Translate this page into:
High expression of LncRNA-MALAT1 in renal tubular cells induced by high glucose and its promotion role on pyroptosis by regulating miR-206
⁎Corresponding author at: Department of Traditional Chinese Medicine, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui 230001, China. yurucui021@163.com (Rucui Yu),
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background
Diabetic nephropathy (DN) is the main reason for renal failure in terminal stage. Studies have shown that long-chain non-coding RNA (LncRNA) improves DN by regulating microRNA (miR). The study intended to investigate the role of MALAT1/miR-206 on high glucose (HG)-induced inflammation, oxidative stress and cell death of renal tubular epithelial cells (RTECs).
Methods
HG-intervened HK-2 cells to establish DN cell model. Transfection effect was confirmed by qRT-PCR and MALAT1, NLRP3, Caspase-1, IL-1β, GSMDD-N and miR-206 were tested. Western blot was performed to test the proteins in NLRP3, Caspase-1, IL-1β and GSMDD-N, and to observe the pyroptosis. Elisa kit was applied to detect TNF-α, IL-6 and MCP-1.
Results
Double luciferase report experiment and RNA immunoprecipitation (RIP) confirmed that MALAT1 could bind to miR-206. HG induced MALAT1 in HK-2 cells increased. Knocking down MALAT1 could reduce TNF-α, IL-6 and MCP-1. In HG-intervened HK-2 cells, the over-expressed miR-206 transfected into the cells was consistent with the changes of inflammatory factors and cytokines after knock-down MALAT1.
Conclusion
Up-regulation MALAT1 could reverse the inhibition of miR-206 on pyroptosis and inflammatory response. MALAT1 can improve HG-induced HK-2 pyroptosis by targeting miR-206, which is expected to be a latent target to treat DN in clinic.
Keywords
MALAT1
miR-206
Pyroptosis
High glucose
Renal tubular cells
1 Introduction
Diabetic nephropathy (DN), as the main reason for renal disease in terminal stage, is also a common complication of diabetic patients (Ioannou, 2017; Kitada and Koya, 2017). Reports show that about 30%–40% of diabetic patients will deteriorate to DN, and about 50% of DN patients will eventually develop to ESRD (Feng et al., 2018; Kanwar et al., 2011; Magee et al., 2017). Patients can only undergo dialysis and kidney transplantation, which undoubtedly increases the burden of patients. At present, there are different opinions on the mechanism of DN, including the regulation of protein kinase C (PKC), transforming growth factor β (TGF-β) and activation of related inflammatory pathways (Al-Onazi et al., 2016; Soetikno et al., 2011). However, these methods have little effect on clinical treatment of DN. Therefore, it is of great significance to further explore the potential mechanism of DN progress and provide new strategies for prevention and treatment of DN.
Non-coding RNA (ncRNA) is frequently investigated, among which long-chain non-coding RNA (LncRNA) has attracted more and more attention in recent years (St Laurent et al., 2015; Bhan et al., 2017). LncRNA with more than 20nt was initially taken as a metabolic waste of transcription process (Hou et al., 2018). However, it has been found in recent years that LncRNA can regulate microRNA(miR) by acting as endogenous competitive RNA (ceRNA) to participate in the occurrence of various diseases (Paraskevopoulou and Hatzigeorgiou, 2016). For example, other studies have found that MALAT1 aggravates renal tubular epithelial fibrosis. LncRNA MALAT1, also known as LINC00047, was the first discovered LncRNA (Nie et al., 2016). Studies have found MALAT1-intervened high glucose (HG) can induce the epithelial-mesenchymal transition and damage in HK-2 cells (Wang et al., 2020).
Pyroptosis, also known as inflammatory cell necrosis, is a newly proposed programmed cell death method (Zhang et al., 2019). Compared with other cell death modes, it is characterized in that the cell membrane ruptures due to the continuous cell expansion, which further releases substances in the cell to activate a strong inflammatory reaction and aggravate the disease progression of patients (Vande Walle and Lamkanfi, 2016; Shi et al., 2017; Chou et al., 2019). LncRNA-miR axis was found to improve oxidative stress and pyroptosis of renal tubular cells (Zhu et al., 2020). However, it is not clear whether MALAT1 has the same effect of inhibiting pyroptosis in renal tubular epithelial cells (RTECs) caused by HG. Therefore, this time, we investigated the role of MALAT1 on pyroptosis of RETCs caused by HG, and to provide potential therapeutic schemes for clinical use.
