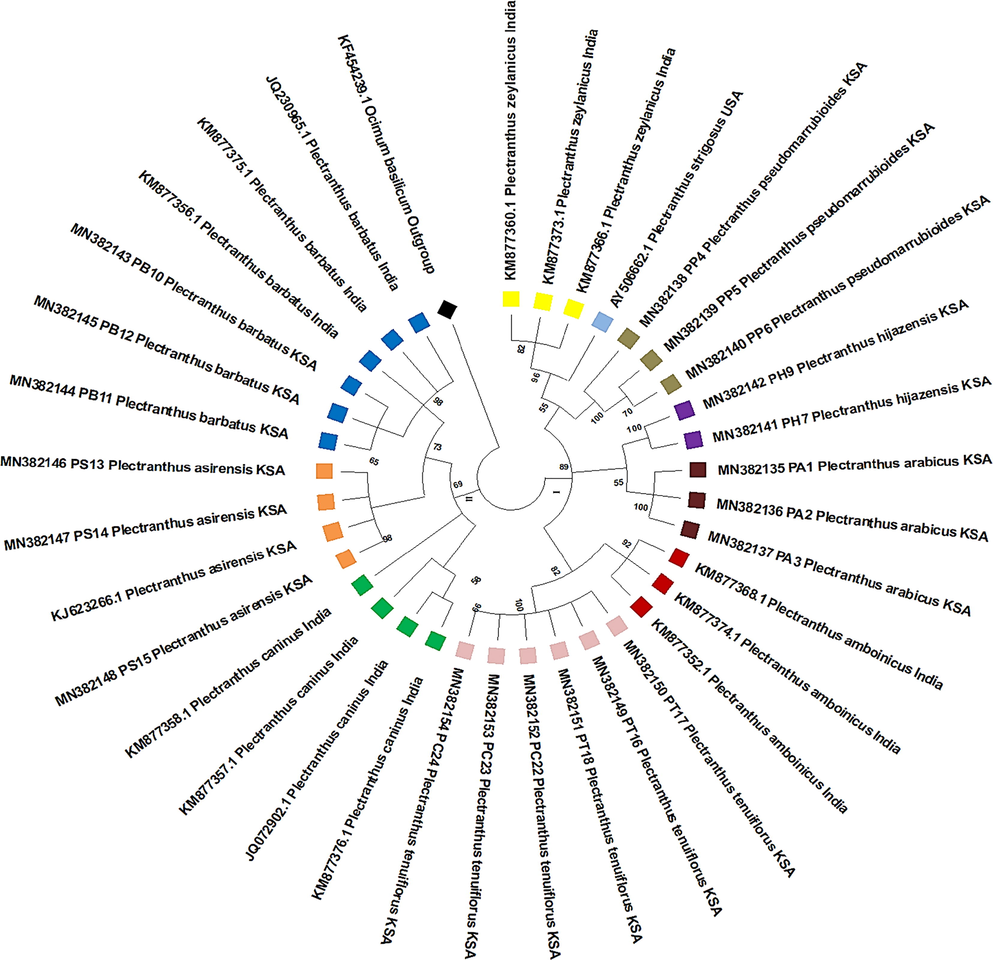
Maximum likelihood tree of ITS2 barcodes showing the phylogenetic relationships among Plectranthus spp. from the Kingdom of Saudi Arabi and other species of Plectranthus originating from different geographic regions. Bootstrap = 1000 replications.
