Translate this page into:
Honey bee gut an unexpected niche of human pathogen
⁎Corresponding author. ishtiaq@kust.edu.pk (Syed Ishtiaq Anjum)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The gut microbiota of honey bees (Apis mellifera) can be symbiotic or pathogenic and therefore, important for bee survival and honey production. To study gut cultivable bacteria of honey bees, 30 honey bee samples were collected from district Kohat of Khyber Pakhtunkhwa Province as there is no information about the diversity of bee gut microbiota from Pakistan. Complete digestive system of the worker bee was dissected and processed for bacterial isolation. A total of 219 bacteria were obtained and characterized by bacterialogical parameters. The human pathogenic bacterial isolates were identified and confirmed on 16S ribosomal DNA sequencing. Combined microbiological practices revealed the presence of following bacterial genera: Bacillus, Enterococcus, Escherichia, Micrococcus, Morganella, Ochrobactrum, Pseudomonas, Salmonella, Shigella, Sphingomonas and Staphylococcus. Two pathogenic bacteria Salmonella enterica and Shigella sonnei causing diseases in man and other animals are confidently characterized. This work suggested that forager Apis mellifera gut acts as reservoir and potential vector of bacterial pathogens.
Keywords
Apis mellifera
Bacterial flora
Colony PCR
16S rDNA sequencing
1 Introduction
A wide range of pathogens as viruses, bacteria and parasites may affect honey bee colony, some are more harmful that leads to colony collapse (Potts et al., 2010; Evans and Schwarz, 2011). The honey bee populations are going to decline in various parts of the world and these could be due to adverse effect of multiple honey bee pathogens. Further, increasing prevalence of parasites and pathogens are among the most significant threats to managed bee colonies. (O’Neal et al., 2018; Cox-Foster et al., 2007). Like two known bacteria (Melissococcus plutonius and Paenibacillus larvae) that effect honey bees brood not adults and causes significant losses to beekeepers around the world. Many aspects of their transmissions, virulence and adult host mortality are poorly documented and still remain obscure (Genersch, 2010).
Contaminated water not only acts a reservoir of pathogenic bacteria (Khalil et al., 1994) but access to polluted water affect the health of insects especially honey bee (Staveley et al., 2014). The usage of reclaimed water sugar solution as a drinking water had negative impacts on the average deaths of the honeybee colonies. Reclaimed water also alters the shape of mid gut of bees (Hananeh, et al., 2014). Water resources are polluted having various animal and plants pathogenic bacteria. Several pathogens, in feces of animals are transported to plants by non-potable irrigation water, fertilizer and insects (Tyler and Triplett, 2008). As honey bee spread plant bacterial pathogens including Erwinia amylovora (fire blight pathogen) and Pseudomonas syringae while pollinating various plants (Pattemore et al., 2014)
A range of techniques has been used so far to characterize the gut bacterial flora of honey bee like culture dependent method and phenotypic screenings. Although biochemical characterization is very helpful in bacterial identification but Sanger-based and next generation DNA sequencing techniques are useful in identification of distinctive set of bacteria present in honey bees gut (Li et al., 2012; Cox-Foster et al., 2007; Engel et al., 2012). So this study aimed to investigate the bacterial communities from the digestive tract of managed honey bee workers, captured in North West districts of Khyber Pakhtunkhwa Pakistan, using bacteriological and molecular techniques. These findings will improve current knowledge on the composition and structure of bee gut microbiota and provide the framework for understanding their contribution to honey bee health and potential application in disease control.
2 Material and methods
2.1 Study area
Kohat is 2545 square kilometers area, located at 33°35′13 N 71°26′29E in Khyber-Pakhtunkhwa province of Pakistan. River Indus is present on the east (Zone 1) where cruciferous vegetation attracts beekeepers to manage farms. On the west (Zone 2) a rich agricultural land irrigated by Tanda dam while on the north and the south (Zone 3) rocky dry hills and slopes with large patches of open croplands provide a center of attraction for migratory beekeeping practices.