2 Methods and materials
2.1 Collection of clinical samples
Forty-one patients with DN from January 2016 to January 2018 were obtained as the patient group, and the other 30 healthy individuals as the healthy control group. Peripheral blood was obtained and centrifuged to obtain serum samples for detection. This research conformed to the Medical Ethics Committee, and the informed consent forms were obtained. The inclusion criteria of DN patients were as follows: the glomerular filtration rate (GFR) was<60 ml/min/1.73 m2, and the urinary protein/creatinine ratio was more than 30 mg/g in patients with positive urine protein (the amount of urine protein was greater than 0.5g/24 h) or the excretion rate of urine albumin was continuously greater than 200μg/ min. The exclusion criteria were as follows: all patients were unable to cooperate with the investigation and statistics, and patients were complicated with malignant tumors. The baseline data of patients were listed in Table 1.
Factor
DN (n = 41)
Healthy control group (n = 30)
P value
Gender
0.770
Male (n = 44)
26
18
Female (n = 27)
15
12
Age
0.705
≥60 years old (n = 35)
21
14
< 60 years old (n = 36)
20
16
Basic diseases
Hypertension (n = 30)
19
11
0.415
Hyperlipidemia (n = 18)
12
6
0.375
COPD(n = 5)
3
2
0.916
Blood glucose (mmol/L)
12.65 ± 4.54
–
HbA1c(%)
8.85 ± 2.23
–
SCr (μmol/L)
93.11 ± 10.21
–
2.2 Cell culture and model construction
HK-2 were obtained from ATCC. The purchased cells were placed in RPMI 1640 including 10% FBS, 100 U/mL penicillin and 100 μg/ mL streptomycin, and cltured with 5% CO2 at 37 °C. After starvation in serum-free for 24 h, we established control group (CG) and HG group respectively to construct HK-2 cells induced by HG. The CG received 5.5 mM glucose for 12 h, 24 h and 48 h, and the HG group received 30 mM glucose for 12 h, 24 h and 48 h.
2.3 Cell transfection
In this study, Lipofectamine 2000 kit was used for transfection. pcDNA3.1-NC, -MALAT1, and si-NC, -MALAT1#1, -MALAT1#2, miR-206-mimics, miR-NC were obtained from Sangon Biotech. The above vectors were transfected into HG cells, while the control group was not transfected. The transfection effect was verified by qRT-PCR. The primer sequences were listed in Table 2.
Gene
Upstream primer (5′–3′)
Downstream primer (5′–3′)
si-MALAT1#1
GGAAGUAAUUCAAGAUCAAGA
UUGAUCUUGAAUUACUUCCGU
si-MALAT1#2
GCGACGAGUUGUGCUGCUAUC
UAGCAGCACAACUCGUCGCUG
pcDNA3.1-MALAT1
GGCGGTACCATGAAACAATTTGGAGAAG
GCGCTCGAGCTAAGTTTGTACATTTTGCC
miR-206-mimics
UGGAAUGUAAGGAAGUGUGUGG
ACACACUUCCUUACAUUCCAUU
miR-NC
UUCUCCGAACGUGUCACGUTT
ACGUGACACGUUCGGAGAATT
2.4 qRT-PCR detection
The collected samples (serum, cells) were extracted using Trizol reagent and RNA integrity was detected. cDNA was obtained by reverse transcription of total RNA (Bio-Rad, USA) using iScript™ Reverse Transcription Supermix kit, TransScript Green miRNA Two-Step qRT-PCR SuperMix, and amplified using TB Green™ Fast qPCR Mix (Takara, Japan) and ABI 7500 PCR (ABI, USA). Finally, the qPCR results were calculated by 2−ΔΔCt. U6 was applied as the internal control of miR, GAPDH for mRNA and LncRNA. The primer sequences were listed in Table 3.
Gene
Upstream primer (5′–3′)
Downstream primer (5′–3′)
MALAT1
AGCCCAAATCTCAAGCGGTGC
TGCATCGAGGTGAGGGGTGA
miR-206
GGCGGTGGAATGTAAGGAAG
GGCTGTCGTGGACTGCG
NLRP3
CACCTGTTGTGCAATCTGAAG
GCAAGATCCTGACAACATGC
Caspase-1
TGGAAGGTAGGCAAGACT
ATAGTGGGCATCTGGGTC
IL-1β
GAAGCTCACCTTTTGACAGTG
TGCATGCTCTCGCTAGGACAG
GSDMD
CCGGAACTCGCTATCCCTGT
ATACCACCCCTTCCCAGGTG
U6
CTCGCTTCGGCAGCACA
AACGCTTCACGAATTTGCGT
GAPDH
AACGGATTTGGTCGTATTGG
TTGATTTTGGAGGGATCTCG
2.5 Elisa detection
After transfecting for 48 h, the cells were collected, and the supernatant was taken to detect TNF-α, IL-6 and MCP-1 (Li et al., 2018).
