Translate this page into:
Genome wide analysis of ATP-binding Cassette (ABC) transporter in the eastern honey bee (Apis cerana Fabricius, 1793)
⁎Corresponding author at: Unit of Bee Research and Honey Production, Biology Department, Faculty of Science, King Khalid University, P.O. Box 9004, Abha 61413, Saudi Arabia. khalidtalpur@hotmail.com (Khalid Ali Khan)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
The ATP-binding Cassette (ABC) transporter family genes play a significant role in transporting substances such as heavy metals, phytohormones, and secondary metabolites across the structural membrane. However, it plays a vital function in the eastern honey bee (Apis cerana), still uncovered. The present study discovered a total of 28 ABC transporters genes in A. cerana genome. The ABC transporters genes phylogenetic tree gradually divide into eight groups, named from ABCA to ABCH. From these groups, ABCG belongs to the largest family containing maximum ABC transporters genes. The domain architecture and number of exon-intron differed from one gene to another gene. The exons ranged from 4 to 30. The sequence motif and alignment analysis established similar structural, functional sites in all A. cerana ABC transporter. In the present study results, 1st motif was observed in all genes except XM_028666551.1. It was observed that the A. cerana genes sequence illustrated synteny with A. dorsata. Furthermore, the biochemical and physical properties, conserved motifs, 3D (three-dimensional) structure prediction, and molecular docking with ATP molecules were also studied. Consequently, the results provided applicable information for a more functional analysis of ABC transporter genes in the honey bee and a reference study for other insects.
Keywords
Docking
Exons
Gene
Honey bee
Phylogeny
Protein interaction
1 Introduction
The highly populated environment is the habitat of honey bees, where they have a strong association and food sharing with their nest-mates (DeGrandi-Hoffman and Chen, 2015). This close association causes widespread pathogens inside the bee colony. Hence, honey bees are at more risk of pathogens. This led researchers to focus on how bees maintain their health and resist the disease. Therefore, the honey bees have followed two strategies for protection against pathogens, such as innate immunity (Danihlík et al., 2015) and social immunity (Goode et al., 2006). In the first strategy, honey bees protect themselves and their colony through innate immunity; for example, the worker bees are the main pollinators, forage on crops and wildflowers. Despite this, foraging caused large exposure of honey bees with harmful pathogens (bacteria /viruses) infection and agricultural pesticides, transmitted from workers to colony bees (Raymann and Moran, 2018). In addition, the honey bees’ innate immunity studies have revealed the younger forager bees have a strong innate immune system than the older foragers (Bull et al., 2012). In the second strategy, the bees’ social immunity studies have revealed the health maintenance function. Worker bees smell the infected and diseased adults and broods, dumped their hive from foreign invaders/dead bodies and clear their bodies (Goode et al., 2006). Six morphologically different A. cerana subspecies are endemic in Asia spread throughout the Asian climatic zone series, and further used for commercial beekeeping and pollination for thousands of years. Due to morphological and behavioral similarities, A. mellifera and A. cerana are close relatives. Despite this, A. cerana contained various distinct features in comparison with A. mellifera. Such as A. cerana workers fanned their hives with pointing heads outward, as opposed to A. mellifera workers bees which ventilated with pointing heads inward; A. cerana foragers are good to accumulate nectars from dispersed flower resources, while in A. mellifera foragers, this character is neglected; A. mellifera used propolis (resinous material) to seal hive apertures and protect against pathogens. Furthermore, A. cerana is an indigenous Asian species that evolved serial striking biological features to conflict with the unfavorable conditions in their habitats. A. cerana foragers under cloudy situation at 7 °C temperature visit many floral species, while A. mellifera goes into a stagnant state at this temperature. In addition, A. cerana also formed a special defense mechanism set to control pathogens, parasites, and predators (Evans and Spivak, 2010).
On comparison further revealed that A. cerana has a hasty, unpredictable, and rapid zigzagging flight that helps to escape from bee-eating and honey birds, while A. mellifera has clumsy and steady flight. A. cerana species is an indigenous host to ectoparasitic mite (Varroa destructor) (Anderson and Trueman, 2000), the most injurious A. mellifera pest, and evolved resistance against pest across a long duration of mutual association significantly. Their worker bees can effectively eradicate mite brood and adults through successive cleaning behavior (Guan_Huang, 2005).
The ABC (ATP-binding cassette) transporters comprise the most significant superfamily protein, present in all organisms (Dean et al., 2001). These transporter proteins share extremely conserved nucleotide-binding domains (NBDs), which consist of signature motifs (the C loop), connecting two walker boxes, Walker A and Walker B (Walker et al., 1982). Additionally, to nucleotide-binding domains (NBDs), the eukaryotic ATP-binding cassette transporter usually contains one/two TMDs (transmembrane domains), comprising six to 11 membrane-spanning alpha-helices and prescribe the substrate specificity. A classic architecture domain of the whole transporter has represented the TMD-NBD-TMD-NBD sequence from N to C terminus; while half transporter contains just one NBD-TMD set. Based on NBDs homology, human transporter proteins ABC have been grouped into seven different sub-families, ABCA-ABCG. The ABCH transporter was firstly identified in Drosophila melanogaster, also present in Zebrafish and arthropods but absent in fungi, plants, and mammals (Tian et al., 2017).
Mainly, the transporter ABC translates membrane-bound proteins that hold many molecules, including sugar, lipids, amino acids, vitamins, peptides, sterols, endogenous metabolites, xenobiotics, hormones, and inorganics across the membrane (Dean et al., 2001). Further, these used energies are released with ATP hydrolysis during NBDs molecular transportation across the plasma membrane. It also performed a function in translation, ribosome assembly, and cell signaling (Sturm et al., 2009). Such as ABCE – ABCF transporter proteins do not work as a transporter but are involved in ribosome assembly, transcription, and translation (Tyzack et al., 2000). Moreover, the ABC transporters in plants are also involved in detoxification, phytoalexin function, and osmolality (Verrier et al., 2008).
