Translate this page into:
Genome-wide analysis and expression profiling of CalS genes in Glycine max revealed their role in development and salt stress
⁎Corresponding author. sfli@szu.edu.cn (Shuangfei Li)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Abiotic stress affects plants' growth and development. Soybean is an important crop of the world, however, its production is affected by abiotic stresses. Callose Synthase is the most crucial enzyme response to environmental and developmental signals. However, in soybean, information on the callose synthase genes is limited. In this study, we analyzed the callose synthase gene family of soybean at the genome-wide scale. We also studied the genes positions, gene structure, evolutionary relations, miRNAs target sites, and expression of CalS genes. Resultantly 24 CalS genes were found in soybean, with diverse chromosomal locations, cis-acting elements, conserved motifs, and gene structures. Further, GmCalS genes were divided into four phylogenetic classes. The evolutionary classification of CalSs was supported by the motif and gene structure analyses. Phytohormones, abiotic stresses, and growth-responsive elements were identified in the promoter of GmCalSs. In addition, the GmCalS genes higher expression in roots, leaves, flowers, and nodules tissues provided their significance in development. Furthermore, the higher expression of GmCalS17 and GmCalS19 genes in response to salt stress indicated their importance against salt stress. These findings will be helpful for further investigation of the CalS genes in other crops.
Keywords
Callose Synthase
Expression
Growth
Regulation
qRT-PCR
Salt
1 Introduction
Callose is generally found in pollen tubes, grains, cell walls, and root hairs and is essential for transporting intercellular water, cell differentiation, and development (Chen and Kim, 2009; Nedukha, 2015). However, it is present in phloem sieve plates and at cell plasmodesmata, where it can regulate the passage of molecules from one cell to another (Ellinger and Voigt, 2014). Callose Synthase is the most crucial enzyme in callose biosynthesis, with numerous transmembrane segments and a hydrophilic center loop. Furthermore, it responds to environmental and developmental signals (Granato et al., 2019). Callose is produced in many distinct places inside the plants and functions as a phloem transport regulator, significantly influencing plants' development and response to stress (Granato et al., 2019). Pollen formation, cold stimulation, mechanical injury, fungal and bacterial infection, and insect infestation alter the CalSs expression (Feng et al., 2021).
The CalSs are essential regulators in the plant vegetative growth. Barratt et al. (2011) examined in Arabidopsis growth retardation was due to AtCalS9, AtCalS10, and AtCalS12 genes loss (Barratt et al., 2011). In addition, during the plant's vegetative growth, the AtCalS7 mutant was responsible for the dwarf phenotype in A. thaliana (Barratt et al., 2011). However, callose lining loss limits the efficiency of phloem transport and stops the process of transportation assimilating, leading to the development and growth retardation (Barratt et al., 2011). Shi et al. (2015) investigated that AtCalS5 maintained normal callose formation during development of pollen (Shi et al., 2015). Slewinski et al. (2012) reported that mutation in Tie-dyed2 (ZmCalS) gene was responsible for yellow leaves in maize (Slewinski et al., 2012). It was discovered that CalS12 was responsible for synthesizing callose at pathogen attack sites (Liu et al., 2018). In addition, Hyaloperonospora arabidopsis and salicylic acid (SA) induce AtCalS1, AtCalS5, AtCalS9, AtCalS10, and AtCalS12 expression (Dong et al., 2008). In Citrus limon ClCalS1 gene silencing causes more susceptibility to Xanthomonas citri (Enrique et al., 2011). Meanwhile, CalSs are regulated in several signaling pathways. However, hormones and transcription factors participate in different biological regulatory mechanisms. For example, ABA treatment boosts the rice callose synthase activity, and plants resist brown planthopper (BPH) by enhancing callose deposition (Liu et al., 2017). Feng et al. (2021) found CalSs important role against drought, salt, heat, and, cold stress in cotton (Feng et al., 2021).
The finding of the CalS gene family in many plants revealed their significance in development and response to environmental stress. To date, 15 CalS genes have been identified in Brassica rapa, 7 in Hordeum vulgare, 12 in Zea mays, 8 in Vitis vinifera, 32 in Brassica napus, 12 in Citrus sinensis, and 12 in Arabidopsis thaliana (Feng et al., 2021). However, callose synthase genes have not been well studied in soybean. Soybeans (Glycine max) are important because of their economic and nutritional worth. This oil and protein-rich plant contains essential amino acids for humans and other animals. Salt and other environmental stresses pose a danger to soybean production all over the world. Thus, soybean research is crucial for enhancing food security and increasing crop yields. This work investigated chromosomal location, cis-acting elements, conserved motifs, gene structure, and miRNA perdition. In addition, CalS genes expression was observed in several tissues. Understanding how the GmCalS genes respond to salt stress is a foundation for investigating other CalS genes in salt-affected crops.
2 Materials and methods
2.1 Identification of CalS genes
Soybean (Glycine max Wm82.a2.v1) CalS genes were found using BLASTP and HMM approaches. Soybean genome sequences were obtained from the Soybean Genome Database (Schmutz et al., 2010). However, AtCalS amino acids were utilized as a query in a BLASTP search. The amino acid sequences of AtCalSs were obtained from TAIR (https://www.arabidopsis.org/) (Lamesch et al., 2012). Moreover, the HMMER 3.13 program (El-Gebali et al., 2019) was also used to search CalS genes. The HMM file for the CalS domain (PF02364) was obtained from the Pfam database. A total of 24 GmCalSs were identified in the soybean genome after screening the presence of the PF02364 domain in sequences. Data for the M. truncatula genome was downloaded from the Phytozome JGI 12.0 dataset (https://phytozome.jgi.doe.gov/pz/portal.html).
2.2 Physicochemical characteristics and subcellular localization
We predicted the physicochemical characteristics of the GmCalS protein using the ProtParam program (https://web.expasy.org/protparam/) (Gasteiger et al., 2005). CELLO (https://cello.life.nctu.edu.tw/) version 2.5 was used to estimate the subcellular localization of GmCalS proteins. Using the TBtools program, the figure of the exons-introns configuration of GmCalSs was created. The MEME website was used to identify the conserved motifs of GmCalS sequences. The TBtools program was used to construct the motifs distribution.
