Translate this page into:
Gene regulators associating the T2DM and periodontitis contributing disease prognostic markers and therapeutic target
⁎Corresponding author. dr.ravipatil@gmail.com (Shankargouda Patil)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Periodontitis can be exacerbated by variety of systemic disorders including type 2 diabetes. T2DM patients exhibited a threefold increased risk of periodontitis. The mechanism through which T2DM induces periodontitis is uncertain. This study aims to identify co-regulatory molecules involved in T2DM periodontitis. Understanding the mechanisms could lead to new prognostic biomarkers and therapeutic targets.
Methodology
Gene expression datasets (GSE156993)with experimental conditions: 1) healthy control 2) periodontitis, and 3). T2DM with periodontitis were assessed. The up-regulated genes in Group1 (healthy control vs periodontitis) and Group2 (healthy control vs T2DM with periodontitis)were identified. Protein interaction network were constructed from up-regulated genes and the topological characteristics network were determined. Each network's hubs containing the transcription factor that was up-regulated, as well as its coregulators were retrieved. Finally, the hubs were enriched based on the gene ontology and molecular pathways to recognize thehubs function.
Results
751 genes in Group1 and 762 in Group2 were noticed up-regulated. Two individual protein interaction network constructed from theseup-regulated genesthat showed complex interactions based on topological analysis. Theextracted hubs from each network displayed a variety of pathways relevant to the immune system, cell cycle, and signal processing. Analysis of hubs showed FOS transcription factor was common in both conditions containing diverse transcriptional coregulators such PKP2, RUNX2, TSC22D3, SUPT6H, and MAP3K7 in periodontitis and IDS, OPTN, PPP1R12A, BATF, and LMNA in T2DM with periodontitis.
Conclusion
Our findings demonstrate FOS has a variety of regulatory patterns, providing evidence of T2M progression to periodontitis. Further experiments are still needed to confirm these transcriptional coregulators as prognostic biomarkers and therapeutic targets.
Keywords
Biomarkers
Diabetes Mellitus
Gene Expression
Periodontitis
Prognosis
1 Introduction
Periodontitis is a chronic inflammatory condition that damages the teeth's supporting tissues, eventually leading to loss of tooth. Periodontal disease was considered as a one of the most common disease across the globe(Vos et al. 2017). Periodontitis is associated with both modifiable and non-modifiable risk factors, which includes genetic factors, systemic diseases, and environmental factors. Notably, periodontal deterioration occurs not just as a result of direct damaging effects bacterial pathogens' but also as a result of a disrupted immune response against pathogens(Van Dyke and Serhan 2003). Despite bacterial infection, investigations of identical twins show that host genetics play a vital role in periodontal disease susceptibility(Michalowicz et al. 2000).Additionally, systemic illness like diabetes mellitus has proven to have great impact on periodontitis.
Type II diabetes mellitus (T2DM) is a metabolic disorders marked by hyperglycemia due to insulin resistance and /or insufficient secretion(Blair n.d.).T2DM has shown to be of the one causative factors for periodontitis. Particularly, patients with T2DM have a threefold elevated risk of periodontitis compared to non-diabetic patients suggesting uncontrolled glycaemicis the key determining player for periodontitis risk(Mealey and Ocampo 2007). Furthermore, study show a “two-way interaction” between periodontal disease and diabetes, with diabetes increasing the risk of periodontal disease and periodontal disease negatively impacting glycemic control and aggravating the incidence of diabetic complications(Grossi and Genco 1998).Although dentists have long recognised the necessity of a diabetes diagnosis in their patients due to the association with periodontitis, the molecular mechanism triggerT2DM- towards periodontal disease is poorly understood.Discovery of linkage molecular mechanisms could lead to new therapeutic targets for individualized periodontitis care in diabetes mellitus patients.