Till now, very little is known about the ABC transporter in the A. cerana genome. So, the current study involved a genome-wide screening of ABC transporter in A. cerana genome. In addition, several bioinformatics analyses were also carried out to explore the basic and advanced features of ABC transporter; including, gene structure, phylogenetic relations, and conserved protein domains. The outcomes of this study will provide the base for further functional analysis of the ABC transporter in A. cerana and can also contribute to a better understanding of their molecular mechanisms.
2 Material and methods
2.1 Identification of the ABC transporters
The A. cerana genome was downloaded from the Honey bee genome database (https://hymenoptera.elsiklab.missouri.edu/). Drosophila melanogaster ABC transporter proteins were used as the query sequence. A local BLASTp similarity search was carried out to find the ABC transporters in the A. cerana genome. Most closely related A. cerana sequences were retrieved and subjected to PfamScan and Batch CDD-NCBI search to validate the presence of ABC transporters conserved domain. The data redundancy was removed, and the identified proteins sequences were used for further downstream analysis, including molecular weight (MW) and isoelectric point (IP), using ProtParam online tool, available at the ExPASy web server.
2.2 Comparative synteny analysis
To study the visualization of the genome’s conservation, a synteny relative analysis was performed by the Circoletto Tool (tools.bat.infspire.org/circoletto/).
2.3 Multiple sequence alignment and phylogenetic analysis
All identified protein sequences were aligned using MUSCLE with 16 iterations. The whole transporter sequence was aligned through ClustalW and extorted to phylogenetic approach analysis with MEGA5 through 1000 replication bootstrap.
2.4 Identification of gene structure and conserved motif analysis
The intron-exon distribution was demonstrated by the gene structure display server. The ABC transporter’s protein conserved motifs were determined through an online MEME server (http://meme-suite.org/) through subsequent parameters, such as no. of motifs: 20 and optimum width range: 6-200. TBtools software (http://github.com/CJ-Chen/TBtools) was utilized to build the distribution of the motifs.
2.5 Protein-protein interaction analysis
To construct the network of protein-protein interaction, the STRING 11.0 (https://string-db.org/) server was used with A. mellifera as the reference genome. The display parameters were set as confidence; 0.9, network; edges evidence, and maximum interaction; 10 interactions. The clustering was carried by MCL cluster with 10 inflations.
2.6 Molecular docking analysis
To find active sites, docking pockets, and comparing different ABC-transporter proteins with ATP molecules, we selected one protein from each group like ABCC, ABCG, and ABCB. Furthermore, the selected genes were ABCC5 (XM_017055838.2), ABCG4 (XM_017063007.2) and ABCB7 (XM_028668368.1). Molecular docking was done using AutoDock Vina and its associated tools. The structure of protein PDB was constructed by eliminating water molecules and heteroatoms while optimizing the interaction inserted polar hydrogen. The three-dimensional grid of size 66 × 56 × 54 Å is used with 3D coordinates (4.402, −8.060, and 8.874) to define the region of interactions. The final prepared models and ligands were docked with AutoDock vina. The binding affinity and RMSD values were also calculated. A binding affinity of −7 kcal/mol was set as the threshold for screening the best-docked models. Models with binding affinity <−7 kcal/mol and 0.00 RMSD value were selected for further detailed study. The selected docked models were visualized in the discovery studio.
3 Results
3.1 Identification and physiological properties ABC transporter genes
The genome-wide recognition of ABC transporter genes in A. cerana identified 28 a total of ABC transporter genes. Although all proteins contain conserved ABC transporter domains, they showed high diversity in their sequences. The details of all 28 ABC transporter proteins, including molecular weight, protein length, and IP, are mentioned in Table.1. The encoded proteins' length was ranged from 145 to 1749 amino acids; the molecular weight was from 16.043 to 200.874 kDa and the IP from 5.97- 9.5.