2.3 Genes location and phylogenetic analysis
The soybean genome generic feature format (GFF) file was used to determine the chromosomal position of the GmCalSs. The genes chromosomal locations were determined with the use of TBTools. To better understand the evolutionary relationships among CalSs, a phylogenetic tree was built using AtCalSs, MdCalSs, and GmCalSs from the three different plant species. Multiple sequence alignment was carried out using the MEGAX program (Kumar et al., 2018). In this analysis, the neighbor-joining (NJ) method was employed to build a phylogenetic tree with 1,000 bootstraps.
2.4 Synteny analysis and Ka/Ks ratios
Circoletto Tool (tools. bat. infspire.org/circoletto/) was used for the synteny analysis. Additionally, the ratios of Ka/Ks were calculated with the help of the KaKs 2.0 Calculator (which may be found at https://sourceforge.net/projects/kakscalculator2/). We computed the estimated divergence time for the duplicated gene pairs using the formula t = Ks/2r' and r = 6.161029 × 109) (Lynch and Conery, 2000).
2.5 Prediction of cis-regulatory elements
The 2 kb sequences upstream of the start codons in the soybean genome were used to determine the cis-regulatory elements in the GmCalS genes promoters. PlantCARE website (Lescot, 2002) was used to analyze the promoter sequences of all GmCalS genes, and TBtools software was used to generate the figure.
2.6 Prediction of miRNAs
To predict miRNAs target sites, the complementary DNA sequences (CDS) of all GmCalSs were submitted to the psRNATarget website (https://www.zhaolab.org/psRNATarget/analysis?function=2) (Dai et al., 2018).
2.7 Expression profiling of GmCalS genes
A publicly accessible database was used for the GmCalSs expression analysis in different tissues. The transcriptome data of tissue expression was obtained from the NCBI SRA website with accession number SRA012188.1. The expression data in the roots, nodules, leaves, and flowers are present in Supplementary Table S6. The fragments per kilobase million (FPKM) were used to compute the transcript abundance. We used the TBtools software to generate an expression heat map.
2.8 qRT-PCR analysis
The gene expression analysis was performed on Williams 82 variety of soybean. These seeds were germinated in a mixture of vermiculite and humus in pots for 15 days. Further, NaCl (250 mM) was applied to soybean plants for 0 h, 1 h, 2 h, 4 h, 8 h, and 12 h. Leaves after the treatment were put in liquid nitrogen at −80 °C for future research. Total RNA was extracted from leaves using Trizol reagent following the manufacturer's instructions (TIANGEN, Beijing, China). The PrimeScriptTM RT Reagent Kit (TaKaRa, Shiga, Japan) was utilized for the cDNA synthesis using 3 µg RNA. In order to make the primers, we used Primer Premier 5. The Actin gene (NC_016089) was used as an internal control. Supplementary Table S1 lists the primers used in this research, and qRT-PCR analysis was performed in three biological replicates.
3 Results
3.1 Identification and characterization of GmCalSs
To identify genes belonging to the CalS family in soybean, BLASTP and HMM approaches were performed. Twelve AtCalS proteins were used as queries for the BLASTP search. Consequently, 24 GmCalS genes containing the CalS domain with Pfam ID PF02364 were found. These genes were referred to as GmCalS1–GmCalS24. Table 1 provides information on all 24 GmCalS genes. GmCalS genes varied in length as the number of amino acids was from 813 (GmCalS23) to 1965 (GmCalS19). The number of exons was from one (GmCalS5 and GmCalS17) to fifty-one (GmCalS12 and GmCalS24) (Table 1). The highest number of introns (50) was found in two genes (GmCalS12 and GmCalS24), while introns were absent from two genes (GmCalS5 and GmCalS17) (Table 1). The 24 GmCalS proteins were predicted with molecular weights ranging from 92.76 kDa (GmCalS23) to 227.93 kDa (GmCalS22), and their isoelectric points ranged from 7.99 (GmCalS22) to 9.6 (GmCalS13). Based on the in silico subcellular localization findings, nine GmCalS proteins were found on the chloroplast, and 15 GmCalS were present in the nucleus (Table 1).