2.2 Sample collection and dissection of the bees
In order to study the cultivable honey bee gut bacteria, 30 unhealthy worker honey bees (bees were unable to fly and crawl on the ground in front of the hive.) were collected from honey bee farms distributed in cruciferous vegetation (Zone 1–3) in district Kohat Khyber Pakhtunkhwa Pakistan. After collection, live bees were transported to the laboratory of Entomology/Bee lab Department of Zoology Kohat University of Science and Technology Kohat, Khyber Pakhtunkhwa Pakistan in the small cages containing sugar powder followed by storage at −4 °C until processing. Before dissection, whole bees were washed in 95% ethanol in conical tubes. The complete digestive systems of bees were aseptically dissected by clipping the stinger with sterile forceps and carefully pulled the whole gut. The dissected guts were macerated with sterile dissection scissors in 0.8% NaCl solution and immediately stored at −80 °C. (Anjum et al; 2018. Ellegaard and Engel, 2019).
2.3 Culturing and identification of bacteria
From the preserved bee gut samples, different dilutions (i.e. 1/10, 1/100 and 1/1000) were made and 100 µl aliquots of the diluted sample were inoculated in LB Agar plates for 24–48 h at 37 °C. The separated colonies in master plates were sub cultured in LB agar plates and incubated at 37 °C. The isolated colonies were characterized by various bacteriological techniques like colony morphology, gram staining followed by various biochemical tests with the help of Bergey’s Manual and API 20 by procedure mentioned elsewhere (Iqbal et al., 2014; Hussain et al., 2013). Further, identification of selected pathogenic bacteria was achieved by 16s rDNA analysis.
2.4 Colony PCR and DNA sequencing
Single isolated bacterial colony was subjected to amplification of the 16S rDNA gene using thermal cycler (BioRed USA) according to Khan et al. (2014). PCR product (10 µl) was analyzed after electrophoresis and 40 µl was purified with PCR clean up kit (Invitrogen Inc. USA). The DNA estimation was carried by using Qubit dsDNA Hs assay Kit USA with Qubit 2.0 Fluorometer (Invitrogen, Life Technology USA). The pure quantified PCR products were sequenced using either forward or reverse primers and Big Dye Terminator Kit (Applied Biosystems Inc., USA) by Genetic Analyzer (ABI 377 Applied Biosystems Inc., USA).
2.5 Sequence analysis