Gene Name
Direction
Location
Number of Amino acid
Molecular weight
Theoretical IP
>XM_017061551.2
F
796074..805212
740
81906.92
8.89
XM_017063007.2
R
567283..579217
642
72854.11
8.89
XM_028665683.1
R
55981..80472
725
81281.68
8.9
>XM_028668147.1
F
5955..20683
551
61745.34
8.54
XM_017050518.2
R
47790..81067
1714
192140.26
6.42
>XM_017061132.2
F
508983..526137
838
95774.71
8.73
XM_017064673.2
F
1538530..1594830
1343
147509.33
7.55
XM_028665983.1
R
1226532..1261847
805
90082.76
7.8
XM_028668645.1
R
217429..225638
819
91363.57
8.73
XM_017061950.2
R
508195..514207
1353
154313.22
7.15
XM_017049887.2
F
1072516..1080221
563
62026.41
8.64
XM_017055525.2
F
460683..463560
632
71734.17
6.45
XM_028669553.1
R
522377..533959
1529
173178.59
8.72
XM_028664559.1
R
161942..171891
1355
154074.8
9.02
XM_028669661.1
R
302610..345481
692
77749.69
5.97
XM_017055838.2
R
157528..169083
1749
200874.45
8.87
XM_017051631.1
R
554..3234
537
60497.85
6.55
XM_017066491.2
R
290738..294566
660
74101.02
7.95
XM_017063262.2
F
670930..675028
608
68433.37
8.2
XM_028665690.1
R
2652612..2671553
1291
147672.18
8.62
XM_017065726.2
F
281223..284775
632
71779.29
6.86
XM_028668368.1
R
357224..358882
145
16043.47
7.75
XM_017053731.2
R
694329..704860
1392
156778.59
8.73
XM_017048648.2
F
313141..317381
664
76784.13
9.5
XM_017066360.2
R
470837..474449
758
84479.03
9.22
XM_028666551.1
F
70816..84595
514
59832.66
8.98
XM_017054061.2
R
1242287..1248580
1180
135486.49
6.24
XM_017059994.2
F
72744..77785
1296
150493.92
6.76
3.2 Multiple sequence alignment and phylogenetic analysis of ABC transporter genes
The multiple sequence alignment demonstrated that many important residues and structural motifs, e.g., L4XXG8, GXXGXGK, LSGG, R45, A49, DEPTXXVD, were conserved throughout the ABC transporters (Fig. 1). All ABC transporters' combined unrooted phylogenetic tree was categorized into eight major groups (ABCA, ABCB, ABCC, ABCD, ABCE, ABCF, ABCG, and ABCH) with one MRP (Fig. 2). The ABCA group consists of only one gene, i.e., XM_017050518.2. Similarly, ABCC has five genes including XM_017055838.2, XM_017061132.2, XM_028668368.1, XM_017049887.2, and XM_017064673.2 and the ABCD has two genes (XM_017048648.2 and XM_017066360.2). The ABCE has three, and ABCF has six ABC transporters.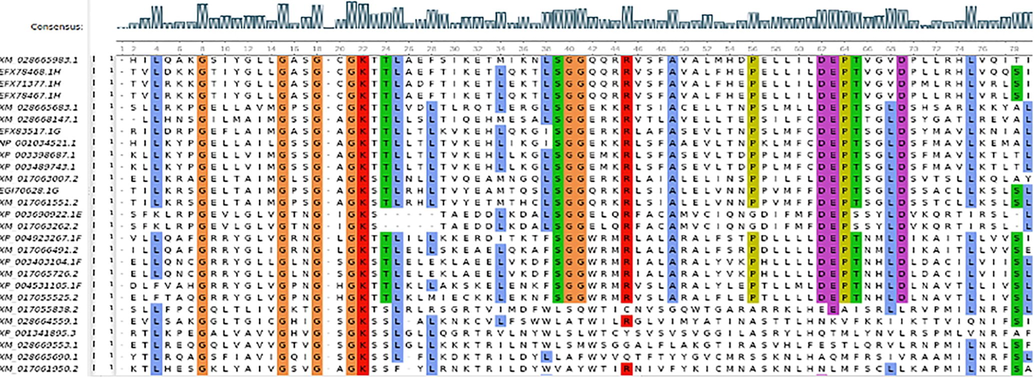
The multiple sequence alignment.
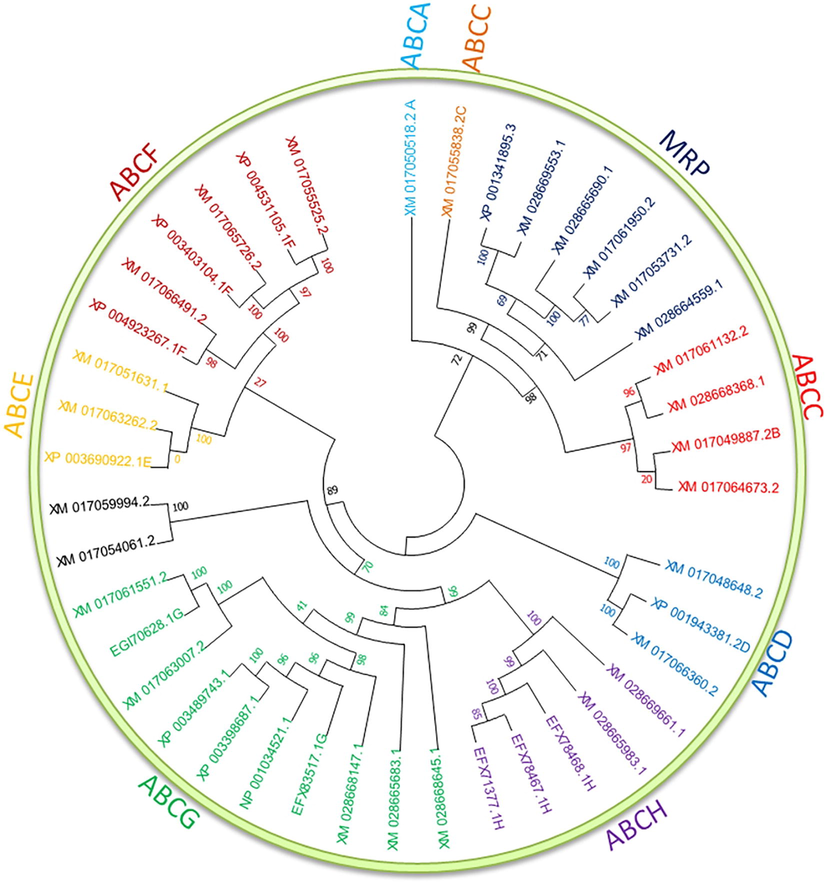
A phylogenetic tree categorized into eight major groups.
3.3 Gene structure organization
As presented in Fig. 2, the number, distribution, and length of introns and exons were not immensely different among the entire gene family. The exons ranged from 4 (XM_028668368.1) to 30 (XM_017055838.2). However, some genes such as XM_017061551.2 and XM_028668147.1 have a similar number of exons (13 exons). In addition, XM_028665983.1 and XM_028669661.1 genes have 14 exons but differ in sequence length (Fig. 3).