Gene Name
Chromosome
Renamed
Strand
Start (bp)
End (bp)
AA
M.W
PI
Subcellular Localization
Exons
Glyma.04G192300
Chr04
GmCalS1
−1
46,344,683
46,403,452
1899
216692.3
8.63
Nucleus
50
Glyma.04G213100
Chr04
GmCalS2
1
48,485,249
48,499,658
1951
224253.27
9.19
Chloroplast
42
Glyma.05G191600
Chr05
GmCalS3
−1
37,711,639
37,726,572
1958
225287.88
8.95
Chloroplast
42
Glyma.06G173500
Chr06
GmCalS4
1
14,595,971
14,646,811
1607
184244.18
8.75
Chloroplast
45
Glyma.06G292500
Chr06
GmCalS5
−1
48,137,063
48,142,021
815
93966.56
8.86
Nucleus
1
Glyma.06G153100
Chr06
GmCalS6
−1
12,495,442
12,500,348
818
93705.89
8.96
Nucleus
14
Glyma.08G308200
Chr08
GmCalS7
−1
42,654,427
42,691,553
1867
218107.77
8.06
Nucleus
42
Glyma.08G156800
Chr08
GmCalS8
−1
12,115,786
12,130,558
1904
219045.95
9.14
Chloroplast
42
Glyma.08G157400
Chr08
GmCalS9
−1
12,162,921
12,193,517
1947
224701.85
9.2
Chloroplast
42
Glyma.08G361500
Chr08
GmCalS10
−1
47,300,299
47,324,252
1958
225711.83
9.15
Nucleus
42
Glyma.08G308700
Chr08
GmCalS11
1
42,739,735
42,764,488
1921
223453.16
8.1
Nucleus
42
Glyma.10G295100
Chr10
GmCalS12
1
51,201,095
51,244,654
1906
219441.26
8.37
Nucleus
51
Glyma.12G113300
Chr12
GmCalS13
1
11,127,377
11,132,579
826
96356.12
9.6
Chloroplast
2
Glyma.13G239300
Chr13
GmCalS14
−1
34,970,872
34,976,701
1446
167624.51
9.23
Chloroplast
2
Glyma.13G297100
Chr13
GmCalS15
−1
39,600,226
39,605,930
1743
203339.97
8.95
Nucleus
2
Glyma.13G261000
Chr13
GmCalS16
1
36,494,874
36,510,778
1965
227443.27
8.88
Nucleus
41
Glyma.15G074000
Chr15
GmCalS17
1
5,659,386
5,666,706
1799
208412.46
9.23
Chloroplast
1
Glyma.15G268800
Chr15
GmCalS18
1
50,529,595
50,548,992
1948
224918.86
9.15
Chloroplast
42
Glyma.15G245800
Chr15
GmCalS19
−1
46,829,270
46,851,722
1965
227404.31
8.88
Nucleus
41
Glyma.18G300200
Chr18
GmCalS20
1
57,789,188
57,820,853
1958
225535.87
9.18
Nucleus
42
Glyma.18G107900
Chr18
GmCalS21
1
12,220,182
12,260,477
1918
223114.09
8.57
Nucleus
42
Glyma.18G109100
Chr18
GmCalS22
1
12,576,818
12,661,446
1958
227935.27
7.99
Nucleus
42
Glyma.20G244800
Chr20
GmCalS23
1
47,478,494
47,490,211
813
92766.88
9.21
Nucleus
18
Glyma.20G244900
Chr20
GmCalS24
1
47,494,663
47,553,301
1905
219179.91
8.57
Nucleus
51
3.2 CalS genes phylogenetic relationships
Here, we construct a phylogenetic tree to understand the evolutionary links between the AtCalS, MtCalS, and GmCalS genes. A. thaliana contains 12 CalS genes and M. truncatula has 15 CalS genes (Supplementary Table S2). The M. truncatula, A. thaliana, and G. max CalS protein sequences were aligned to generate an unrooted phylogenetic tree. The 51 CalSs genes from the three plant species were divided into four groups (Fig. 1). The 19 CalSs in Group A were as follows: 7 from A. thaliana, 5 from M. truncatula, 7 from G. max. Group B included 7 G. max CalSs, 4 M. truncatula CalSs, 1 A. thaliana CalS. However, 1 CalSs was found in A. thaliana, 2 in M. truncatula, and 5 in G. max in Group C. Furthermore, 3 CalSs were found in A. thaliana, 3 in M. truncatula, and 5 in G. max in Group D. CalSs in the same group may perform similar functions. Significantly, GmCalS genes showed consistent distribution across all groups. Group A and B had the highest number of GmCalSs (7), followed by C and D (5). It was also discovered that the GmCalSs have the strongest evolutionary ties to the M. truncatula species.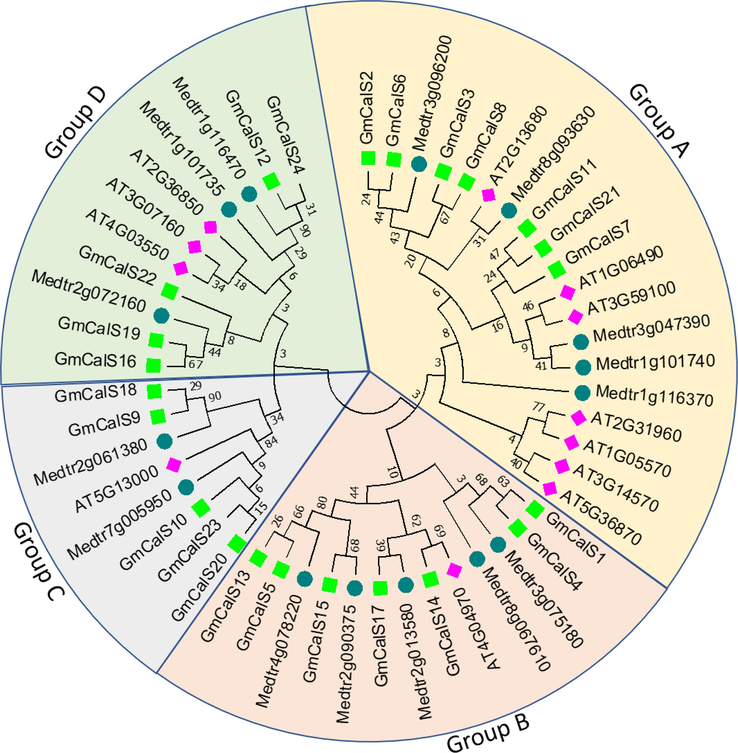
Evolution of the CalS gene in Medicago truncatula, Arabidopsis thaliana, and Glycine max investigated using a neighbour-joining phylogenetic tree. CalS genes were divided into four groups, each represented by a different color, in the plants A. thaliana (12), Medicago truncatula (15), and G. max (24).