2.6 Western blot test
The transfected cells were obtained and lysed with protein extraction reagent, then the protein was tested using BCA kit, 35 μg protein sample was taken, 10%SDS-PAGE gel electrophoresis was performed, then the protein was moved to PVDF, which was rinsed and sealed with 5% defatted milk at 25 °C for 1 h. Then primary anti-NLRP3, Caspase-1, Pro-Caspase-1, IL-1β, Pro-IL-1β, GSDMD-N, GAPDH (Abcam, USA) were put into the membrane and incubated. The membrane was cultured at 4 °C for 1 h to counter HRP(1:5000) coupled secondary antibodies. The band was observed using enhanced chemiluminescent reagent (Thermo Fisher Science, Inc.).
2.7 Double fluorescein report
Complementary DNA fragments including MALAT1-WT or MALAT1-mut fragments were treated with the downstream of the luciferase gene in psi-CHECK2. The miR-206-mimicsMALAT1-WT or MALAT1-mut reporter vector was co-transfected with the reagent (Invitogen, USA). Forty-eight hours after transfection, fireflies and renin luciferase activities were tested.
2.8 RNA immunoprecipitation (RIP)
RIP was applied to explore the role of MALAT1 and miR-206 on latent binding protein Ago2 using EZMagna RIP kit. HK-2 was lysed and cultivated with protein A magnetic beads, which were conjugated at 4℃. After 6 h, the beads were rinsed, and placed with 0.1% SDS/0.5 mg/ml protease K at 55℃ for 30 min to wipe off proteins. At last, the immunoprecipitated RNA was analyzed using qRT-PCR to prove the exist of MALAT1 and miR-206.
2.9 Statistical analysis
GraphPad 7 was applied to visualize the required figures and for analysis. The measurement data was represented as Meas ± SD. Independent sample t test was applied for inter-group comparison, one-way ANOVA for multi-group comparison, which was represented as F. LSD-t test was applied for post-event pairwise comparison, repeated measurement ANOVA for multi-time analysis, Bonferroni for back testing, and Pearson test for the correlation of genes. When P < 0.05, the difference was statistically evident.
3 Results
3.1 MALAT1 expression elevated in HG-intervened HK-2 cells and DN patients
qRT-PCR detection revealed that MALAT1 in DN patients was evidently higher than that of healthy individuals (Fig. 1A), and we further detected HK-2 cultured with different levels of HG, and found that MALAT1 in HK-2 increased continuously in a concentration-dependent manner (Fig. 1B).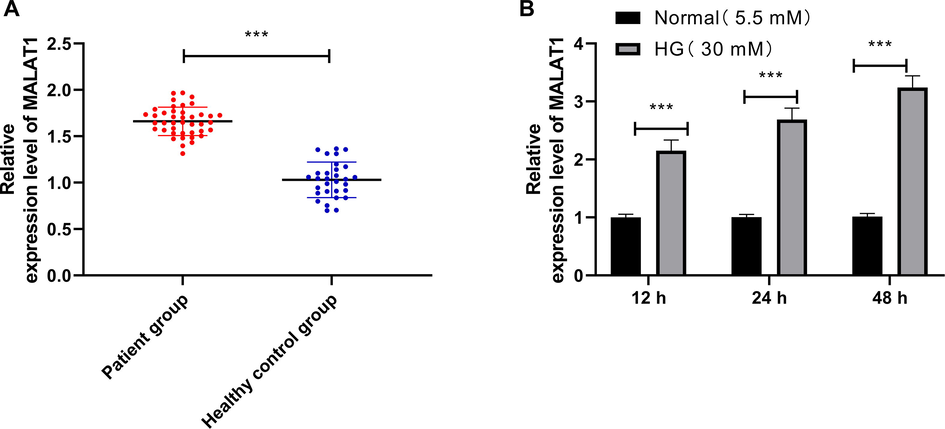
MALAT1 in HG-induced HK-2 cells and DN patients.