Distribution of exons, introns, and UTR (untranslated regions) in Apis cerana ABC transporters gene sequences.
3.4 Conserved motif analysis
The ABC transporter proteins structure was also investigated amino acid sequences. The MEME motif analysis identified several common and unique motifs in A. cerana ABC transporters. Commonly shared motifs usually tended in the same groups indicating similar functions. In the present study results 1st motif was observed in all genes except XM_028666551.1. motif_2nd was present in 26 genes out of 28 genes (excluding XM_028668645.1. and M_028668368.1), motif_3rd was observed in XM_017061132.2, XM_017064673.2, XM_017061950.2, XM_028664559.1, XM_017055838.2, XM_028665690.1, XM_028668368.1 and XM_017053731.2 while motife_4th was observed in XM_017061132.2, XM_017064673.2, XM_017061950.2, XM_017049887.2, XM_028669553.1, XM_028664559.1, XM_017055838.2, XM_028665690.1 and XM_017053731.2. Motifs_5th, was observed in seven genes (XM_017061950.2, XM_028669553.1, XM_028664559.1, XM_017055838.2, XM_028665690.1, XM_017053731.2 and XM_017048648.2), motif_6th was observed in eight genes. Motif_7 and motif_8th was present in eight genes. In summary, some motifs were family-specific, some group-specific, some clade-specific, and some texa specific. The length of motifs was also varied motif to motif like motif_1st had 32 amino acids (aa), motif_2nd had 20 amino acids. We found the motif_3rd, 5th and 10th motifs had 50 amino acids while motifs 4th and 9th had 41aa.Motif 6th had 47 while 7th and 8th motif had 21 amino acid (Fig. 4).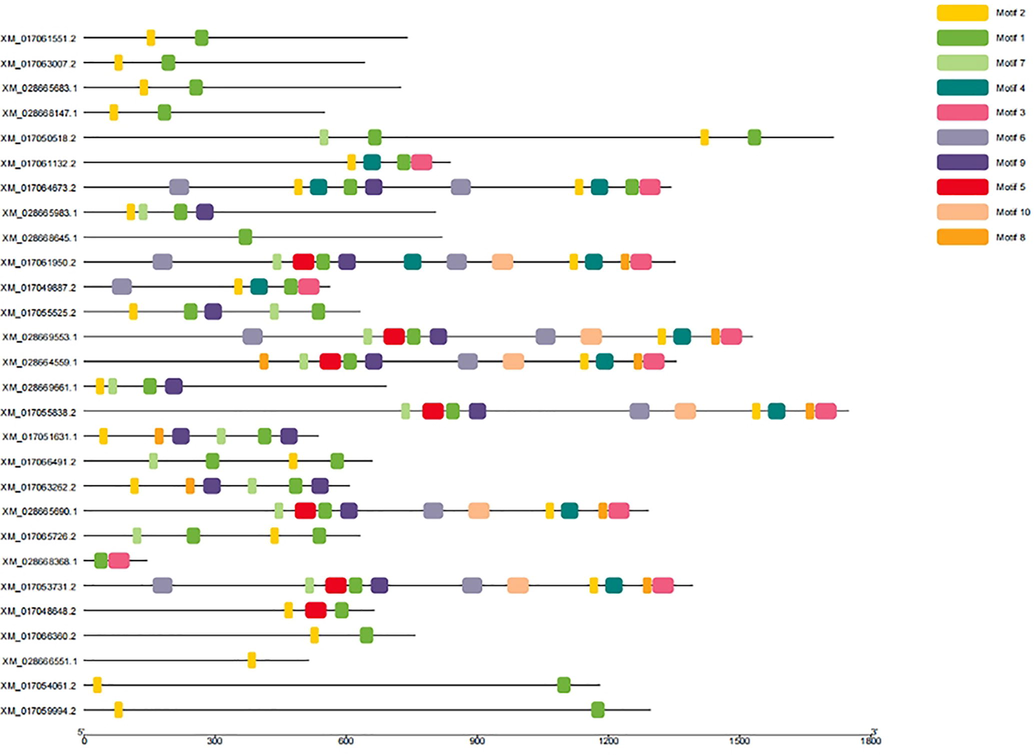
De novo MEME motifs’ distributions in the Apis cerana ABC transporters proteins.