3.3 Gene structures and conserved motifs analysis
To understand the evolution of the soybean CalSs, we examined the GmCalSs exon–intron structures. According to the findings, introns were between 0 and 50 and exons from 1 and 51. Overall, there are two genes with a single exon and no intron; three genes with two exons and a single intron; one gene with thirteen fourteen exons; one gene with seventeen introns and eighteen exons; two genes with forty-one exons and forty introns; eleven genes with forty-two exons and forty-one introns; and one gene with forty-five exons and forty-four introns; one gene with fifty exons and forty-nine introns; one gene with fifty-one exons and fifty introns (Fig. 2B). In addition, GmCalS gene members of the same Class had remarkably similar gene structures, consistent with the phylogenetic groups to which they belonged. In addition, the protein sequences were also analyzed to determine the motifs. The CalS genes have a conserved motif ranging from 4 to 10. Ten conserved motifs were found in this research, and information on these motifs can be found in Supplementary Table S3. Similar patterns of motif distribution were also seen within the group. Motifs 1, 2, 3, 4, 5, 8, and 9 were found in the GmCalS6 gene, whereas motifs 4, 8, 6, and 9 were found in the GmCalS23 gene. Furthermore, motifs 4, 5, 6, 8, 9, and 10 were observed in gene GmCalS13 while gene GmCalS5 had 1, 2, 3, 4, 5, 8, and 9 (Fig. 2A). However, the GmCalS14 gene contains 1, 2, 3, 4, 5, 6, 8, 9, and 10 motifs. It was also shown that all ten motifs were found in GmCalS1, GmCalS2, GmCalS3, GmCalS4, GmCalS5, GmCalS7, GmCalS8, GmCalS9, GmCalS10, GmCalS11, GmCalS12, GmCalS15, GmCalS16 GmCalS17, GmCalS18, GmCalS19, GmCalS20, GmCalS21, GmCalS22, GmCalS24 genes.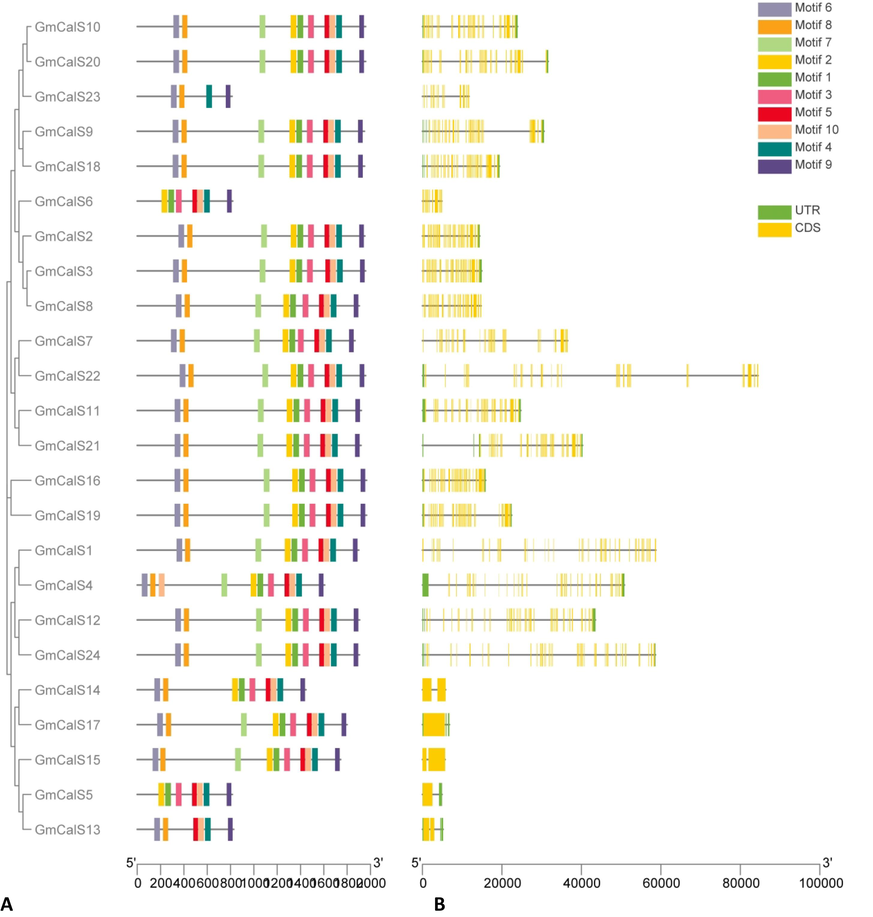
Analysis of GmCalS gene structure and conserved motifs.Based on their evolutionary relationships, the GmCalS genes were separated into four categories.(A) Conserved motif compositions of GmCalS (B). Gene structure of GmCalS genes. Exons are shown by the yellow color, UTRs, and introns by the black line. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4 Chromosomal locations and synteny analysis
We determined the chromosomal position of 24 GmCalSs genes and found that only ten chromosomes contained GmCalSs genes. Most chromosomes (Chr05, Chr10, and Chr12) contained just a single gene, but Chr04 had two genes. Further, Chr06, Chr13, Chr15, and Chr18 each had three, and Chr08 had five genes (Fig. 3). In our results, chromosomes Chr01, Chr02, Chr03, Chr07, Chr09, Chr11, Chr14, Chr16, Chr17, and Chr19 were found without any GmCalS gene. A study of the synteny among G. max, A. thaliana, and M. truncatula revealed a connection with the expression of genes and their evolution, functions, duplications, and triplications. It was discovered that the sequences of numerous Cals genes found in M. truncatula showed synteny with the CalS genes found in soybean. In addition, there were synteny links between the CalS genes of soybean and A. thaliana (Fig. 4).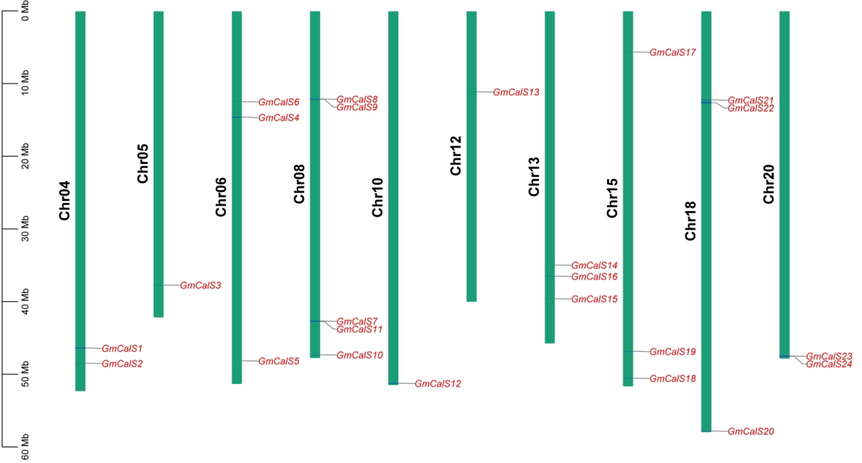
GmCalS Gene Position on the Chromosome. Green represented chromosomes, whereas red represented genes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
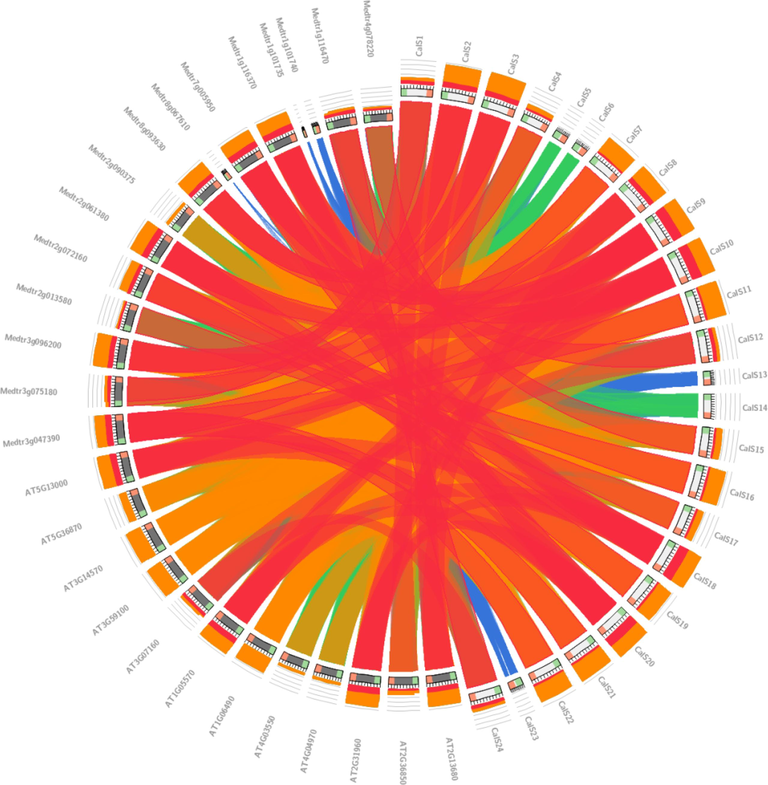
Identification of the synteny between the CalS sequences of A. thaliana, M. truncatula, and G. max.