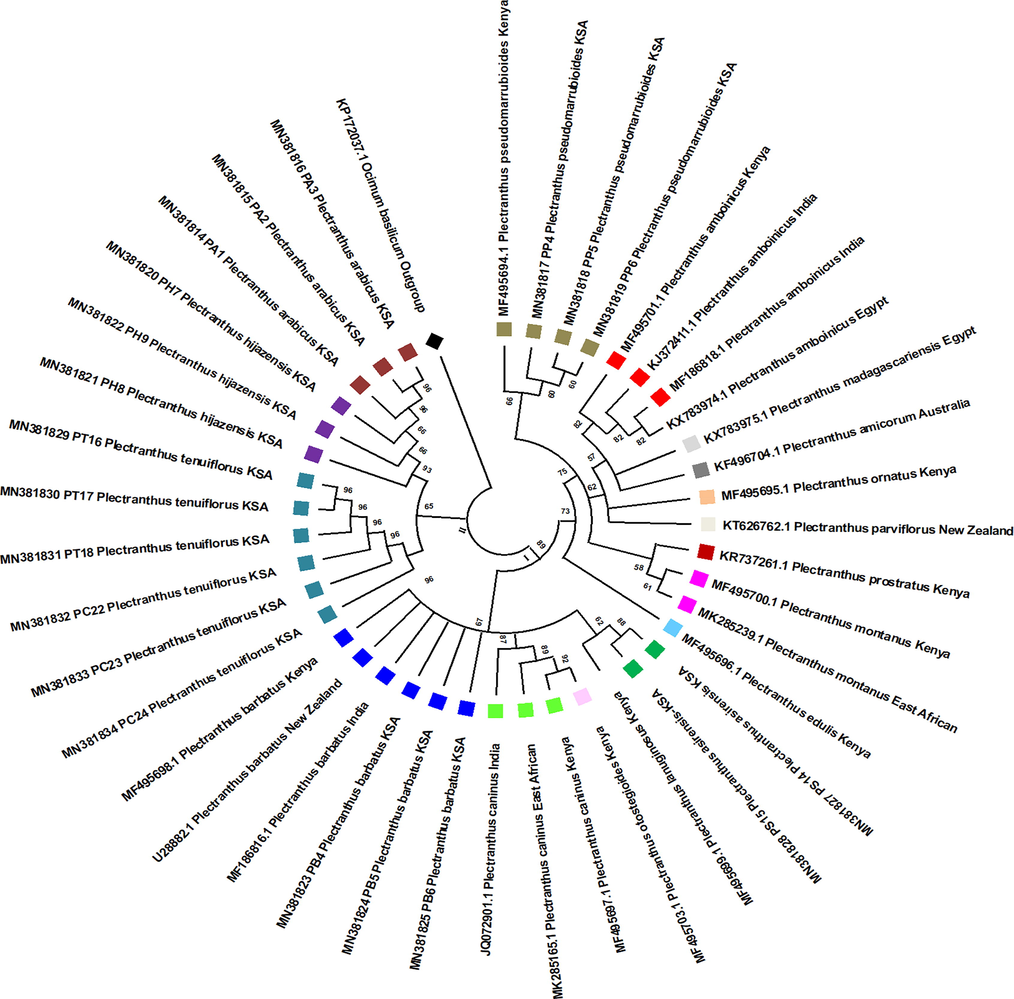
Maximum likelihood tree of rbcL barcodes showing the phylogenetic relationships among Plectranthus spp. from the Kingdom of Saudi Arabia and other species of Plectranthus originating from different geographic regions. Bootstrap = 1000 replications.
Overall, ML produced the best trees in terms of forming monophyletic groups for each species and nodes with high support values. Further, the ML-ITS2 tree showed clades with high support values and branches and good separation of species. The ML tree based on the ITS2 locus data (Fig. 4) divided the sequences into two main clades: Ⅰ and II. Clade I, with a high supporting value (89%), was divided in three main sub-clades, and clade II was also divided into three main branches. The phylogenetic tree constructed from rbcL gene data (Fig. 5) was also split into two clades (I and II). Clade I, with a high support value (89%), was divided into two main branches. Clade II was also divided into two branches. In general, Figs. 4 and 5 show that some species appeared to be highly related to each other as they always appeared in the same clade. For example, P. barbatus P. caninus, P. asirensis, P. otostegioides, and P. lanuginosus appeared to be highly related, as well as P. pseudomarrubioides, P. ornatus, P. montanus, P. amboinicus, P. parviflorus, and P. edulis. Furthermore, P. tenuiflorus, P. hijazensis, and P. arabicus always appeared together in one clade.
4 Discussion
An initial objective of this project was to identify and discriminate between Plectranthus species in Saudi Arabia using a DNA barcoding approach. The present study focused on six Plectranthus species native to Saudi Arabia. The analyses included the first sequences obtained from these six Plectranthus species de novo.