Systems biology has recently made a substantial contribution to understanding complicated diseases by combining biological data from a number of molecular biological platforms. Such integration can be used to assess molecular linkages between the diseases using relevant molecular data from microarray and high throughput sequencing studies to identify diagnostic biomarkers and therapeutic targets(Vijayashree Priyadharsini 2019). Currently, several microarray or sequencing data have provided the global gene-expression for various diseases that are being deposited and available for further research use. Herein, the microarray gene expression data was used to identify possible molecular regulatory connections between diabetes mellitus and periodontitis. We demonstrated a few significant regulatory molecules that were specific and common between the conditions. Our identified regulatory molecules provide putative molecular connective between periodontitis and diabetes mellitus that contribute in defining several previously unknown and unknown cross-talk genes, molecular function and pathways.
2 Materials and methods
2.1 Data resource and microarray data
The Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) was used to obtain the gene expression microarray dataset by search with relevant key terms such as diabetes, periodontitis, periodontal disease, hyperglycemia, tooth bacterial infection and T2DM. Use of these search terms was done individual or with combination in the NCBI GEO database. Among the various dataset, GSE156993 was found suitable and relevant to solve the objective of this study. GSE156993contains peripheral blood mononuclear cells (PBMCs) gene expression profile data of healthy control (n = 6); periodontitis (n = 6); and T2DM with periodontitis (n = 5). Each expression data included in our study were represented with their age, gender, fasting plasma glucose and HbA1C levels. Accordingly, the average and standard deviation of each ensemble between the groups were shown in Table.1.
Parameter
Healthy control (Average ± SD)
Periodontitis (Average ± SD)
T2DM with Periodontitis (Average ± SD)
Age (Years)
41.14 ± 3.18
45.50 ± 6.94
48.4 ± 9.68
Gender (count)
Male: 2
Female:4Male: 2
Female:4Male:1 Female:4
Fasting plasma glucose (mg/dL)
87 ± 8.8
88.5 ± 4.6
274.2 ± 48.4
HbA1C levels (%)
5.4 ± 0.19
4.7 ± 0.4
12.02.4 ± 1.85
2.2 Gene expression analysis
The expression array was pre-processed to remove the noise removed from the original microarray data. Affy-module in the R package was used for the standardisation and background correction of raw data(Gautier et al. 2004). Each microarray chip had 54,675 probes, an average expression level was considered ifone or more probes were assigned to same gene. After data normalization and transformation the differentially expressed genes (DEGs) between the conditions Group1) healthy control vs periodontitis; Group2) healthy control vs T2DM with periodontitis were determined using limma module in R package(Diboun et al. 2006). Further, the statistical significance of gene expression for each group was assessed by student t-test and the obtained P-value < 0.05 was considered as statistical significance.
2.3 Protein interaction network
Next, protein–protein interaction (PPI) networks were built using the significant up-regulated genes in Groups 1 and 2 analyses, respectively. We use our own proprietary database comprising a huge range of protein interaction data from potential resources such as BIND(Isserlin, El-Badrawi, and Bader 2011), IntAct (Kerrien et al. 2012), BioGRID(Chatr-aryamontri et al. 2015), DIP(Salwinski 2004), and HPRD(Keshava Prasad et al. 2009). For the network construction, each up-regulated gene wasextended to one node of immediateneighbouring protein out-side the up-regulated gene set.Using the interaction data, the PPI network was created and visualized using Cytoscape version 3.8.2 ( https://www.cytoscape.org)(Shannon et al. 2003). Each extended network provides the functional proteomes behaviour of group1 and group2 conditions. Furthermore, the network analyser module of cytoscape was used to assess the topological features such as node degree, betweenness centrality, stress centrality, and closeness centrality for the network(Anirudhan et al. 2021).
2.4 Regulatory hubs from the network
Data on transcription factor was obtained in the Network Analyst database (https://www.networkanalyst.ca/). From each network, the transcription factors (TFs) that were over-represented in microarray analysis, as well as their linked proteins (coregulators) were extracted. These retrieved transcription factors and their associated proteins were referred to as hubs. The hubs were chosen to connect minimum two proteins to each of the transcription factor.