4 Knocking down MALAT1 hindered HG-intervened inflammatory factors in HK-2
We constructed si-MALAT1 sequence to observe MALAT1 on HG-intervened HK-2. According to the silencing role of si-MALAT1, we selected si-MALAT1#2 with significant effect for transfection (Fig. 2A, B). Further observation of the concentration of inflammatory factors revealed that TNF - α, IL-6 and MCP-1 in HK-2 intervened by HG increased evidently, but TNF-α, IL-6 and MCP-1 in HK-2 intervened by HG were evidently inhibited after transfection of si-MALAT1#2 (Fig. 2C-E). This suggested that inhibition of MALAT1 could reduce the inflammatory reaction in HK-2.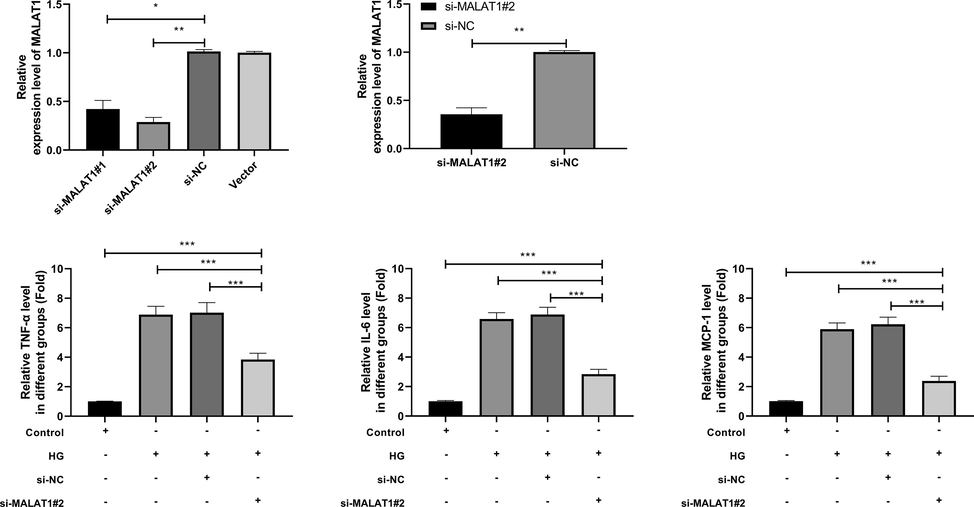
Effect of MALAT1 knock-down on HG-induced inflammatory response of HK-2 cells.
4.1 Knocking down MALAT1 inhibited HG-induced pyroptosis of HK-2 cells
Further experiments were carried out to determine MALAT1 on HG-intervened HK-2 pyroptosis. We tested cell pyroptosis protein-related proteins and found that the relative expression levels of NLRP3, caspase1, IL-1β and GSDMD-N protein and mRNA elevated evidently (Fig. 3A, B). However, we also found that those were declined by transfection of si-MALAT1#2, suggesting that MALAT1 could improve HG-intervened pyroptosis of HK-2.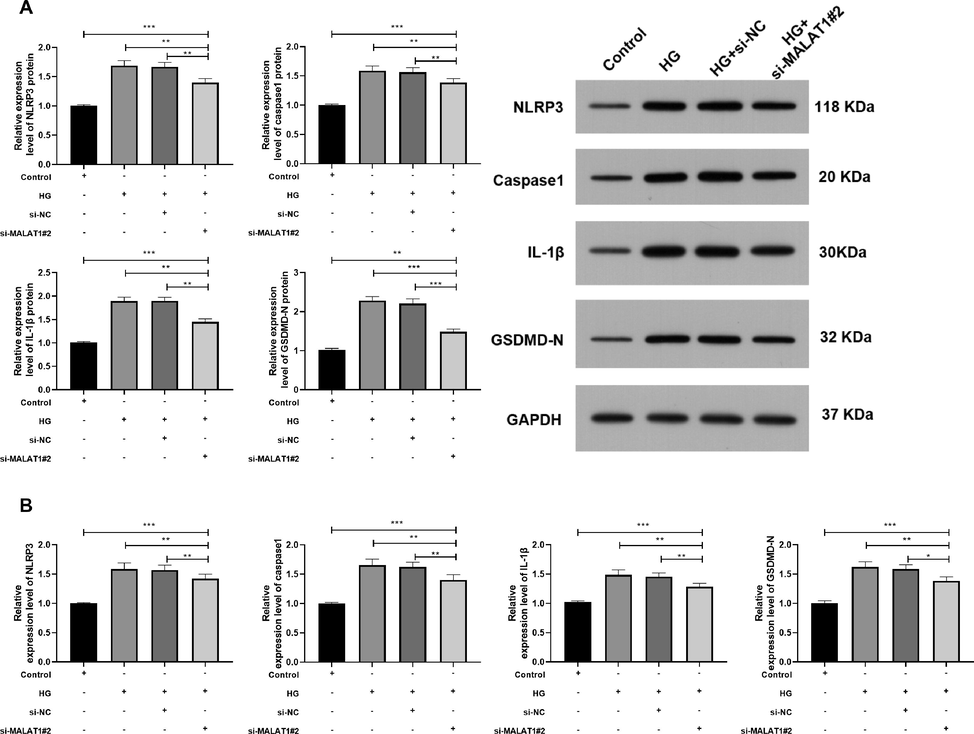
Role of MALAT1 knock-down on HG-induced pyroptosis of HK-2 cells.