4.1 DNA extraction and PCR challenges
In the Plectranthus genus, there are many issues surrounding DNA isolation and purification, which hinder the acquisition of high-quality nrDNA. These include degradation of DNA owing to endonucleases, co-isolation of highly viscous polysaccharides, inhibitor compounds such as polyphenols, and presence of secondary compounds that directly or indirectly interfere with enzymatic reactions (Manikandan et al., 2017). In the present study, three methods of DNA extraction were compared, and the method described in (Sahu et al., 2012) was found to show the best performance in terms quantity and quality of DNA extracted and successful PCR product collection, particularly with the ITS2 and rbcL loci. This finding is consistent with that of (Manikandan et al., 2017).
It was observed that in dried, old, or degraded samples, fewer PCR products and less successful sequencing were observed, especially for the ITS1, matK, and trnH-psbA loci. The findings of the present study showed that the ITS2 and rbcL loci represented the highest PCR success rates (100%) as well as the highest sequencing success rates (95% and 100%, respectively; Table 2). These observations are consistent with those of (Al-Juhani, 2019), who reported that the rbcL and ITS2 regions had higher rates of amplification and sequencing success, even when using old, dry, or degraded specimens.
4.2 DNA barcodes and species resolution
Al-Qurainy et al., (2014) studied Saudi Arabian P. asirensis and reported the possibility of using nrDNA-ITS and the rbcL and rpoC1 chloroplast loci for DNA barcoding in Plectranthus species. The present study showed that, in Plectranthus, ITS2 exhibited a higher discrimination rate at the genus level (85.7%) and species resolution (28.5%) than rbcL, which showed a discrimination rate of 71.4% at the genus level and species resolution of 14.2% (Fig. 1). The ITS2 locus is, therefore, recommended to identify herbal medicinal plants (Chen et al., 2010), for the following reasons: (i) it is located within the nuclear genome, which has a different rate of evolution than the plastid genome, (ii) it provides higher species resolution, and (iii) it has a much shorter sequence, allowing for higher recovery from processed plant materials found within herbal products.
Moreover, a possible explanation for the relatively low species-level identification in the present results is the lack of sufficient reference data; several species of Plectranthus do not have reference sequence data available in GenBank. There were no reference sequences available for P. hijazensis, P. arabicus, or P. tenuiflorus, for most of the markers selected for examination. Therefore, BLAST revealed closely related species but not specific species and was only able to successfully identify to the genus level, with low species resolution. Thus, the voucher specimens of the present study are the first ever sequences for the aforementioned species deposited in GenBank and are publicly available. (Hollingsworth, 2011) argued that species with a single sample are potentially distinguishable (i.e., for successful species identification) if the sequence is unique.
In general, the minimum interspecific distances were greater than the maximum intraspecific distances for the rbcL and ITS2 barcodes (Table 3). The barcoding gap in the ITS2 region was obvious and larger than that in rbcL, where the minimum interspecific distance for ITS2 was 0.139 and the maximum intraspecific distance was 0.000. Species discrimination is considered successful if the minimum interspecific K2P distance is larger than the maximum intraspecific distance for that species (CPBG China Plant BOL Group, 2011); this condition applies to the samples in the current study as well (Fig. 2).
The secondary structures of the ITS2 are shown in Fig. 3. Similar ITS2 secondary structures were observed across all Plectranthus spp. However, differences were observed in the morphology of the four helices among species. Variation in the secondary structure of ITS2 has previously been reported as a tool for identifying species at the molecular morphological characteristics level (Keller et al., 2010). Liu et al. (2019) noted a clear difference between the ITS2 structures of true “Gaoben” samples and the structures of three adulterant species and recommended using ITS2 as a minibarcode to distinguish between closely or distantly related plant species used in Chinese medicine. Hence, the ITS2 secondary structure could be used to validate Plectranthus species identification.
4.3 Phylogeny and taxonomy of Plectranthus spp.
The results of the present study demonstrated that P. asirensis, P. caninus, P. otostegioides, P. barbatus, and P. lanuginosus were grouped together in clade II of the phylogenetic tree constructed using ITS2 data and in clade I of the rbcL phylogenetic tree. This is also consistent with the observations of (Al-Qurainy et al., 2014) and (Musila et al., 2017), who showed that P. asirensis, P. caninus, and P. barbatus are grouped together in trees built based on the nrITS gene, matK, and rbcL.