2.5 Enrichment analysis
The obtained hubs were subjected to GO enrichment and REACTOME pathway analysis. DAVID ( https://david.abcc.ncifcrf.gov/: Database for Annotation, Visualization and Integrated Discovery) was utilised in this investigation. DAVID provides the functional annotation of the hubs with biological process, molecular functions and cellular localization. We interested inbiological process (BP) and molecular functions (MF) of the regulatory hubs. Likewise, REACTOME pathways enrichment was used to demonstrate the involvement of regulatory hubs in the molecular pathways. Both, functional annotation (DAVID) and molecular pathways (KEGG) were selected appropriate to the regulatory hubs based on cut-off threshold p < 0.05.
3 Results
3.1 Identified DESs of periodontitis with and without T2DM
The GSE156993 dataset was processed using limma programme in R. The differentially genes with a p-value < 0.05 were considered for subsequent analysis. As a result in 1331DEGs were identified in Group1 analysis, of which 751 were up-regulated and 580 were down-regulated in periodontitis compared to control (Group 1). Likewise, the analysis of Group 2 showed 762 were up-regulated and 917 were down-regulated in T2DM with periodontitis. Among the DEGs, only the up regulated gene from both group 1 and 2 analysis were used for further analysis.
3.2 Protein network of periodontitis and T2DM with periodontitis
The network constructed from the group1 with751 up-regulated genes showed, extended interaction of 13,568 containing 1471 proteins (Fig. 1).Similarly, the protein interaction network generated from762 up-regulated genes of group2 contains 14,504 interaction contributed by 1495 proteins (Fig. 2). Each network provide the protein interactome behaviour upon the dysregulation of genes in respective condition. The protein interaction network was visualized and the network properties were determined by Cytoscape software version 3.6.1. The topological analysis of group1 and group2 presented complex network properties as demonstrated in Table 2. To be noted, group2 network was with more number of proteins, and interaction and with highest network degree compared to the group1 network (Table 2).