3.5 Ka/Ks calculation
In order to calculate the molecular evolution rate, Ka/Ks for each duplicated gene pair estimated. When the Ka/Ks ratio was more than 1, it was considered that purifying selection was occurring among the duplicated genes; when it was less than 1, it was supposed that neutral selection was occurring; and when it was equal to 1, it was assumed that positive selection was occurring (Zaynab et al., 2021). Our results show that purifying selection was applied to most GmCalS duplicated genes during duplication. If the Ks values of GmCalS genes are higher than 0.52, the deviation time may be more than 100 million years ago (MYA). More intriguingly, the Ks value for the duplicated gene pair (GmCalS5/GmCalS13) was 0.632, indicating that the duplication event happened at about 51.31 MYA (Table 2).
Seq_1
Seq_2
Ka
Ks
Ka_Ks
Time(MYA)
Selection pressure
Duplication Type
GmCalS9
GmCalS18
0.016278265
0.088621853
0.183682292
7.192130789
Purifying
Segmental Duplication
GmCalS3
GmCalS8
0.015983369
0.090813741
0.176001653
7.37001412
Purifying
Segmental Duplication
GmCalS7
GmCalS22
0.104444264
0.171631656
0.608537294
13.92881419
Purifying
Segmental Duplication
GmCalS11
GmCalS21
0.013356072
0.091962241
0.145234299
7.463220911
Purifying
Segmental Duplication
GmCalS1
GmCalS4
0.020875212
0.099036766
0.210782446
8.037355928
Purifying
Segmental Duplication
GmCalS12
GmCalS24
0.019005594
0.099437946
0.191130193
8.06991384
Purifying
Segmental Duplication
GmCalS14
GmCalS17
0.0209955
0.124397777
0.168777129
10.09553578
Purifying
Segmental Duplication
GmCalS5
GmCalS13
0.32513289
0.632252677
0.514245177
51.31063956
Purifying
Segmental Duplication
GmCalS16
GmCalS19
0.011121102
0.105115001
0.105799383
8.530636742
Purifying
Segmental Duplication
3.6 Prediction of miRNAs
We discovered miRNAs targeting GmCalS genes to understand the miRNA-arbitrated post-transcriptional modification of GmCalSs. These miRNAs are part of different families. Supplementary Table S4 has the data of all miRNA-targeted sites/genes. According to the findings, gma-miR159 targeted a total of seven genes (GmCalS1, GmCalS4, GmCalS12, GmCalS14, GmCalS17, GmCalS20, and GmCalS24). The microRNAs, gma-miR172 targeted five genes GmCalS12, GmCalS16, GmCalS19, GmCalS20 and GmCalS21 genes; gma-miR171 targeted GmCalS1, GmCalS3, GmCalS4, GmCalS5, GmCalS7, GmCalS9, GmCalS10, GmCalS11,GmCalS14, GmCalS16, GmCalS17, GmCalS19, GmCalS20, and GmCalS21 genes; gma-miR395 targeted GmCalS11, and GmCalS22 genes; gma-miR394 targeted GmCalS7, GmCalS11, and GmCalS18 genes; gma-miR167 targeted GmCalS3, and GmCalS8 genes; gma-miR169 targeted GmCalS2, GmCalS3 GmCalS4, GmCalS7, GmCalS8, GmCalS11, GmCalS12, GmCalS14, GmCalS16, GmCalS17, GmCalS19, GmCalS21, GmCalS22,and GmCalS24 genes. It has been discovered that several common genes, such as GmCalS1, GmCalS10, GmCalS12, GmCalS14, and GmCalS20, are targeted by mostly different miRNAs.
3.7 Promoter analysis of GmCalS genes
The cis-regulatory elements in the promoters of GmCalS genes were studied to understand their regulatory functions in response to abiotic stress and during soybean growth. Supplementary Table S5 shows the GmCalS genes cis-elements details. Overall, we observed abiotic stress, phytohormones, and growth-responsive elements (Fig. 5). Several abiotic stress-responsive components including anaerobic, temperatures, light, and drought were found in GmCalS promoters. These components include the Box 4 motif, GT1 motif, GA motif, ARE motif, MBS motif, TC-rich repeats, and LTR motif. Similarly, the TCA-element, P-box/GARE-motif, ABRE, and TGACG-motif are responsible for the responses to five different phytohormones (salicylic acid, auxin, gibberellin, methyl jasmonate, and abscisic acid). It was found that some of the elements are unique to certain genes and are distributed inconsistently. In addition, we identified four elements associated with development, including meristem expression, endosperm expression, circadian regulation and zein metabolism. These elements include the O2-site, circadian, CAT-box, and GCN4 motif, which perform a dynamic role in the various phases of growth and development of soybean. It is possible to conclude that differential gene expression for GmCalSs may occur during different phases of development, and abiotic stress.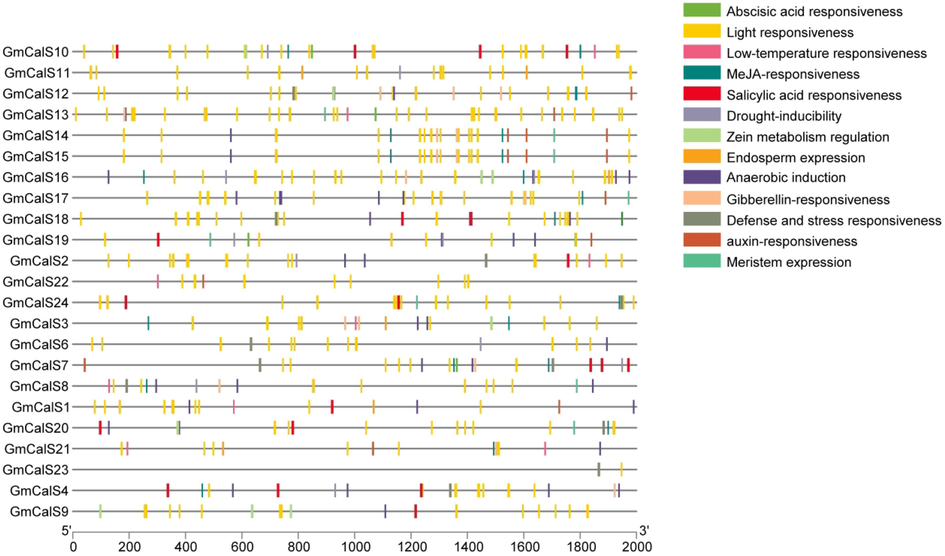
The promoter regions of the GmCalS genes are linked to cis-elements that respond to a variety of hormones and stresses. Supplementary Table S5 provides further information.