4.2 MALAT1 targeted miR-206
ceRNA is currently an important pathway for LcnRNA to participate in body functions. We made common prediction through StarBase, LncBase, miRcode and miRDB to explore the MALAT1 downstream targeted miR, and finally found 5 common miRs, miR-216b-5p, miR-1297, miR-206, miR-613 and miR-338-3p (Fig. 4A). Subsequently, we detected the difference of miR in HG-intervened HK-2 and found that the difference of miR-206 was the most significant (Fig. 4B). Fig. 4c indicates the specific binding site of MALAT1 and miR-206. miR-206 in serum of DN patients showed a downward trend (Fig. 4D), and miR-206 and MALAT1 in serum of DN patients showed a negative correlation (Fig. 4E), suggesting that MALAT1 might target miR-206. We further performed RIP and double luciferase reports to verify this. RIP results showed that both MALAT1 and miR-206 could be precipitated by Ago2 antibody (Fig. 4F), and double luciferase report showed that miR-206-mimics could hinder MALAT1-WT fluorescence activity (Fig. 4G). These suggested that MALAT1 can target miR-206.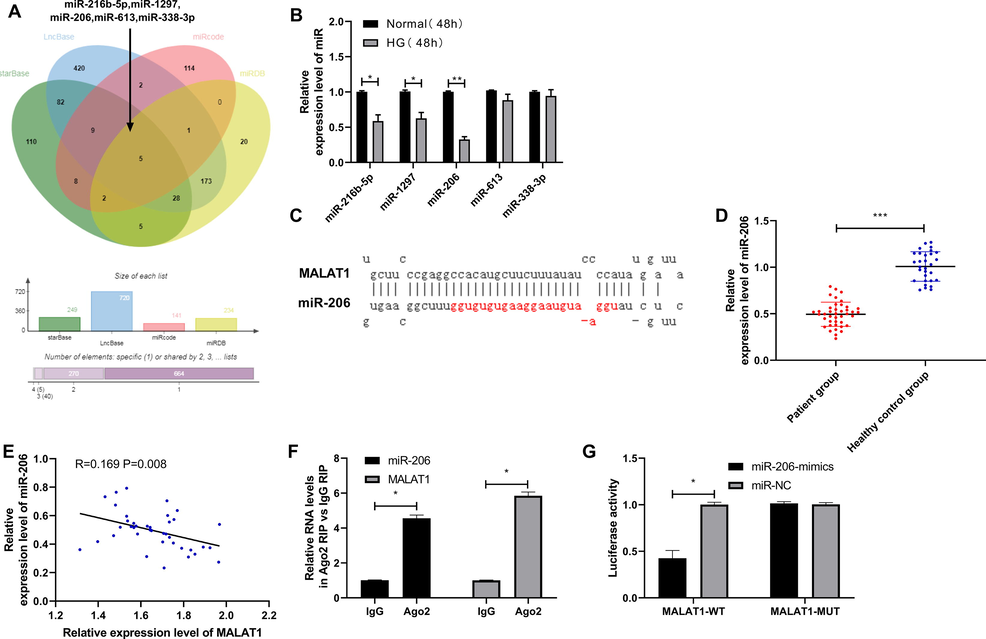
Targeted binding of MALAT1 and miR-206.