The network constructed from the group1 with 751 up-regulated genes showing extended 13,568 interaction with 1471 proteins.
The network constructed from the group2 with 762 up-regulated genes showing extended 14,504 interaction with 1495 proteins.
Topological Parameter
Periodontitis (group1)
T2DM with Periodontitis (group2)
Average Shortest Path Length
3.46
4.12
Betweenness Centrality
0.017
0.019
Closeness Centrality
0.362
0.172
Clustering Coefficient
0.032
0.217
Network Degree
2.17
4.87
Neighbourhood Connectivity
7.62
13.21
Topological Coefficient
0.312
0.224
3.3 Core regulatory hubs of the network
Each network was segmented into small-network known as hub containing one transcription factor with its immediate regulating proteins. These immediate regulating proteins may recognized as transcriptional coregulators that interacts with the transcription factor to increase the rate of transcribing genes. From the group1 network, nine hubs were extracted containing the TF and its immediate coregulators. Among these hubs, threewere retained containing the ESR1, FOS, and SP1, transcription factors that were up-regulated while group1 gene expression analysis. For instance, FOS was up regulated and interacts with72 coregulators in the network. Among these coregulators, FOXO3, SOS1, TADA3, IGF1R, MED16, TCF20, SRC, and SMAD4 were upregulated in periodontitis (Figs. 3-5). Similarly from the group 2 network, four hubs with up regulated TFs such as FOS, TAL1, ITGAL, and TGIF1 were extracted. The TFs noticed interacting with the multiple proteins in our group 2 network (Figs. 6-9).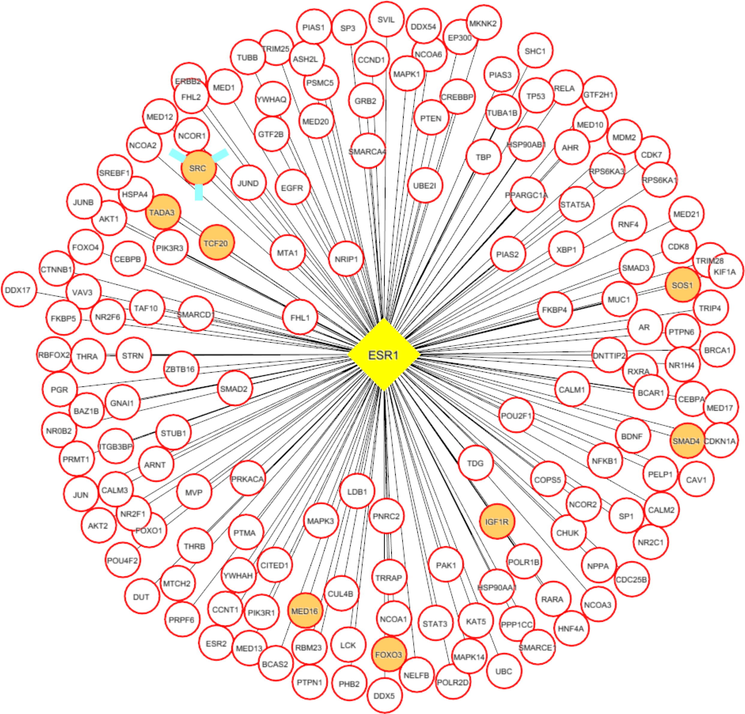
ESR1 (transcription factor- yellow colored node) hub with its interacting proteins, the orange node represent up-regulated genes encoding protein in expression analysis.
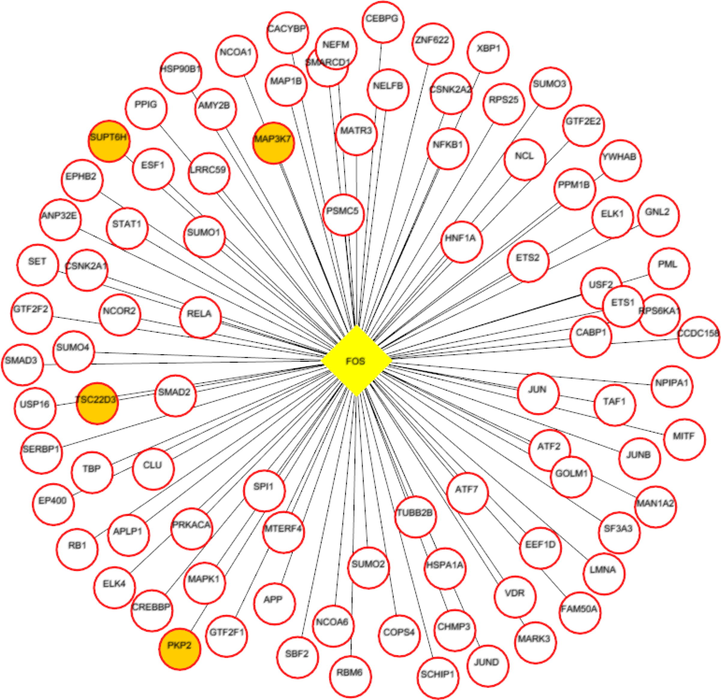
FOS (transcription factor- yellow colored node) hub with its interacting proteins, the orange node represent up-regulated genes encoding protein in expression analysis.