3.5 Comparative synteny analysis identified ABC transporters
The comparative synteny study analysis among A. cerana, A. dorsata, and A. mellifera established a significant relationship in gene evolution, function, triplication, duplication, and expression. It was observed that the A.cerana XM_028668147.1 gene sequence illustrated synteny with the A. dorsata 0298gene sequence. A. cerana gene sequence XM_028668147.1 demonstrated synteny with A. mellifera gnl|Amel_HAv3.10256 (Fig. 5).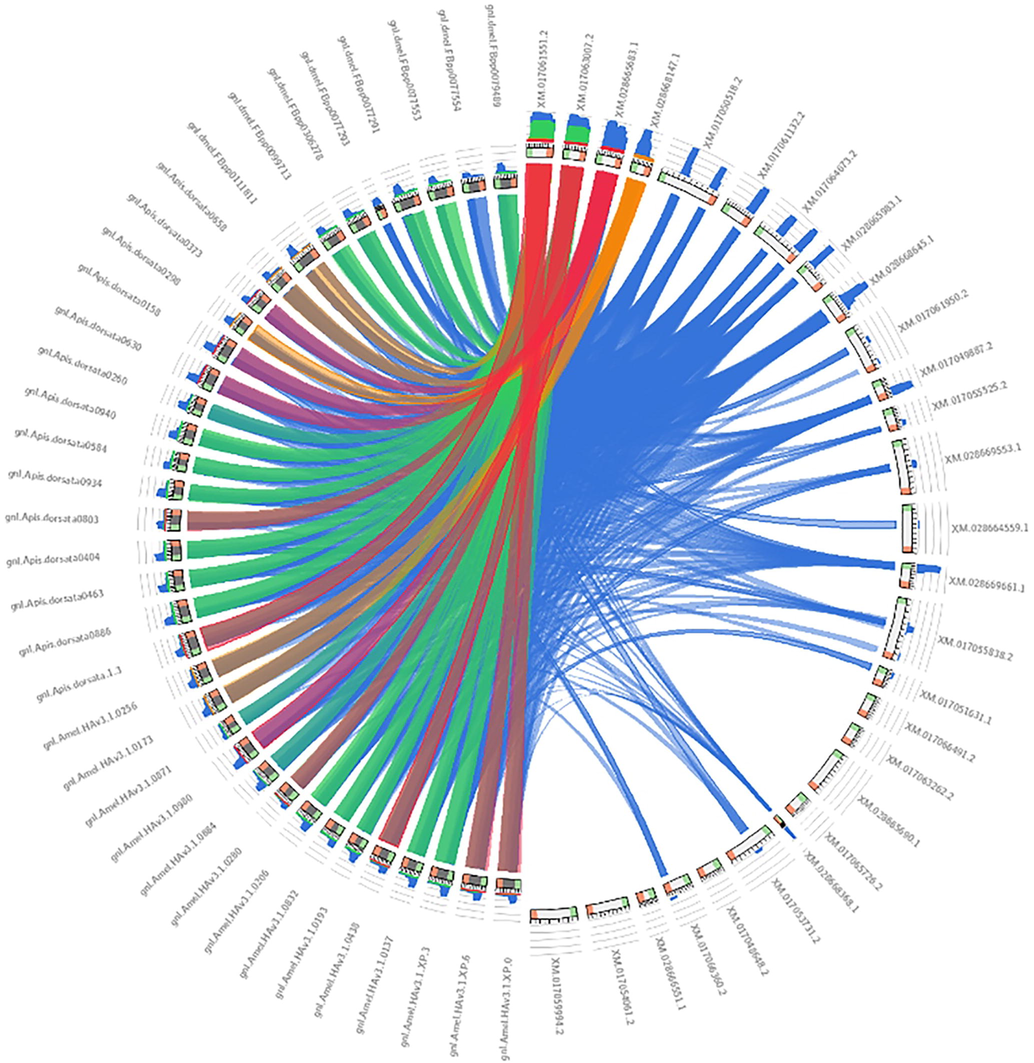
Synteny analysis among Apis cerana, A. dorsata, and A. mellifera.
3.6 Protein-protein interaction network analysis
The PPI network identified the hubs protein of ABC transporters. The ABCC5; XM_017055838.2, ABCG4; XM_017063007.2 and ABCB7; XM_028668368.1 demonstrated strong interaction network among all ABC transporters. In addition XM_017063262.2 had highest number of interaction with other ABC proteins. The XM_017063262.2 interacted with XM_017048648.2, XM_017061132.2, XM_017066491.2, XM_028668368.1, XM_017064673.2, XM_017065726.2 and XM_017055838.2. Similarly ABCC5;XM_017055838.2 showed interaction with ABCB7;XM_028668368.1, ABCG4;XM_017063007.2, XM_017055525.2, and XM_017065726.2 (Fig. 6).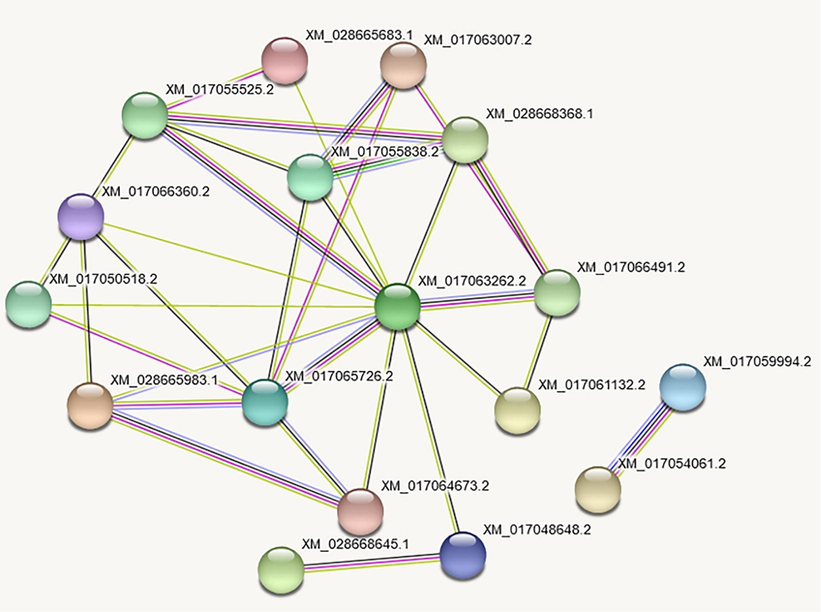
The PPI network identified the hubs protein of ABC transporters.