The 16S rDNA sequence of Salmonella spp and Shigella spp were compared through BLAST with known 16S rRNA gene sequences in the GenBank and analysed using sequence scanner v1.0 (Applied Biosystems) and Chromas Lite 2.1.1 (Technelysium Pty Ltd) software packages mentioned elsewhere (Gantner et al., 2011; Martínez-Hernández et al., 2013).
3 Results discussions
A total of 219 bacterial isolates from gut of 30 worker honeybees were obtained and the presence of 11 genera were confidently identified (Fig. 1). 23% Salmonella and 18% Shigella were characterized up to species level by using bacteriological parameters combined with 16s rDNA gene sequencing. BLAST searching revealed that 87% of the obtained sequences were identical to Salmonella enterica and 84% of Shigella sonnei (Table 1). Generally, gut bacterial diversity of honey bee collected from three zones were almost similar but the Enterococcus spp in zone 1 and zone 2 and Pseudomonas spp in zone 3 were prevalent (Fig. 2). Five bacterial genera, Bacilli, Staphylococci, Escherichia, Pseudomonas and Micrococci (Fig. 1) appeared recurrently and may contribute the major flora of bee gut. Colonies PCR of the selected pathogenic bacterial isolates were amplified, except few colonies of Salmonella and Shigella (Fig. 3). The isolates KHT03 and KHT30 produced low quality PCR product (Fig. 3) which were not subjected to BLAST analysis. *Percent recovery was calculated as (number of isolated colonies from which Salmonella/Shigella were recovered/total number of colonies isolated) × 100. And Percent confirmation was calculated as (number of colonies confirmed as Salmonella enterica/Shigella sonnei /number of typical colonies picked) × 100.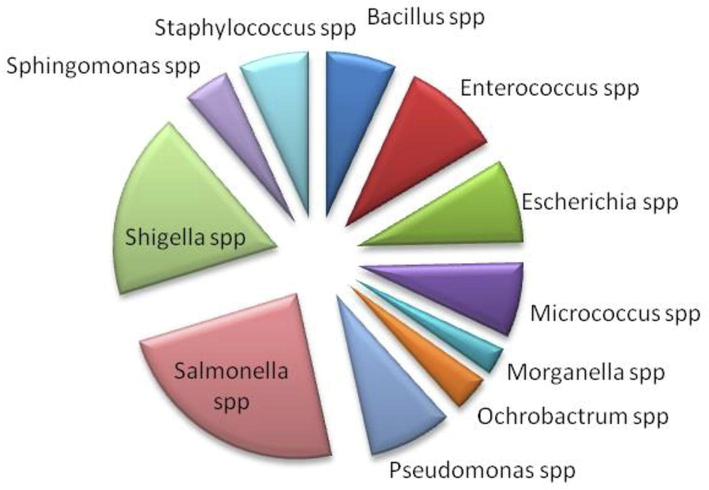
Diversity of bacteria harboring in the guts of worker bees in Kohat.
Selected samples
Number (%)recovered
No. colonies
No. (%)confirmed
sequence homology
Accession no. (GenBank)
Salmonella spp.
52(23)
40
35 (87)
Salmonella enterica subsp. DSM 9220
NR 044372.1
Shigella spp.
40(18)
33
28 (85)
Shigella sonnei strain CECT 4887
NR 104826.1
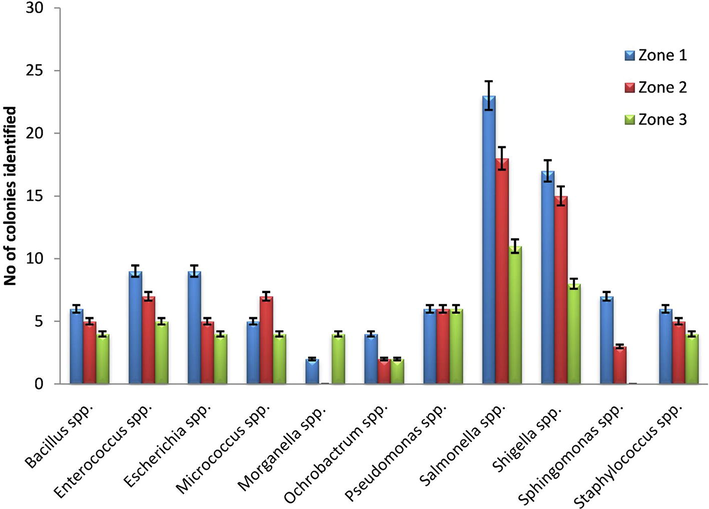
Guts bacterial isolates of honeybee collected from different regiones in Kohat.
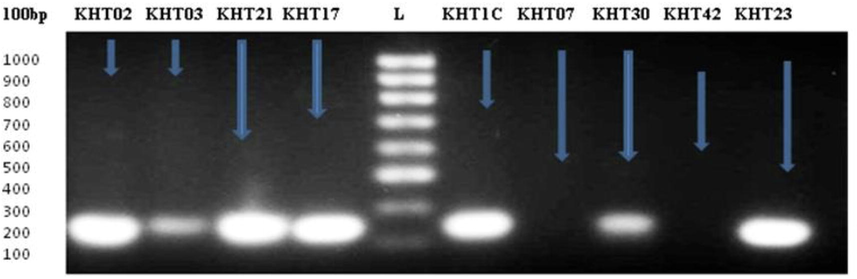
Showing bands of 16s rDNA gene amplification, two lanes (KHT02 and KHT23) visualizing Shigella sonnei DNA fragments while three lanes (KHT21, KHT17 and KHT1C) separating Salmonella enterica DNA fragments, 100 bp DNA ladder (L).