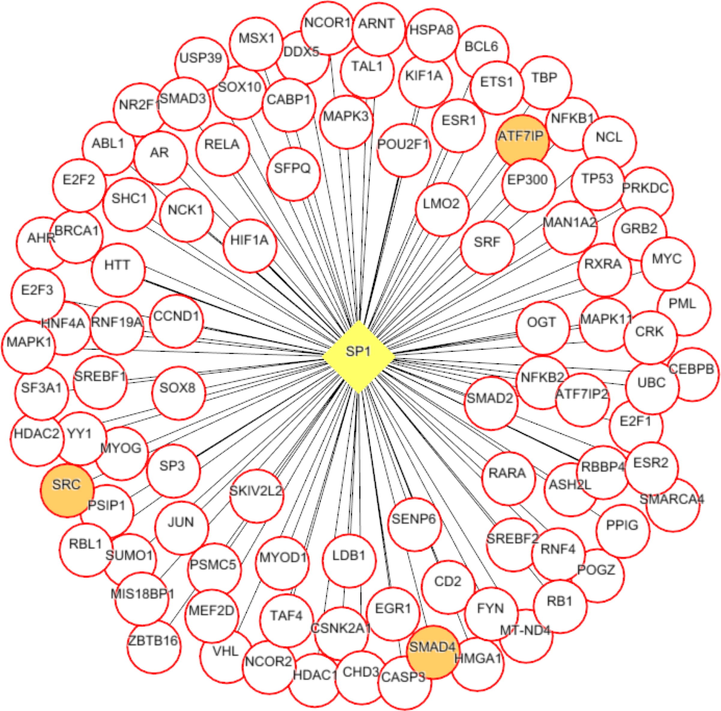
SP1 (transcription factor- yellow colored node) hub with its interacting proteins, the orange node represent up-regulated genes encoding protein in expression analysis.
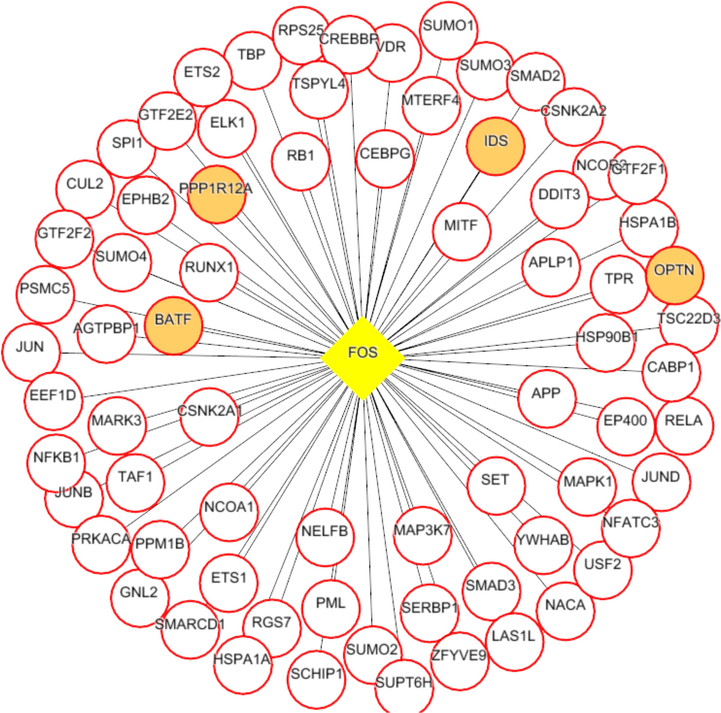
FOS (transcription factor- yellow colored node) hub with its interacting proteins derived from group2 network, the orange node represent up-regulated genes encoding protein in expression analysis.
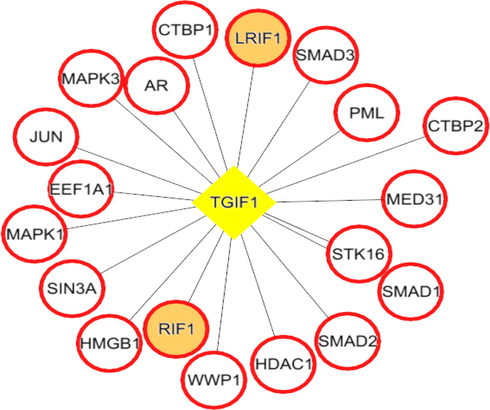
TGIF1 (transcription factor- yellow colored node) hub with its interacting proteins derived from group2 network, the orange node represent up-regulated genes encoding protein in expression analysis.