Furthermore, we showed that three species, P. tenuiflorus, P. hijazensis, and P. arabicus, were always grouped together in the same clade in the trees constructed based on the ITS2 and rbcL loci. This observation broadly supports the findings of (Abdel Khalik and Osman, 2017), who used RAPD and ISSR markers to study Plectranthus species native to Saudi Arabia and found that these species are clustered together.
Comparisons with the results of (Paton et al., 2018), who utilized the trnL-F, rps16, and trnS-G plastid genomes, confirmed the validity of the findings of the present study. The species studied herein were classified into two branches within the Coleus clade. Furthermore, our results showed that P. hijazensis and P. asirensis were separated in two clades with high bootstrap values, which supports the recommendation of (Abdel Khalik and Osman, 2017) to separate these two species into different subgenera.
The findings of the present study support the observations of (Musila et al., 2017), as P. aegyptiacus (P. tenuiflorus) was grouped into the same clade as P. pseudomarrubioides and P. amboinicus in the tree constructed based on the ITS2 locus. Nevertheless, in the tree based on the rbcL data, P. edulis was grouped with P. pseudomarrubioides, P. ornatus, P. montanus, P. amboinicus, P. amicorum, P. madagascariensis, and P. parviflorus.
The present outcomes are consistent with those in previous studies (Frigerio et al., 2019) that have recommended DNA barcoding (trnH-psbA, ITS, and rbcL 1-B) as a useful tool for identifying medicinal and aromatic plants. Our findings also support the observations of Al-Juhani (2019), who showed that ITS2 and rbcL produce species resolutions of up to 77% for 100 species growing on drylands in KSA, most of which are used in traditional medicine. Han et al., (2016) used ITS2 DNA barcoding to verify and detect adulterants of 295 herbal medicine species in China, confirming that the ITS2 region could be used to identify most of the samples (87.7%) and detect adulterants (Han et al., 2016). Furthermore, (Vassou et al., 2016) used the rbcL region to create an API-RDBL library of 374 medicinal plants used in the Indian Pharmacopoeia and mentioned the usefulness of rbcL DNA barcoding in authenticating herbal drugs and detecting adulterants.
Our results are also in line with the results of (Gao et al., 2010), who highlighted the use of the ITS2 region to identify 114 samples of the Fabaceae family, which contains many important medicinal plants, and confirmed the power of the ITS2 region as a tool to authenticate herbal medicines. Furthermore, (Lv et al., 2020) indicated that ITS2 can identify 98% of medicinal specimens from the Apocynaceae family at the species and genus levels.
Finally, (Le et al., 2020) confirmed the effectiveness of ITS2 barcoding in detecting, identifying, and classifying the genetic relationships in agricultural varieties of palm used in botanical gardens and orchards. Together, these findings strongly support the use of the ITS2 region as a tool to distinguish between varieties of Plectranthus, considering that several species of Plectranthus spp. are used ornamentally and in gardens.
5 Conclusion
This investigation showed that short‐region ITS2 DNA barcodes had strong discriminatory powers. We recommend using ITS2 as a tool to validate the identity of medical Plectranthus species. Molecular processing of the whole‐chloroplast genome, as well as next generation sequencing techniques, are promising modern tools that would be fruitful areas for future research into Plectranthus spp. Further information on both chloroplast and nuclear genes, in addition to micro-morphological outcomes, could provide more definitive evidence to solve problems and distinguish between closely related species and establish a greater degree of accuracy on the taxonomic status of Plectranthus species.
The newly recorded P. hijazensis species is endemic to Saudi Arabia and was found to be genetically distinct from the other studied Plectranthus species. Therefore, the species should be the focus of future research in different fields of chemotaxonomy, micro-morphology, and scanning electron microscope (SEM) studies. Our result also emphasizes the urgent need to intensify efforts to preserve the local natural diversity.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Comparative Anatomy of Stems and Leaves of Plectranthus L. (Lamiaceae) in Saudi Arabia and Systematic Implications. Microsc. Res. Tech.. 2016;79:583-594.