4.3 Elevation of miR-206 could hinder the occurrence of inflammatory response to cells after over-expression of MALAT1 and improve cell apoptosis
At the end of the study, we first tested the efficiency after transfection (Fig. 5A), and then constructed co-transfected cells to determine that MALAT1 could regulate miR-206 to affect HG-intervened inflammatory response and pyroptosis caused by HK-2 cells. After that, we found through Elisa that TNF-α, IL-6 and MCP-1 were decreased after miR-206-mimics transfection (Fig. 5B), while qRT-PCR and WB experiments found that NLRP3, caspase1-1, IL-1β and GSDMD-N protein and mRNA were evidently inhibited after miR-206-mimics transfection (Fig. 5B, C). However, the above results were reversed after co-transfection of pcDNA3.1-MALAT1 and miR-206-mimics, and there was no difference compared with miR-NC + pcDNA3.1, which suggested that MALAT1 could participate in pyroptosis of RTECs by regulating miR-206.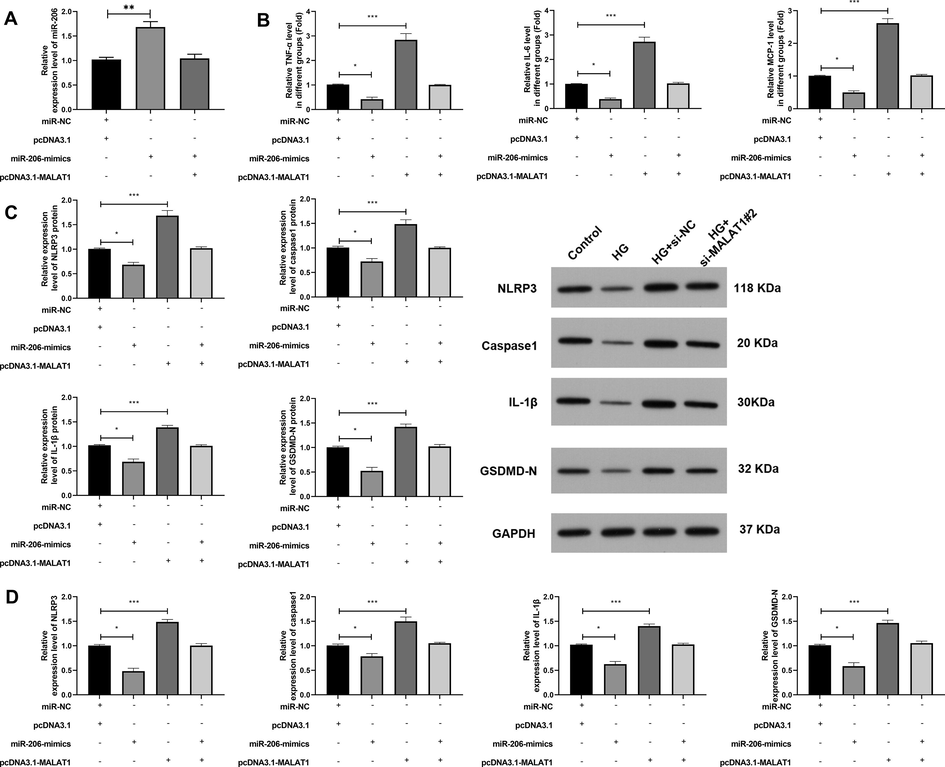
MALAT1 participated in pyroptosis of renal tubular epithelial cells through miR-206.
5 Discussion
DN patients are often accompanied by epithelial dysfunction, inflammation, and hypertrophy. However, the mechanism of DN is still unclear and there is no effective treatment method, so it’s particularly important to explore new therapeutic targets for DN (Zhang et al., 2018; Wang et al., 2019). LncRNA has played an important regulatory role in many diseases in recent years. Especially in metabolic diseases, Liu et al. (Liu et al., 2019) have found that LncRNA MALAT1/ microRNA-382-3p/Resistin axis can effectively reduce insulin resistance in type 2 diabetes (Li et al., 2018). MALAT1 is located on chromosome 11q13.1 as one of the members of LncRNA. Previous studies have found that (Liu et al., 2019; Zhao et al., 2020; Zhang et al., 2020). MALAT1 has increased expression in various tumors. And the study found that MALAT1 expression increased in diabetic patients. However, the relevant mechanism of MALAT1 in DN patients is still unclear. We detected the serum in DN patients and HG-intervened HK-2 and found that MALAT1 was higher than that of the CG, and the inflammation and pyroptosis were evidently reduced by knocking down MALAT1. It was suggested that MALAT1 might be a latent target to treat DN.