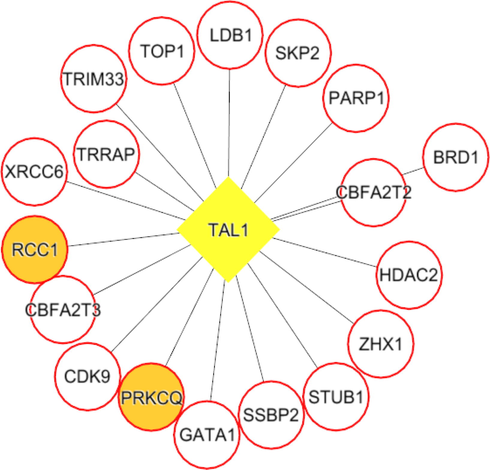
TAL1 (transcription factor- yellow colored node) hub with its interacting proteins derived from group2 network, the orange node represent up-regulated genes encoding protein in expression analysis.
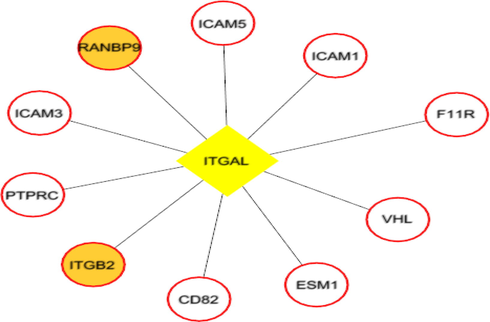
ITGAL (transcription factor- yellow colored node) hub with its interacting proteins derived from group2 network, the orange node represent up-regulated genes encoding protein in expression analysis.
3.4 Functional assessment of the regulatory hubs
The results of Gene Ontology analysis using DAVID revealed that the group1 hubs were significantly enriched in BP, and MF, involving in regulation of transcription, regulation of cell cycle, mRNA transcription from RNA polymerase II promoter, cell differentiation, insulin receptor signaling, apoptotic process and cellular response to oxidative stress (Supplementary File 1); Likewise, the group 2 hubs genes were significantly enriched participate intranscription factor binding, regulation of cell cycle, inflammatory response, transcription coactivator binding, integrin binding, NF-kappaB binding, cellular response to lipopolysaccharide, cell–cell adhesion. (Supplementary file 2). Additionally, the pathway analysis revealed significantly association with Generic Transcription Pathway, ESR-mediated signaling, Signaling by Interleukins pathway, Signaling by Receptor Tyrosine Kinases, Signal Transduction, Cytokine Signaling in Immune system for group1 hub (Supplementary file 1) and involvement in Generic Transcription Pathway, Signaling by TGF-beta Receptor Complex, Cellular responses to stress, Toll Like Receptor 10 (TLR10) Cascade, Cellular responses to stimuli, Cytokine Signaling in Immune system and MAP kinase activation of group2 hubs(Supplementary file 2).
3.5 Transcriptional regulators of the hubs
Next, the hubs from the group 1 and 2 were looked for the presence of common TFs. FOS transcription factor noticed in common and up-regulated in both the conditions(group1 and 2). Additionally, these common TF noticed with unique coregulators across each condition. For instance in group1 condition, the hub with FOS transcription factor interacts with PKP2, TSC22D3, SUPT6H, and MAP3K7 that were up-regulated in expression analysis. Whereas the FOS transcription factor hub from group2 interacts with up-regulated IDS, OPTN, PPP1R12A and BATF proteins.
4 Discussion
The vast majority of people with type 2 diabetes seek medical treatment in order to prevent future complications. A periodontitis is one of the complications that can occur with type 2 diabetes. Patients with T2DM are at a three times greater risk of developing periodontitis. An early diagnosis or the ability to predict the likelihood of periodontitis will be of important for the early precaution and disease treatment. Understanding the molecular pathogenesis betweenT2DM and periodontitis will opens up new avenue for the development of diagnostic biomarker and novel therapeutic strategies.