- [CrossRef] [Google Scholar]
- Genetic Analysis Of Plectranthus L. (Lamiaceae) In Saudi Arabia Based On RAPD And ISSR Markers. Pak. J. Bot.. 2017;49:1073-1084.
- [Google Scholar]
- A new species of Plectranthus (Lamiaceae) from Saudi Arabia. Turk. J. Bot.. 2016;40:506-513.
- [CrossRef] [Google Scholar]
- A Systematic Revision of the Genus Plectranthus L. (Lamiaceae) in Saudi Arabia Based on Morphological, Palynological, and Micromorphological Characters of Trichomes. Am. J. Plant Sci.. 2016;7:1429-1444.
- [CrossRef] [Google Scholar]
- Medicinal plants in southwestern Saudi Arabia. Econ. Bot.. 1987;41:354-360.
- [CrossRef] [Google Scholar]
- Evaluation of the capacity of the DNA barcode ITS2 for identifying and discriminating dryland plants. GMR. 2019;18:18133.
- [Google Scholar]
- Selection of DNA barcoding loci and phylogenetic study of a medicinal and endemic plant, Plectranthus asirensis J.R.I. Wood from Saudi Arabia. Genet. Mol. Res.. 2014;13:6184-6190.
- [CrossRef] [Google Scholar]
- Testing the efficacy of DNA barcodes for identifying the vascular plants of Canada. PLoS ONE. 2017;12:e0169515
- [Google Scholar]
- CBOL, Plant Working Group., 2009. A DNA barcode for land plants. Proceedings of the National Academy of Sciences of the United States of America.
- Flora of the Kingdom of Saudi Arabia Illustrated. National Agriculture Research Center, Riyadh: Ministry of Agriculture and Water; 2000.
- Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLos One. 2010;5:e8613
- [CrossRef] [Google Scholar]
- Comparative analysis of a large dataset indicates that ITS should be incorporated into the core barcode for seed plants. PNAS. 2011;108:19641-19646.
- [Google Scholar]
- Collenette, S., 1999. Wild Flowers of Saudi Arabia. National Commission for Wildlife Conservation and Development (NCWCD). Riyadh.
- A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull.. 1987;19:11-15.
- [Google Scholar]
- DNA barcoding to trace Medicinal and Aromatic Plants from the field to the food supplement. J. Appl. Bot. Food. Qual.. 2019;92:33-38.
- [CrossRef] [Google Scholar]
- Identification of medicinal plants in the family Fabaceae using a potential DNA barcode ITS2. J. Ethnopharmacol.. 2010;130:116-121.
- [CrossRef] [Google Scholar]
- Distribution of Exudate Flavonoids in the Genus Plectranthus. Biochem. Syst. Ecol.. 2010;38:335-341.
- [CrossRef] [Google Scholar]
- The short ITS2 sequence serves as an efficient taxonomic sequence tag in comparison with the full-length ITS. BioMed Res. Int.. 2013;5:1-7.
- [CrossRef] [Google Scholar]
- An authenticity survey of herbal medicines from markets in China using DNA barcoding. Sci. Rep.. 2016;6:18723.
- [CrossRef] [Google Scholar]
- 5.8S-28S rRNA interaction and HMM-based ITS2 annotation. Gene. 2009;430:50-57.
- [CrossRef] [Google Scholar]
- Including RNA secondary structures improves accuracy and robustness in reconstruction of phylogenetic trees. Biol. Direct.. 2010;5:1-12.
- [Google Scholar]
- A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE. 2007;2:e508
- [CrossRef] [Google Scholar]
- MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol.. 2016;33:1870-1874.
- [CrossRef] [Google Scholar]
- The utility of DNA barcodes to confirm the identification of palm collections in botanical gardens. PLoS ONE. 2020;15:e0235569
- [CrossRef] [Google Scholar]
- Sampling strategy and potential utility of indels for DNA barcoding of closely related plant species: A case study in Taxus. Int. J. Mol. Sci.. 2012;13:8740-8751.
- [CrossRef] [Google Scholar]
- Molecular Authentication of the Medicinal Species of Ligusticum (Ligustici Rhizoma et Radix, “Gao-ben”) by Integrating Non-coding Internal Transcribed Spacer 2 (ITS2) and Its Secondary Structure. Front. Plant Sci.. 2019;10:429.