Indeed, honey bee populations all over the world are at high risk of decline due to unidentified reasons however a high load of parasites and microbial pathogens especially bacteria strongly connected with the disappearing of bee population (Olofsson and Vásquez, 2008; Gilliam, 1997; Di Prisco et al., 2013; Core et al., 2012). Additionally, pathogens are not passive microbes when they enter the arthropod vector but actively influence vector gene expression that can manipulate the local environment (Abraham et al., 2017)
The present study demonstrated 219 bacterial isolates of 11 bacterial genera, identified (Fig. 1) from worker honey bee gut, collected from the different bee yards in Kohat. The worker bee harbored some well-known pathogenic or potentially pathogenic bacteria species, including Shigella sonnei and Salmonella enterica. These bacteria are known for various diseases such as diarrheal illnesses, nausea, vomiting, and fever in human being (Greeley, 2013; Karlsson et al., 2013; Holman et al., 2014). The Diptera flies as a carrier of pathogenic bacteria like Salmonella enterica have previously been reported (Holt et al., 2007), however the presence of the human pathogen in bee gut are quit alarming. Although it has been reported that honey bee spread plant bacterial pathogens including Erwinia amylovora and Pseudomonas syringae while pollinating various plants (Pattemore et al., 2014).).
Enterococcus spp in the guts of Apis nigrocincta based on an identity BLAST search of its COI region also revealing the richness of the group in the gut Apis spp. (Lombogia et al; 2020). Recently, bacterial isolates (Enterococcus spp.) were isolated from Egyptian honey bee’s intestinal tract and were expected that honey bee has the potential to be a source of new bacteria. (Elzeini et al; 2020). Pseudomonas spp. Enterococcus spp. were also recovered from ventriculum of Apis mellifera ligustica in Italy (Kačániová et al; 2020). Consequently, prevalence of the Enterococcus spp Enterococcus spp in bee guts collected from Zone 1–3 are in accordance of the recent published reports. However, differences and richness may be due to the type strains, procedures, and condition of cultivation. Similarly, Pseudomonas spp were also most abundant bacteria followed by Bacillus sp in drinking water of the study areas (Hussain et al; 2013).
The high incidence of bacteria presents in bee’s gut is a public health risk, as the synanthropic behavior of bees may conducive to disseminate through a wide variety of routes (Menasria et al., 2014). Worker honey bees forage in different sites where sugar food is prepared, processed, stored and thus it may increase likelihood of the risk of bacterial transmission. The lack of sufficient food is partially a management issue in bee keeping practices (Mattila and Otis, 2007). Beekeepers usually feed sugar solution during starvation but quality and diversity of sugar sources can affect number of bees (Pernal and Currie, 2001). Much like the human gut microbiota, many bee gut bacteria are specific to the bee gut and can be directly transmitted between individuals through social interaction. (Zheng et al., 2018). Apart from bee sociability, the main risk factor of transmission is water as in the study area it is highly contaminated with human pathogenic bacteria (Hussain et al., 2013). It is suggested that these pathogenic bacteria in bee gut are transferred from foraging sites or sugar supplements through contaminated water.
Moreover, it has been reported that sugar feeding like HFCS (high fructose corn syrup and SS (Sucrose syrup) decrease life span of worker bees (Sammataro and Weiss, 2013) and also colony collapse disorder (CCD) is an alarming colony mortality, correlated with the deleterious effects of sugar feeding (Van Engelsdorp et al., 2007; VanEngelsdorp and Meixner, 2010). But it is observed in the current study that beekeepers always use feeder continuously without sterilization for sugar feeding during starvation to honey bees. It is obvious that the feed or the water may be contaminated with pathogens like bacteria and may be ultimately resulted in bee’s population declining and also a source of transmission. However, there is a need of research to correlate bacteria present in bee gut to honey bee morbidity and mortality and to evaluate the sugar solutions used for bee feeding, hive tools, feeder and water as source of transmission of these bacteria.