Despite the fact that researchers have made significant progress in understanding the molecular linkage between T2DM and periodontitis by themeans of inflammatory mechanism. Type II diabetes has been associated to higher levels of inflammatory markers (Dandona 2004). Hyperglycemia has been shown to activate pathways that promote inflammation, oxidative stress, and apoptosis in cells (Brownlee 2005). It has been established that patients with diabetes have higher levels of IL-6 and TNF- alpha(Dandona 2004). Likewise, the periodontitis patients exhibit high levels of inflammatory markers including IL-6 and TNF- alpha leading to activate multiple pathways similar to T2DM. Xiangchun Li et al., 2019 compared to normal dental pulp with dental pulp acquired from teeth that had periodontitis revealed symptoms of inflammation, oxidative stress, and showed elevated apoptosis and autophagy(X. Li et al. 2019). Also, inflammatory molecules have been shown to disrupt intracellular insulin signalling, which may play a role in insulin resistance that can worsen the T2DM condition as a result of periodontal disease(Hotamisligil 2000; Rotter, Nagaev, and Smith 2003).
Considering enamours molecular association between the T2DM and periodontitis, there is still a lack of understanding in generegulatory mechanism by the means of transcription factors thatlinking both the conditions. In this work, we used a high-throughput technique to detect DEGs from the periodontitis patients (n = 6) and the T2DM patients presenting periodontitis (n = 5) were compared to the normal healthy control (n = 6). We used PBMC gene expression profile retrieved from the repository and analysed to generate protein interaction network. The network was segmented into hubs containing transcription factors and coregulators. The protein in the hubs were functionally enriched and most contributing regulatory hubs integrating T2DM and periodontitis were determined. In the gene expression analysis of group 1 datasets showed 751 overexpressed genes that were used to construct the proteinnetwork containing 13568. Likewise the complex network was generated from the 762 up regulated genes. Analysis based on the network properties, suggest group 2 network was highly complex based on the topological parameters (Table.2).
Both the network was segmented to obtain nine hubs from each of the protein interaction network. The periodontitis (group1) hubscontainstranscription factor such as ESR1, FOS, FOXD1, NFIC, NOBOX, RUNX2, SF1, SP1, and ZEB1 that regulate multiple genes.Of the nine, three regulatory hubs were selected presence of TFs up-regulation on gene expression analysis.For instance,the ESR1transcription factorwas overexpressed with 26 coregulatorsin the periodontitis network. Among the 26 interacting proteins, SOS1, TADA3, IGF1R, MED16, TCF20, and SRC were up-regulated in periodontitis condition. ESR1 is aestrogen receptor 1, highly expressed in periodontitis gingiva, associated withvarious oral conditions (Arid et al. 2019; Dalledone et al. 2019; Shang et al. 2016). Polymorphisms in ESR1 gene noticed associated with chronic periodontitis(Zhang et al. 2010). Similarly, the other transcriptionfactors suchFOSand SP1 regulate multiple genes, of which PKP2, TSC22D3, SUPT6H, MAP3K7, SMAD4, and ATF7IP were up regulated in periodontitis. On the other hand, we assess the nine hubs of group 2 network. Among these, four were selected containing the FOS, TAL1, ITGAL, and TGIF1 transcription factors containing coregulators such IDS, OPTN, PPP1R12A, BATF, PRKCQ, RCC1, LMNA, ITGB2, RIF1, LRIF1, and RANBP9 and noticed altered in group2 (T2DM patients with periodontitis) gene expression analysis.