- [Google Scholar]
- Identification of medicinal plants within the Apocynaceae family using ITS2 and psbA-trnH barcodes. Chin. J. Natural Med.. 2020;18:594-605.
- [CrossRef] [Google Scholar]
- Discriminatory power of rbcL barcode locus for authentication of some of United Arab Emirates (UAE) native plants.3. Biotech.. 2017;7:144.
- [CrossRef] [Google Scholar]
- Optimizing the pure genomic DNA isolation procedure for Plectranthus amboinicus DNA – a prerequisite for further genomic studies. J. Appl. Adv. Res.. 2017;2:249-255.
- [CrossRef] [Google Scholar]
- DNA barcoding and taxonomy in Diptera: A tale of high intraspecific variability and low identification success. Syst. Biol.. 2006;55:715-728.
- [CrossRef] [Google Scholar]
- The nuclear internal transcribed spacer (ITS2) as a practical plant DNA barcode for herbal medicines. J. Appl. Res. Med. Aromat. Plants. 2016;3:94-100.
- [CrossRef] [Google Scholar]
- DNA barcoding: an efficient tool to overcome authentication challenges in the herbal market. Plant Biotechnol. J.. 2016;14:8-21.
- [CrossRef] [Google Scholar]
- DNA barcoding of five medicinal plants from Siwa Oasis, Egypt. KMITL. 2016;16:49-56.
- [Google Scholar]
- Phylogeny of Ten Kenyan Plectranthus Species in the Coleus Clade Inferred from Leaf Micromorphology, rbcL and matK Genes. J. Bot. 2017:1-16.
- [CrossRef] [Google Scholar]
- Variation of karyotype and nuclear DNA content among four species of Plectranthus L’ Héritier, 1788 (Lamiaceae) from Brazil. Comp. Cytogenet.. 2015;9:549-563.
- [CrossRef] [Google Scholar]
- Plant DNA barcodes and the influence of gene flow. Mol. Ecol. Mol. Ecol. Resour.. 2012;12:575-580.
- [Google Scholar]
- Phylogeny and evolution of basils and allies (Ocimeae, Labiatae) based on three plastid DNA regions. Mol. Phylogenet. Evol.. 2004;31:277-299.
- [CrossRef] [Google Scholar]
- Phylogenetic study of Plectranthus, Coleus and allies (Lamiaceae): taxonomy, distribution and medicinal use. Bot. J. Linn. Soc.. 2018;188:355-376.
- [CrossRef] [Google Scholar]
- DNA extraction protocol for plants with high levels of secondary metabolites and polysaccharides without using liquid nitrogen and phenol. Int. Sch. Res. Notices 2012:1-6.
- [CrossRef] [Google Scholar]
- A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA. 2005;11:361-364.
- [Google Scholar]
- Assessing product adulteration in natural health products for laxative yielding plants, Cassia, Senna, and Chamaecrista, in Southern India using DNA barcoding. Int. J. Legal Med.. 2015;129:693-700.
- [CrossRef] [Google Scholar]
- Genome-based approaches to the authentication of medicinal plants. Planta Med.. 2008;74:603-623.
- [CrossRef] [Google Scholar]
- Creation of reference DNA barcode library and authentication of medicinal plant raw drugs used in Ayurvedic medicine. BMC Complement. Altern. Med.. 2016;16:9-15.
- [CrossRef] [Google Scholar]
- An integrated system for identifying the hidden assassins in traditional medicines containing aristolochic acids. Sci. Rep.. 2015;5:11318.
- [CrossRef] [Google Scholar]
- Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS One. 2010;5:e13102
- [CrossRef] [Google Scholar]
- Barcode ITS2: A useful tool for identifying Trachelospermum jasminoides and a good monitor for medicine market. Sci. Rep.. 2017;7:5037.
- [CrossRef] [Google Scholar]
- DNA Barcoding: A Tool for Standardization of Herbal Medicinal Products (HMPS) Of Lamiaceae from Pakistan. Pak. J. Bot.. 2017;48:2167-2174.
- [Google Scholar]