3.7 Three-dimensional (3D) protein modeling and molecular docking analysis
Based on the PPI results, we selected three ABC transporters from three ABC groups like ABCG4, ABCC5, and ABCB7. The Swiss model predicted the 3D structures demonstrated that ABCG4 (XM_017063007.2) consists of two chains, A and B, while the other two proteins, ABCC5 (XM_017055838.2) and ABCB7 (XM_028668368.1), had only a single chain. The ribbon and surface structure of ABCG4 presented the dimer formation of the two chains making a complex structure for their functions. We can also conclude that the different ABC transporters groups had different 3D structures, which shows their functional diversity (Fig. 7).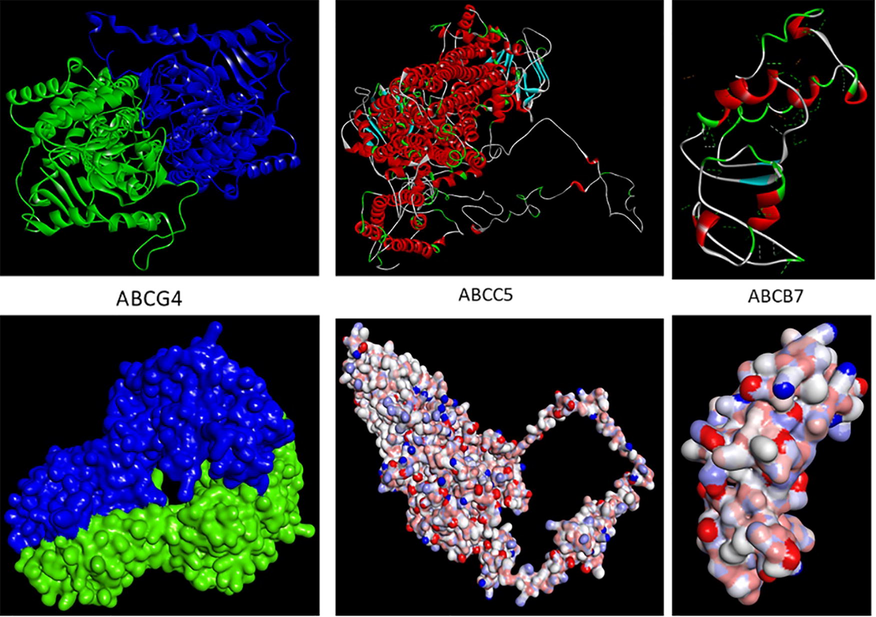
Three dimensional structure of ABC transporters.
Furthermore, these proteins structures were also evaluated for their interaction with ATP molecules. The transmembrane proteins (ABCC5; XM_017055838.2, ABCG4; XM_017063007.2 and ABCB7; XM_028668368.1) docked with the ATP and their binding affinity are given Figure. Of these proteins, the ABCG4 showed the lowest binding affinity with −7.3 kcal/mol and interacted with ATP by residues Asn114, Asn116 Glu117, Phe118, Arg119, Arg356 Gly359, and Arg457. Similarly, ABCC5 had the binding affinity of −7.4 kcal/mol and interacted with ATP by Pro1271, Ser1273, Gln1490, Asp1491, Arg1554, Leu1556, Val1570 residues. The ABCB7 showed the highest binding affinities with −7.5 Kcal/mol and residues Arg69, Thr72, Ser77, Val79, Asp89, Ser90, Asp91, and Glu92. (Figure). The comparison among the three groups of ABC-transporter protein, e.g., ABCB, ABCC, and ABCG, demonstrated different active sites and binding affinity with ATP molecules. IN summary, the ABCB group has a strong interaction with ATP molecules, followed by ABCC and ABCG (Fig. 8)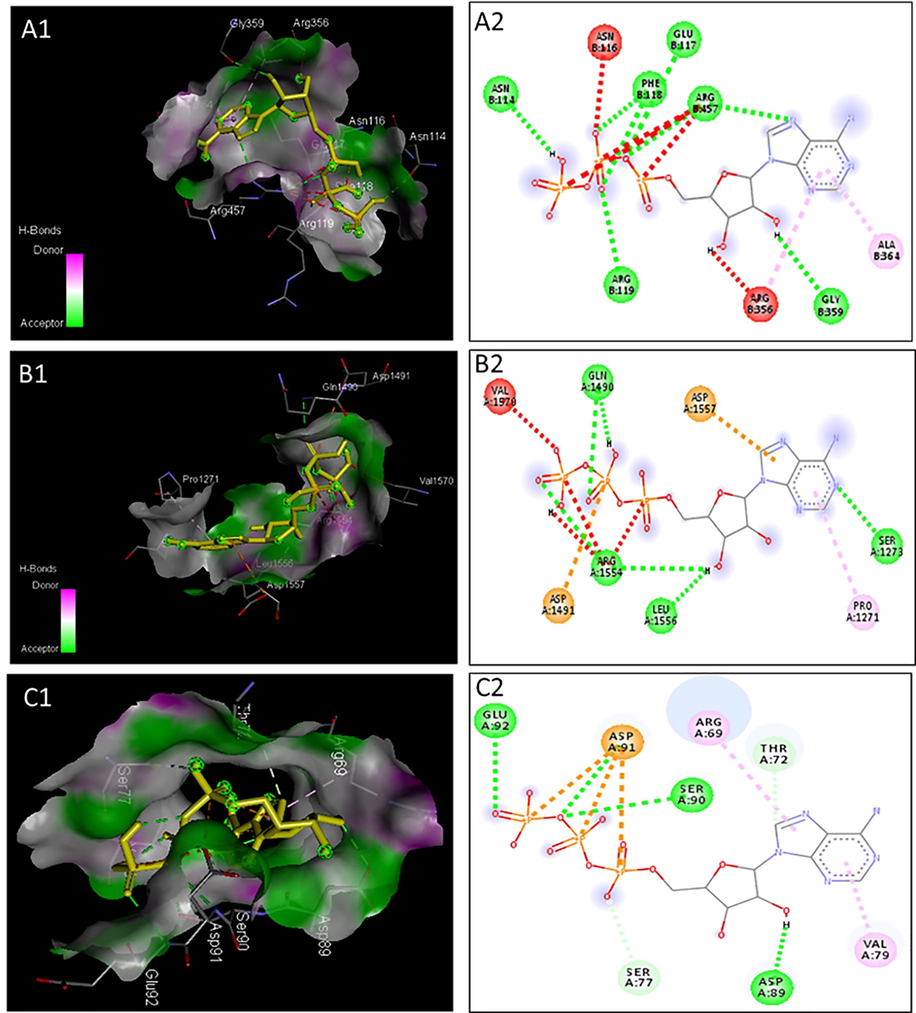
Molecular docking of Apis cerana ABC transporters.