3.8 GmCalS genes expression in various tissues
This study utilized RNA-seq data to explore the GmCalS genes expression in flowers, leaves,nodules and roots. The findings demonstrated that several genes had higher expression in several tissues (Fig. 6; Supplementary Table S6). The results revealed that in leaves some genes including GmCalS5, GmCalS7, GmCalS9, GmCalS10, GmCalS11, GmCalS12, GmCalS13, GmCalS14, GmCalS15, GmCalS17, GmCalS18, GmCalS19, GmCalS21, and GmCalS24 display higher expression. However, GmCalS3, GmCalS5, GmCalS7, GmCalS8, GmCalS9, GmCalS10, GmCalS11, GmCalS12, GmCalS13, GmCalS14, GmCalS15, GmCalS16, GmCalS17, GmCalS18, GmCalS19, GmCalS21, and GmCalS24 genes display higher expression in roots. In nodule, GmCalS5, GmCalS7, GmCalS8, GmCalS9, GmCalS10, GmCalS11, GmCalS12, GmCalS13, GmCalS14, GmCalS15, GmCalS16, GmCalS17, GmCalS18, GmCalS19, GmCalS21 and GmCalS24 genes display higher expression. Several genes showed higher expression in flower, such as GmCalS2, GmCalS5, GmCalS7, GmCalS8, GmCalS9, GmCalS10, GmCalS11, GmCalS12, GmCalS13, GmCalS14, GmCalS15, GmCalS16, GmCalS17, GmCalS18, GmCalS19, GmCalS21 and GmCalS24 (Fig. 6; Supplementary Table S6). It was noted that certain genes demonstrated modest expression levels in various tissues (Fig. 6; Supplementary Table S6). Most genes seem to have a potential role in the growth of soybean.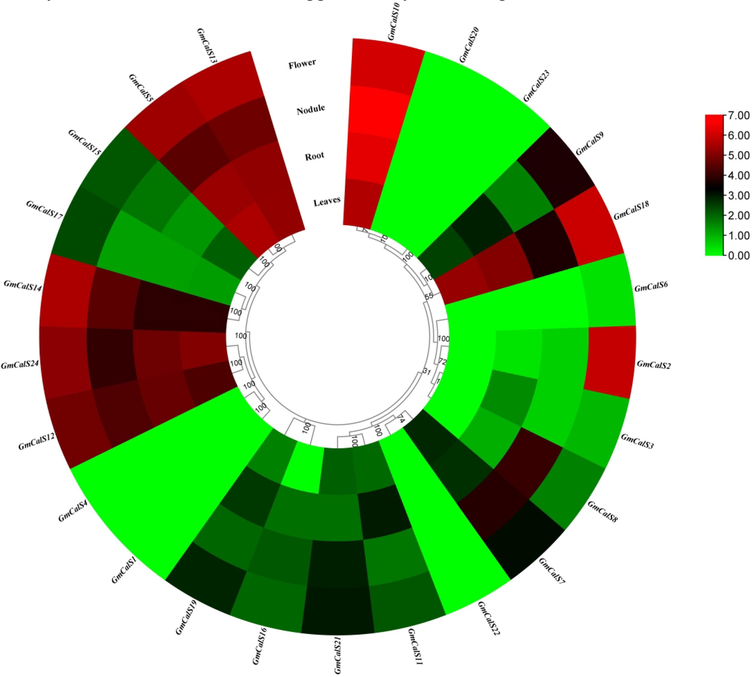
Expression profiling of GmCalS genes in various tissues. The red, black, and green colors display high to low expression levels. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.9 GmCalS genes expression under salt stress using RT-qPCR
In this study, GmCalS genes expression against salt in was observed. The qRT-PCR study was conducted to analyze GmCalS genes expression against salt stress at various time intervals (Fig. 7). According to the findings of the expression study, GmCalS-17, and GmGmCalS-19 exhibited higher expression at the 12 h. Further, stress-induced expression patterns give essential information on the significance of GmCalS genes in dealing with abiotic stress challenges.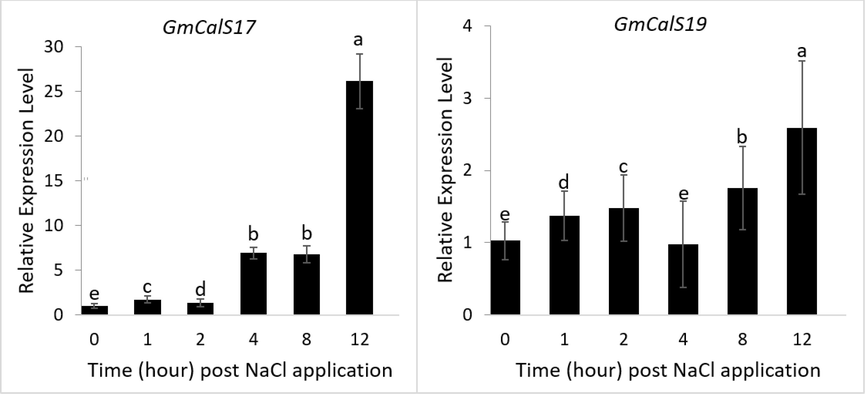
After Salt treatment the expression of the GmCalS genes were observed. Expression levels of two genes were studied after being exposed to NaCl for varying amounts of time (0, 1, 2, 4, 8, and 12 h) using quantitative real-time PCR (qRT-PCR) analysis. Soybean Actin used as the reference gene for normalising qRT-PCR results.