ceRNA is currently one of the important ways for LncRNA to participate in the life process (Song et al., 2020). Studies have found that LncRNA can participate in the progress of diabetes by regulating downstream binding miR. For example, LncRNA TUG1 can improve diabetic nephropathy by hindering miR-21 to elevate TIMP3 (Salmena et al., 2011). The above studies have confirmed that MALAT1 can improve HG-induced HK-2 cell inflammation and pyroptosis, but the mechanism is still unclear. For this reason, we predicted that miR can be combined with the downstream of MALAT1, and common prediction of 5 potential miRs were found through online websites of StarBase, LncBase, miRcode and miRDB. Then we detected HG-intervened HK-2 and found that miR-206 had the most significant difference among these miRs (Wang et al., 2019; Li et al., 2014; Paraskevopoulou et al., 2016; Jeggari et al., 2012). miR-206 is located on human 6p12.2 chromosome. miR-206 was found to be declined in lung cancer, liver cancer and breast cancer. In addition, miR-206 was found in islets and regulated glucokinase activity (Chen and Wang, 2020; Wu et al., 2019; Yunqiao et al., 2014). miR-206 in serum of DN patients was decreased, and correlation analysis found revealed that miR-206 and MALAT1 had a negative correlation, which suggested that there was a regulatory relationship between them. Subsequently, we verified the targeting relationship between MALAT1 and miR-206 through RIP and bifluorescin reports respectively. A newly discovered apoptosis pattern of focal cell death is usually characterized by the release of many inflammatory factors after cell rupture, which makes organs subject to inflammatory storms, thus causing disease aggravation (Samaeekia et al., 2017). Previous studies have shown that LncRNA UCA1 inhibits HK-2 cell apoptosis in DN by targeting miR-206 (Wu et al., 2018). However, there is no relevant research to prove the relationship between miR-206 and pyroptosis. In order to study the anti-pyroptosis role of miR-206 on HK-2 cells in DN, we detected NLRP3, caspase1, IL-1β and GSDMD-N in cells respectively, which are markers of pyroptosis and are important indicators for observing pyroptosis. Previous studies have shown that NLRP3 can activate pro-caspase1 to crack it into caspase1, thus activating pro-IL-1β to accelerate the secretion of IL-1β, while a large amount of secreted IL-1β induces other pro-inflammatory factors, causing organs to produce inflammatory cascade reactions and destroy tissues(Yan et al., 2018; Hughes and O'Neill, 2018). Experiments also revealed that up-regulation of miR-206 effectively hindered pyroptosis and reduce the occurrence of inflammatory reactions. However, we also found that over-expressed MALAT1 and elevated miR-206 co-transfected cells reversed the pyroptosis, which suggested that MALAT1 can mediate miR-206 to regulate HG-induced pyroptosis of HK-2 cells and may be applied as a latent therapeutic target in clinic.
Through the above research, we have confirmed that MALAT1 can mediate miR-206 to regulate HG-induced pyroptosis of HK-2 cells, but we still have certain limitations in this study. Firstly, this study has not conducted further research on the target genes downstream of miR-206. Secondly, the samples in our research are few and the role of MALAT1 in DN has not been analyzed. We will perform further tests to improve our conjectures. To sum up, MALAT1 can improve HG-induced HK-2 pyroptosis by targeting miR-206 and might be a latent target for DN.
Acknowledgements
We would like to thank the National Natural Science Foundation of China for providing financial assistance for our project “Study on the Role of LncRNA UCA1 inhibition pyrocytosis in Diabetic Nephropathy regulated by Yiqi Yangyin Tongluo Decoction” (project Approval Number: 81904155).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Diabetic nephropathy: is it always there? Assumptions, weaknesses and pitfalls in the diagnosis. Hormones (Athens). 2017;16:351-361.
- [Google Scholar]
- Kitada, M. and Koya, D. (2017) Diabetic nephropathy. Nihon Jinzo Gakkai Shi. 59, 38-42.
- Dysregulation of lncRNAs GM5524 and GM15645 involved in highglucoseinduced podocyte apoptosis and autophagy in diabetic nephropathy. Mol. Med. Rep.. 2018;18:3657-3664.
- [Google Scholar]
- A glimpse of various pathogenetic mechanisms of diabetic nephropathy. Annu. Rev. Pathol.. 2011;6:395-423.
- [Google Scholar]
- Diabetic Nephropathy: a Tangled Web to Unweave. Cardiovasc. Drugs Ther.. 2017;31:579-592.
- [Google Scholar]
- Ruboxistaurin attenuates diabetic nephropathy via modulation of TGF-beta1/Smad and GRAP pathways. J. Pharm. Pharmacol.. 2016;68:219-232.
- [Google Scholar]
- Curcumin attenuates diabetic nephropathy by inhibiting PKC-alpha and PKC-beta1 activity in streptozotocin-induced type I diabetic rats. Mol. Nutr. Food Res.. 2011;55:1655-1665.