4 Discussion
Honey bee A. cerana is one of China native bee species. A. cerana and A. mellifera are close relatives, A. cerana showed different characters compared to A. mellifera. Therefore, A. cerana illustrated strong resistance against Varroa destructor (mite) and getting nectar from flower resources.
All ABC transporter genes were classified into eight groups: ABCA, ABCB, ABCC, ABCD, ABCE, ABCF, ABCG, and ABCH. ABCC proteins subfamily has multiple functions, including ion transport, cell surface receptors, and a wide range of substrate translocation from drug to endogenous compounds (Fukuda and Schuetz, 2012; Tian et al., 2017). The human ABCC subfamily members mentioned to MRPs (multidrug resistance-associated proteins) act as sulfonylurea receptors and are involved in potassium channel regulation, transmembrane cystic fibrosis conductance regulation, and chloride channel construction. ABCD family members belong to half-transporter and import acylCoAs and fatty acid into cell organelle (Theodoulou et al., 2006). It is also involved in peroxisome-related development in Caenorhabditis elegans (Petriv et al., 2002). Further, the ABCE transporter family in yeast and human plays a vital function in translation initiation (Zhou et al., 1993). While the other two transporters, ABCE and ABCF, were not described in invertebrates. In penultimate larvae (Tribolium castaneum) significant mortality occurred when knocked down TcABCE-3A (ABCE) gene with RNAi approach (Broehan et al., 2013). In addition, A. cerana containing ABCG transporter protein showed the largest member of ABC subfamily (include 23 members), similar findings were also reported in Diuraphis noxia (Nicholson et al., 2015), Cimex lectularius (bed bug), Anopheles gambiae (Roth et al., 2003), Drosophila (Dean et al., 2001), A. mellifera (Liu et al., 2011), and Pediculus humanus (body louse in humans) (Lee et al., 2010). Sturm et al. (2009) reported that in the D. melanogaster and D. pulex genome, the ABCG transporter expansion result from specific lineage gene duplication (Sturm et al., 2009). The ABCE and ABCF subfamilies consist of unusual ABC transporter proteins, identified through NBDs pair link without TMDs. Thus, their structure was oblique their significance in the biological process rather than transportation. The ABCE1 is considered one of the vital conserved proteins in evolutionary relation and is observed in all organisms except eubacteria. Due to its important role in ribosome synthesis and translation, ABCE1 is vital for entire life stages (Barthelme et al., 2011). The RNase L/ABCE1 of Homo sapiens was primarily identified as an RNase L inhibitor. A. cerana has ten ABCG transporters. The ABCGs have a distinct reverse domain architecture with NBD-TMD (NBD restricted on N-terminus and other is TMD localized at C-terminus). In metazoans, the ABCG transporter considers half-transporters, while after dimerization, it works as functional transporters. Moreover, the ABCGs transporters in fungi and plants are full-transporters and known as PDRs (pleiotropic drug resistance protein) (Kovalchuk and Driessen, 2010). In this study, due to their duplicated structure domain, the PDRs yeast was not integrated with the phylogenetic analyses tree. Similar to other groups, A. cerana has five ABCH transporter, which contains inverse half-transporter that share similar architecture domains identical to ABCG transporter. It was first identified in D. melanogaster, which was only annotated in zebrafish and arthropods (Dean and Annilo, 2005).The ABCH transporter is not studied in mammals, plants, fungi, and C. elegans (Dean et al., 2001).It was studied in zebrafish but not identified in fungi, cod, and catfish (Liu et al., 2013).
The D. melanogaster RNAi screening illustrated that CG9990 D. melanogaster gene silencing was lethal (Mummery-Widmer et al., 2009). The ABCH (Px014955) was abundantly up-regulated in two insecticide-resistant strains of P. xylostella, (You et al., 2013). In T. castaneum, TcABCH-9C knockdown resulted in a hundred percent larval mortality and significantly reduced hatching and fecundity rate. Additionally, the T. castaneum larvae injected with dsRNA of TcABCH-9C showed it is involved in waterproof barrier formation in epicuticle (Broehan et al., 2013). The ABCH subfamily expression in cotton bollworm Manduca sexta and Helicoverpa armigera was vastly induced after treating secondary metabolites (Koenig et al., 2015). The ABC transporter gene structural analysis will be useful in protein functional analysis. Evolutionary background disclosed that exon-intron sequence had figured the evolutionary tree of gene family (Zaynab et al., 2021). Later on, it corresponds to prior scientific approaches retained in some plant genes that may contain no introns or short introns at the period of evolution (Zaynab et al., 2021).
Moreover, the firm gene structure may show a quick-expression response to endogenous and exogenous stimuli. Overall structural analysis suggested that ABC transporter genes showed a similar intron/exon with homogenous functional characters because they originated from duplication events during evolutionary activity. The MEME motif analysis identified several common and unique motifs in A. cerana ABC transporters. Commonly shared motifs usually tended in the same groups indicating similar functions (Cao et al., 2005).