4 Discussion
Plants have contact with their surroundings therefore subjected to abiotic and biotic stresses. Abiotic stress factors affect plants' anatomy, physiology, biochemistry, and morphology, significantly reducing their growth and development (Nadarajah, 2020). Several studies have reported the function of callose in development of plant and against stresses (Piršelová and Matušíková, 2013; Verma and Hong, 2001). Due to callose significance, callose synthase has been studied in several plants. Eight CalSs were identified from Vitis vinifera, 15 CalSs from Chinese cabbage, 12 CalSs from Arabidopsis thaliana, 7 CalSs from Hordeum vulgare, 32 CalSs from Brassica napus and 12 CalSs in Citrus sinensis. The CalS genes in soybean have not been described. The sequences availability of the soybean genome provides resources for identifying CalS genes in the soybean genome (Schmutz et al., 2010). We found 24 GmCalS genes, which is higher than the number of CalS genes in Arabidopsis. This is evidence of a genome duplication event in the evolutionary process of G. max. Gene structure analysis demonstrated that genes from the same group had identical exon–intron patterns. Exon counts varied from 1 to 51, and intron counts from 0 to 50. Feng et al. (2021) reported a similar gene structure pattern in cotton, where the exons counts ranged from 1 to 51 (Feng et al., 2021). Results revealed that genes within same group had a similar structure. These results are consistent with findings in cotton (Feng et al., 2021) and Brassica napus (Liu et al., 2018), demonstrating that similarities in the structure of genes and motifs organization were found in the same class genes. Further, GmCalS genes functions in the response to environmental stress was revealed by the prediction of cis-elements in their promoters. We focus on three distinct cis-elements classes: phytohormones, abiotic stress, and plant growth and development-responsive elements. Furthermore, abiotic and phytohormonal stresses are regulated by cis-elements in the GmCalS genes. Previous research had uncovered cis-regulatory elements associated with abiotic stresses and phytohormones. The promoters of GmCalSs were discovered to have many hormone-responsive elements, suggesting their involvement in GmCalSs regulation. One of the plant's signal molecules was salicylic acid (SA) (Loake and Grant, 2007) increased the expression of AtCalS1/5/9/10/12 in Arabidopsis thaliana. The hormone abscisic acid (ABA), involved in callose synthesis, was crucial in responding to multiple stresses (Liu et al., 2017). Furthermore, ABA biosynthesis up-regulated PtCalS1 expression and blocked plasmodesmata to maintain the dormant state of Populus tomentosa (Tylewicz et al., 2018). Adding methyl jasmonate (MeJA) induced callose deposition in grape leaves. Callose deposition was sped up when Cationic peroxidase 3 (OCP3) expression was suppressed (Repka et al., 2004). OCP3 functions negatively on the JA pathway. In conclusion, callose deposition was governed by ABA, JA, and SA. However, the presence of ABA, SA, and JA-responsive elements in GmCalSs promoters suggests that these hormones regulate the expression of CalSs in soybean. The effects of salt stress treatment were also evaluated by analyzing the expression of GmCalS genes. Higher expression was observed for a few genes against salt stresses. The previous studies reported that CalS genes' expression was increased in response to stress. The GhCalS3 gene in cotton was up-regulated in response to cold, NaCl, and polyethylene glycol stress (Feng et al., 2021). CalS1 and CalS8 are significant genes in Arabidopsis that regulate biotic and abiotic stress responses (Cui and Lee, 2016). These findings indicate that CalS genes significantly influence plant hormone signaling pathways and abiotic stress tolerance. Plants' ability to cope with stress is directly affected by miRNAs (Villanueva et al., 2016). According to the results of this study, the identified mRNAs target GmCalS genes belonging to several families. Similarly, miR156 functions under various abiotic stress conditions in numerous plant species were reported (Arshad et al., 2017; Cui et al., 2016). miR167 was discovered as a key factor in coping with a diverse variety of stimuli (Khraiwesh et al., 2012). In grapevine, miRNA159 expression patterns were discovered. The findings revealed that miRNA159 was participated in gibberellin-induced parthenocarpy (Wang et al., 2018). According to Li et al. (2016), gma-miR172 overexpression in A. thaliana displays enhanced tolerance against drought and salt (Li et al., 2016). Also, miRNA-target genes expression validation is important to understand their function in soybean. This study analyzed the expression of 24 GmCalS genes in nodules, flowers, leaves, and roots using RNA-seq data. However, the results indicate that CalS genes exhibit distinct expression patterns in different developmental tissues. Tissue-specific expression patterns in Brassica napus were studied by using qRT-PCR data. According to the findings, BnCalS genes had elevated expression in the bud, silique, flower, leaf, stem, and root (Liu et al., 2018). Transcriptome-based expression results revealed that GhCalS genes had higher expression in various tissues (Feng et al., 2021). Researchers examined tissue-specific expression in Pyrus bretschneideri and found that the PbrCalS5 gene had higher expression in the pollen tube of pear (Cao et al., 2022). These findings are consistent with the findings of present study, where CalS genes displayed higher expression in the examined tissues (nodules, flowers, leaves, and roots), indicating that CalS may have a significant role in the development of soybean.
5 Conclusion
We identified 24 CalS genes in the soybean genome. Furthermore, chromosomal location, cis-acting elements, conserved motifs, gene structure, and miRNA perdition were analyzed. We discussed the GmCalS gene's expression in response to salt stress. However, we find GmCalS17 and GmCalS19 genes were enhanced by salt stress. In addition, several GmCalS genes were highly expressed in various tissues (roots, leaves, flowers, and nodules). These findings provide a foundation to understand the mechanism of stress resistance in soybean and establish a base for future investigation of the GmCalS genes and its function against salt stress.