Presence of human pathogenic bacteria in bee gut may be the main cause of declining of bee population. The same were proved while injecting various pathogenic bacteria (Salmonella enterica, Staphylococcus aureus, Listeria monocytogenes, Pseudomonas aeruginosa and Serratia marcescen) to honey bees Apis mellifera. Moreover, the infected model bee with S. aureus also act as a vector to other hive mate and 50% of them died within 24 hrs (Ishii et al., 2014). Similar studies were carried out on Drosophilla melanogestar infected with P. aeruginosa and same results were obtained (Linder et al., 2008). Serratia marcescens, an opportunistic pathogen of many plants and animals, including humans, is a virulent opportunistic pathogen of honey bees, which could contribute to bee decline (Raymann et al., 2018). This indicates that human pathogenic bacteria may be the main cause of honey bee mortality and bee population declining.
4 Conclusion
It is concluded that honey bee gut is an alternate habitat for human pathogenic bacteria as high load of these bacteria recovered from the alimentary canal of honey bees. Furthermore, Salmonella and Shigella may play major role in mortality of honey bee in the study area.
Acknowledgements
We acknowledge the financial support provided by the higher education commission of Pakistan. The authors also extend their appreciation to the Researchers supporting project No. (RSP-2020/94) King Saud University, Riyadh Saudi Arabia. Fahad Aldakheel extends his appreciation to the College of Applied Medical Sciences Research Center at King Saud University, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Characterization of gut bacterial flora of Apis mellifera from north-west Pakistan. Saudi J. Biol. Sci.. 2018;25(2):388-392.
- [Google Scholar]
- Pathogen-mediated manipulation of arthropod microbiota to promote infection. Proc. Natl. Acad. Sci.. 2017;114(5):E781-E790.
- [Google Scholar]
- A new threat to honey bees, the parasitic phorid fly Apocephalus borealis. PLoS ONE.. 2012;7:e29639
- [Google Scholar]
- A metagenomic survey of microbes in honey bee colony collapse disorder. Science.. 2007;318:283-287.
- [Google Scholar]
- Neonicotinoid clothianidin adversely affects insect immunity and promotes replication of a viral pathogen in honey bees. Proc. Natl. Acad. Sci... 2013;110:18466-18471.
- [Google Scholar]
- Genomic diversity landscape of the honey bee gut microbiota. Nat. Commun.. 2019;10(1):1-13.
- [Google Scholar]
- Isolation and identification of lactic acid bacteria from the intestinal tracts of honey bees, Apis mellifera L., in Egypt. J. Apic. Res. 2020:1-9.
- [Google Scholar]
- Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci.. 2012;109:11002-11007.
- [Google Scholar]
- Bees brought to their knees: microbes affecting honey bee health. Trends Microbiol.. 2011;19:614-620.
- [Google Scholar]
- Novel primers for 16S rRNA-based archaeal community analyses in environmental samples. J. Microbiol. Methods. 2011;84:12-18.
- [Google Scholar]
- Honey bee pathology: current threats to honey bees and beekeeping. Appl. Microbiol. Biotechnol.. 2010;87:87-97.
- [Google Scholar]
- Identification and roles of non-pathogenic microflora associated with honey bees. FEMS Microbiol. Lett.. 1997;155:1-10.
- [Google Scholar]
- Greeley, R., 2013. Waterborne Outbreak of Shigella sonnei Associated with a State Park--New Jersey 2012, 2013 CSTE Annual Conference, Cste.
- Does reclaimed water induce morphological changes in mid guts of honeybees (Apis mellifera syriaca) Veterinary Sci. Dev.. 2014;4
- [Google Scholar]
- A community outbreak of Salmonella enterica serotype Typhimurium associated with an asymptomatic food handler in two local restaurants. J. Environ. Health. 2014;77:18-20.
- [Google Scholar]
- Isolation of Salmonella enterica serovar Enteritidis from houseflies (Musca domestica) found in rooms containing Salmonella serovar Enteritidis-challenged hens. Appl. Environ. Microbiol.. 2007;73:6030-6035.
- [Google Scholar]
- Biochemical characterization and identification of bacterial strains isolated from drinking water sources of Kohat, Pakistan. African J. Microbiol. Res.. 2013;7:1579-1590.