5 Conclusion
The genome-wide study identified twenty-eight ABC transports encoding genes in the A. cerana (Ac) genome. The phylogenetic analysis revealed that AcABCs are also divided into eight groups similar to other organisms. Of these groups, ABCGs were the largest group in A. cerana genome. Different additional computational analyses provided essential data for further functional validation. The sequence associated with structural and functional motifs also showed some common and unique features in ABCs. These identifications might present new insight for advanced functional analysis on the ABC gene group in the A. cerana genome.
Acknowledgments
The author appreciates the support of the Research Centre for Advanced Materials Science (RCAMS) at King Khalid University Abha, Saudi Arabia through project number KKU/RCAMS/G002-21. I also thank Dr. Madiha Zaynab for her continous assistannce and useful suggestions during the preparation of this manuscript.
Declaration of Competing Interest
The author declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Varroa jacobsoni (Acari: Varroidae) is more than one species. Exp. Appl. Acarol.. 2000;24:165-189.
- [Google Scholar]
- Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proceed. Natl. Acad. Sci.. 2011;108(8):3228-3233.
- [Google Scholar]
- Functional analysis of the ATP-binding cassette (ABC) transporter gene family of Tribolium castaneum. BMC Genomics. 2013;14:1-19.
- [Google Scholar]
- A strong immune response in young adult honeybees masks their increased susceptibility to infection compared to older bees. PLoS Pathogens. 2012;8(12):e1003083.
- [Google Scholar]
- Identification of an RNA silencing suppressor from a plant double-stranded RNA virus. J. Virol.. 2005;79(20):13018-13027.
- [Google Scholar]
- Antimicrobial peptides: a key component of honey bee innate immunity: Physiology, biochemistry, and chemical ecology. J. Apicult. Res.. 2015;54(2):123-136.
- [Google Scholar]
- Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu. Rev. Genomics Hum. Genet.. 2005;6(1):123-142.
- [Google Scholar]
- The human ATP-binding cassette (ABC) transporter superfamily. J. Lipid Res.. 2001;42(7):1007-1017.
- [Google Scholar]
- Nutrition, immunity and viral infections in honey bees. Curr. Opin. Insect Sci.. 2015;10:170-176.
- [Google Scholar]
- Socialized medicine: individual and communal disease barriers in honey bees. J. Invertebr. Pathol.. 2010;103:S62-S72.
- [Google Scholar]
- ABC transporters and their role in nucleoside and nucleotide drug resistance. Biochem. Pharmacol.. 2012;83(8):1073-1083.
- [Google Scholar]
- Hygienic behavior of the honey bee (Apis mellifera) is independent of sucrose responsiveness and foraging ontogeny. Hormones Behav.. 2006;49(3):391-397.
- [Google Scholar]
- Harm of introducing the western honeybee Apis mellifera L. to the Chinese honeybee Apis cerana F. and its ecological impact. Acta Entomol. Sin.. 2005;3:015.
- [Google Scholar]
- The plastic response of Manduca sexta to host and non-host plants. Insect Biochem. Mol. Biol.. 2015;63:72-85.
- [Google Scholar]
- Phylogenetic analysis of fungal ABC transporters. BMC Genomics. 2010;11(1):177.
- [CrossRef] [Google Scholar]
- Lee, S. H., Kang, J. S., Min, J. S., Yoon, K. S., Strycharz, J. P., Johnson, R., Mittapalli, O., Margam, V. M., Sun, W., Li, H. M., 2010. Decreased detoxification genes and genome size make the human body louse an efficient model to study xenobiotic metabolism. Insect Mol. Biol. 19, 599–615.
- Genome-wide identification, characterization and phylogenetic analysis of 50 catfish ATP-binding cassette (ABC) transporter genes. PloS one. 2013;8(5):e63895.
- [Google Scholar]
- Genome-wide identification and characterization of ATP-binding cassette transporters in the silkworm, Bombyx mori. BMC Genomics. 2011;12:1-15.
- [Google Scholar]
- Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458(7241):987-992.
- [Google Scholar]
- The genome of Diuraphis noxia, a global aphid pest of small grains. BMC Genomics. 2015;16:1-16.
- [Google Scholar]
- RNA interference of peroxisome-related genes in C. elegans: a new model for human peroxisomal disorders. Physiol. Genomics. 2002;10(2):79-91.
- [Google Scholar]
- The role of the gut microbiome in health and disease of adult honey bee workers. Curr. Opin. Insect Sci.. 2018;26:97-104.
- [Google Scholar]
- Identification of the Anopheles gambiae ATP-binding cassette transporter superfamily genes. Mol. Cells. 2003;15:150-158.
- [Google Scholar]
- Theodoulou, F. L., Holdsworth, M., and Baker, A. (2006). Peroxisomal ABC transporters. Febs Lett. 580, 1139–1155.
- Genome-wide analysis of ATP-binding cassette (ABC) transporters in the sweetpotato whitefly, Bemisia tabaci. BMC Genomics. 2017;18:1-16.
- [Google Scholar]
- ABC50 interacts with eukaryotic initiation factor 2 and associates with the ribosome in an ATP-dependent manner. J. Biol. Chem.. 2000;275(44):34131-34139.
- [Google Scholar]
- Plant ABC proteins–a unified nomenclature and updated inventory. Trends Plant Sci.. 2008;13(4):151-159.
- [Google Scholar]
- Distantly related sequences in the alpha-and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J.. 1982;1(8):945-951.
- [Google Scholar]
- A heterozygous moth genome provides insights into herbivory and detoxification. Nature genetics. 2013;45(2):220-225.
- [Google Scholar]
- Mitogen-activated protein kinase expression profiling revealed its role in regulating stress responses in potato (Solanum tuberosum) Plants. 2021;10(7):1371.
- [CrossRef] [Google Scholar]
- Expression cloning of 2–5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell. 1993;72:753-765.
- [Google Scholar]