Funding
Shenzhen Science and Technology Program (KCXST20221021111206015 and KCXFZ20201221173404012). Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2024R357), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. This work was supported by the Decree No. 220 by the Government of the Russian Federation (Mega-grant No 220-2961-3099).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- An insight into microRNA156 role in salinity stress responses of Alfalfa. Front. Plant Sci.. 2017;8
- [CrossRef] [Google Scholar]
- Callose synthase GSL7 is necessary for normal phloem transport and inflorescence growth in Arabidopsis. Plant Physiol.. 2011;155:328-341.
- [CrossRef] [Google Scholar]
- PbrCalS5, a callose synthase protein, is involved in pollen tube growth in Pyrus bretschneideri. Planta. 2022;256:22.
- [CrossRef] [Google Scholar]
- Callose synthesis in higher plants. Plant Signal. Behav.. 2009;4:489-492.
- [CrossRef] [Google Scholar]
- Arabidopsis callose synthases CalS1/8 regulate plasmodesmal permeability during stress. Nat. Plants. 2016;2:16034.
- [CrossRef] [Google Scholar]
- OsSGL, a novel DUF1645 domain-containing protein, confers enhanced drought tolerance in transgenic rice and Arabidopsis. Front. Plant Sci.. 2016;7
- [CrossRef] [Google Scholar]
- psRNATarget: a plant small RNA target analysis server (2017 release) Nucleic Acids Res.. 2018;46:W49-W54.
- [CrossRef] [Google Scholar]
- Expression of callose synthase genes and its connection with Npr1 signaling pathway during pathogen infection. Planta. 2008;229:87-98.
- [CrossRef] [Google Scholar]
- The Pfam protein families database in 2019. Nucleic Acids Res.. 2019;47:D427-D432.
- [CrossRef] [Google Scholar]
- Callose biosynthesis in arabidopsis with a focus on pathogen response: what we have learned within the last decade. Ann. Bot.. 2014;114:1349-1358.
- [CrossRef] [Google Scholar]
- Novel demonstration of RNAi in citrus reveals importance of citrus callose synthase in defence against Xanthomonas citri subsp. citri: Novel demonstration of RNAi in citrus defense. Plant Biotechnol. J.. 2011;9:394-407.
- [CrossRef] [Google Scholar]
- Genome-wide analysis of the CalS gene family in cotton reveals their potential roles in fiber development and responses to stress. PeerJ. 2021;9:e12557.
- [Google Scholar]
- Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M.R., Appel, R.D., Bairoch, A., 2005. Protein Identification and Analysis Tools on the ExPASy Server, in: Walker, J.M. (Ed.), The Proteomics Protocols Handbook. Humana Press, Totowa, NJ, pp. 571–607. https://doi.org/10.1385/1-59259-890-0:571.
- Callose synthase family genes plays an important role in the Citrus defense response to Candidatus Liberibacter asiaticus. Eur. J. Plant Pathol.. 2019;155:25-38.
- [CrossRef] [Google Scholar]
- Khraiwesh, B., Zhu, J.-K., Zhu, J., 2012. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 1819, 137–148. https://doi.org/10.1016/j.bbagrm.2011.05.001.
- MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol.. 2018;35:1547-1549.
- [CrossRef] [Google Scholar]
- The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res.. 2012;40:D1202-D1210.
- [CrossRef] [Google Scholar]
- PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res.. 2002;30:325-327.
- [CrossRef] [Google Scholar]
- Overexpression of soybean miR172c confers tolerance to water deficit and salt stress, but increases ABA sensitivity in transgenic Arabidopsis thaliana. EXBOTJ. 2016;67:175-194.
- [CrossRef] [Google Scholar]
- Mechanisms of callose deposition in rice regulated by exogenous abscisic acid and its involvement in rice resistance to Nilaparvata lugens Stål (Hemiptera: Delphacidae): Callose regulated by ABA and its involvement in rice resistance to BPH. Pest. Manag. Sci.. 2017;73:2559-2568.
- [CrossRef] [Google Scholar]
- Characterization of callose deposition and analysis of the callose synthase gene family of Brassica napus in response to Leptosphaeria maculans. IJMS. 2018;19:3769.
- [CrossRef] [Google Scholar]
- Salicylic acid in plant defence—the players and protagonists. Curr. Opin. Plant Biol.. 2007;10:466-472.
- [CrossRef] [Google Scholar]
- The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151-1155.
- [CrossRef] [Google Scholar]
- ROS homeostasis in abiotic stress tolerance in plants. IJMS. 2020;21:5208.
- [CrossRef] [Google Scholar]
- Callose: Localization, functions, and synthesis in plant cells. Cytol. Genet.. 2015;49:49-57.
- [CrossRef] [Google Scholar]
- Callose: the plant cell wall polysaccharide with multiple biological functions. Acta Physiol. Plant. 2013;35:635-644.
- [CrossRef] [Google Scholar]
- Methyl Jasmonate is a potent elicitor of multiple defense responses in grapevine leaves and cell-suspension cultures. Biol. Plant.. 2004;48:273-283.
- [CrossRef] [Google Scholar]
- Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178-183.
- [CrossRef] [Google Scholar]
- GLUCAN SYNTHASE-LIKE 5 (GSL5) plays an essential role in male fertility by regulating callose metabolism during microsporogenesis in rice. Plant Cell Physiol.. 2015;56:497-509.
- [CrossRef] [Google Scholar]
- Tie-dyed2 encodes a callose synthase that functions in vein development and affects symplastic trafficking within the phloem of maize leaves. Plant Physiol.. 2012;160:1540-1550.
- [CrossRef] [Google Scholar]
- Photoperiodic control of seasonal growth is mediated by ABA acting on cell-cell communication. Science. 2018;360:212-215.
- [CrossRef] [Google Scholar]
- Editorial: Signaling through WD-repeat proteins in plants. Front. Plant Sci.. 2016;7
- [CrossRef] [Google Scholar]
- Spatio-temporal expression of miRNA159 family members and their GAMYB target gene during the modulation of gibberellin-induced grapevine parthenocarpy. J. Exp. Bot.. 2018;69:3639-3650.
- [CrossRef] [Google Scholar]
- Mitogen-activated protein kinase expression profiling revealed its role in regulating stress responses in potato (Solanum tuberosum) Plants. 2021;10:1371.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2023.103049.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