- [Google Scholar]
- The Landscape of long noncoding RNA classification. Trends Genet.. 2015;31:239-251.
- [Google Scholar]
- LncRNA TNRC6C-AS1 regulates UNC5B in thyroid cancer to influence cell proliferation, migration, and invasion as a competing endogenous RNA of miR-129-5p. J. Cell. Biochem.. 2018;119:8304-8316.
- [Google Scholar]
- LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett.. 2016;371:99-106.
- [Google Scholar]
- LncRNA GAS5 exacerbates renal tubular epithelial fibrosis by acting as a competing endogenous RNA of miR-96-5p. Biomed. Pharmacother.. 2020;121:109411
- [Google Scholar]
- lncRNA MALAT1 mediated high glucose-induced HK-2 cell epithelial-to-mesenchymal transition and injury. J. Physiol. Biochem.. 2019;75:443-452.
- [Google Scholar]
- Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem. Sci.. 2017;42:245-254.
- [Google Scholar]
- Sirtuin-1 ameliorates cadmium-induced endoplasmic reticulum stress and pyroptosis through XBP-1s deacetylation in human renal tubular epithelial cells. Arch. Toxicol.. 2019;93:965-986.
- [Google Scholar]
- Silencing of KCNQ1OT1 decreases oxidative stress and pyroptosis of renal tubular epithelial cells. Diabetes Metab Syndr Obes.. 2020;13:365-375.
- [Google Scholar]
- Melatonin prevents endothelial cell pyroptosis via regulation of long noncoding RNA MEG3/miR-223/NLRP3 axis. J. Pineal Res.. 2018;64
- [Google Scholar]
- suPAR as a marker of diabetic nephropathy in patients with type 2 diabetes. Int. J. Clin. Exp. Med.. 2019;12:4218-4225.
- [Google Scholar]
- Clinical effect of mineralocorticoid receptor antagonist combined with angiotensin receptor blocker or angiotensin converting enzyme inhibitor in treating diabetic nephropathy: a systematic review and meta-analysis. Int. J. Clin. Exp. Med.. 2018;11:5395-5408.
- [Google Scholar]
- Exercise reduces insulin resistance in Type 2 diabetes mellitus via mediating the lncRNA MALAT1/MicroRNA-382-3p/resistin axis. Mol. Ther. Nucleic Acids. 2019;18:34-44.
- [Google Scholar]
- Long non-coding RNA MALAT1 promotes cell proliferation, migration and invasion in cervical cancer by targeting miR-625-5p and AKT2. Panminerva Med. 2020
- [Google Scholar]
- lncRNA MALAT1 modulates oxaliplatin resistance of gastric cancer via sponging miR-22-3p. Onco Targets Ther.. 2020;13:1343-1354.
- [Google Scholar]
- Long non-coding RNA MALAT1 regulates proliferation, apoptosis, migration and invasion via miR-374b-5p/SRSF7 axis in non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci.. 2020;24:1853-1862.
- [Google Scholar]
- A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-358.
- [Google Scholar]
- LncRNA TUG1 ameliorates diabetic nephropathy by inhibiting miR-21 to promote TIMP3-expression. Int. J. Clin. Exp. Pathol.. 2019;12:717-729.
- [Google Scholar]
- starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res.. 2014;42:D92-97.
- [Google Scholar]
- DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res.. 2016;44:D231-238.
- [Google Scholar]
- miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28:2062-2063.
- [Google Scholar]
- miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res.. 2020;48:D127-D131.
- [Google Scholar]
- SNHG14 confers gefitinib resistance in non-small cell lung cancer by up-regulating ABCB1 via sponging miR-206-3p. Biomed. Pharmacother.. 2019;116:108995
- [Google Scholar]
- MicroRNA-206, down-regulated in hepatocellular carcinoma, suppresses cell proliferation and promotes apoptosis. Hepatogastroenterology. 2014;61:1302-1307.
- [Google Scholar]
- miR-206 inhibits stemness and metastasis of breast cancer by targeting MKL1/IL11 pathway. Clin. Cancer Res.. 2017;23:1091-1103.
- [Google Scholar]
- Nicotine promotes atherosclerosis via ROS-NLRP3-mediated endothelial cell pyroptosis. Cell Death Dis.. 2018;9:171.
- [Google Scholar]
- Downregulation of lncRNA UCA1 inhibits proliferation and invasion of cervical cancer cells through miR-206 expression. Oncol. Res.. 2018;9
- [Google Scholar]