- [Google Scholar]
- Culturable aerobic and facultative anaerobic intestinal bacterial flora of black cobra (Naja naja karachiensis) in Southern Pakistan. ISRN Veterinary Sci.. 2014;2014:5.
- [Google Scholar]
- Establishment of a bacterial infection model using the European honeybee, Apis mellifera L. PloS One. 2014;9:e89917
- [Google Scholar]
- Outbreak of infections caused by Shigella sonnei with reduced susceptibility to azithromycin in the United States. Antimicrob. Agents Chemother.. 2013;57:1559-1560.
- [Google Scholar]
- In vitro antagonistic effect of gut bacteriota isolated from indigenous honey bees and essential oils against paenibacillus larvae. Int. J. Mol. Sci.. 2020;21(18):6736.
- [Google Scholar]
- Flies and water as reservoirs for bacterial enteropathogens in urban and rural areas in and around Lahore, Pakistan. Epidemiol. Infect.. 1994;113:435-444.
- [Google Scholar]
- Isolation and 16s rDNA sequence analysis of bacteria from dieback affected Mango Orchards in Southern Pakistan. Pak. J. Bot.. 2014;46:1431-1435.
- [Google Scholar]
- The prevalence of parasites and pathogens in Asian honeybees Apis cerana in China. PLoS ONE. 2012;7:e47955
- [Google Scholar]
- The effects of temperature on host–pathogen interactions in D. melanogaster : who benefits? J. Insect Physiol.. 2008;54:297-308.
- [Google Scholar]
- Lombogia, C. A., Tulung, M., Posangi, J., & Tallei, T. E. (2020). Bacterial Composition, Community Structure, and Diversity in Apis nigrocincta Gut. International Journal of Microbiology, 2020.
- Resistance and inactivation kinetics of bacterial strains isolated from the non-chlorinated and chlorinated effluents of a WWTP. Int. J. Environ. Res. Public Health. 2013;10:3363-3383.
- [Google Scholar]
- Dwindling pollen resources trigger the transition to broodless populations of long-lived honeybees each autumn. Ecol. Entomol.. 2007;32:496-505.
- [Google Scholar]
- Bacterial load of German cockroach (Blattella germanica) found in hospital environment. Pathogens Global Health. 2014;108:141-147.
- [Google Scholar]
- Detection and identification of a novel lactic acid bacterial flora within the honey stomach of the honeybee Apis mellifera. Curr. Microbiol.. 2008;57:356-363.
- [Google Scholar]
- Evidence of the role of honey bees (Apis mellifera) as vectors of the bacterial plant pathogen Pseudomonas syringae. Australas. Plant Pathol.. 2014;43:571-575.
- [Google Scholar]
- The influence of pollen quality on foraging behavior in honeybees (Apis mellifera L.) Behav. Ecol. Sociobiol.. 2001;51:53-68.
- [Google Scholar]
- Declines of managed honey bees and beekeepers in Europe. J. Apic. Res.. 2010;49:15-22.
- [Google Scholar]
- Comparison of productivity of colonies of honey bees, Apis mellifera, supplemented with sucrose or high fructose corn syrup. J. Insect Sci.. 2013;13
- [Google Scholar]
- A causal analysis of observed declines in managed honey bees (Apis mellifera) Hum. Ecol. Risk Assess.: Int. J.. 2014;20:566-591.
- [Google Scholar]
- Interactions between pesticides and pathogen susceptibility in honey bees. Curr. Opin. Insect Sci.. 2018;26:57-62.
- [Google Scholar]
- Plants as a habitat for beneficial and/or human pathogenic bacteria. Annu. Rev. Phytopathol.. 2008;46:53-73.
- [Google Scholar]
- A survey of honey bee colony losses in the US, fall 2007 to spring 2008. PLoS ONE. 2007;3:e4071.
- [Google Scholar]
- A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. J. Invertebr. Pathol.. 2010;103:S80-S95.
- [Google Scholar]