Functional enrichment analysis of the selected hubs were majorly involvement in immune process, cell cycle, cell death, cell–cell communication and cellular response to stimuli. Particularly, coregulators in the selected group1 hubs were associated with MAPK family signaling cascades, cytokine signaling in immune system, extra-nuclear estrogen signaling, nuclear receptors signaling and receptor tyrosine kinases signaling pathways. These molecular pathways are association with periodontitis. In periodontal disease condition, the host recognize the external stimulus of bacterial lipopolysaccharide, which activates MAPK cascade that results in the generation of inflammatory cytokines. favouring osteoclastogenesis and increased net bone resorption in the periodontal environment(Q. Li, Valerio, and Kirkwood 2012). Simultaneously, the functional enrichment study of group2 hubs showed involvement interleukins signaling, cell cycle and integrin cell surface interactions in T2DM with periodontitis. Notably, the molecular pathway across the both condition looks similar, whereas the contributing coregulators to the pathways were different between group1 and group2 hubs. On hubs integration, FOS transcription factor was noticed common between the both condition with coregulatorssuch as PKP2, TSC22D3, SUPT6H, and MAP3K7 in periodontitis and IDS, OPTN, PPP1R12A, BATF, LMNA in T2DM with periodontitis these results suggest FOS follows divers regulatory pattern based on the comorbidity. Overall our study demonstrates the potential contribution of the transcription factors and its regulatory molecules specific between the conditions. Our identified FOS with their regulating molecules under T2DM can provide prognostic information on periodontitiscomorbidity based on its specific regulatory patterns, which describes its crucial function based molecular function and pathways. However, there are a few things that need to be taken into consideration before drawing any conclusions about the study: 1) These regulators need to be verified by proteomic techniques; 2) This study contained a small number of gene expression samples. Additional studies with larger sample sizes are required to confirm the likelihood that these predicted regulators as prognostic biomarkers.
5 Conclusion
Combining experimental transcriptomic data from diabetes mellitus and periodontitis revealed potentially shared molecular regulators. FOS transcription factor has been identified which showed up-regulation in T2DM and periodontitis conditions. FOS follows diverse pattern of regulation that are leading to the multiple pathways that crucial to both the conditions. Hence FOS and its coregulators can act as prognostic moleculesand mayconsider as therapeutic targets.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Interdependence of metals and its binding proteins in parkinson’s disease for diagnosis. npj Parkinson’s Disease. 2021;7(1):3.
- [Google Scholar]
- Oestrogen receptor alpha, growth hormone receptor, and developmental defect of enamel. Int. J. Paediatr. Dent.. 2019;29(1):29-35.
- [Google Scholar]
- Blair, Meg. “Diabetes Mellitus Review.” Urologic nursing 36(1): 27–36.
- The BioGRID interaction database: 2015 update. Nucleic Acids Res.. 2015;43(D1):D470-D478.
- [Google Scholar]
- Estrogen receptor gene is associated with dental fluorosis in brazilian children. Clin. Oral Invest.. 2019;23(9):3565-3570.
- [Google Scholar]
- Inflammation: the link between insulin resistance, obesity and diabetes. Trends Immunol.. 2004;25(1):4-7.
- [Google Scholar]
- Microarray analysis after RNA Amplification can detect pronounced differences in gene expression using Limma. BMC Genomics. 2006;7(1):252.
- [Google Scholar]
- Affy–analysis of affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307-315.
- [Google Scholar]
- Periodontal disease and diabetes mellitus: a two-way relationship. Ann. Periodontol.. 1998;3(1):51-61.
- [Google Scholar]
- Molecular mechanisms of insulin resistance and the role of the adipocyte. Int. J. Obes. (Lond). 2000;24(S4):S23-S27.
- [Google Scholar]
- Isserlin, Ruth, Rashad A. El-Badrawi, and Gary D. Bader. 2011. “The Biomolecular Interaction Network Database in PSI-MI 2.5.” Database 2011.
- The IntAct molecular interaction database in 2012. Nucleic Acids Res.. 2012;40(D1):D841-D846.
- [Google Scholar]
- Human protein reference database–2009 update. Nucleic Acids Res.. 2009;37(Database):D767-D772.
- [Google Scholar]
- Severe periodontitis may influence cementum and dental pulp through inflammation, oxidative stress, and apoptosis. J. Periodontol.. 2019;90(11):1297-1306.
- [Google Scholar]
- MAPK usage in periodontal disease progression. J. Signal Transduction. 2012;2012:1-17.
- [Google Scholar]
- Mealey, Brian L., and Gloria L. Ocampo. 2007. “Diabetes Mellitus and Periodontal Disease.” Periodontology 2000 44(1): 127–53.
- Evidence of a substantial genetic basis for risk of adult periodontitis. J. Periodontol.. 2000;71(11):1699-1707.
- [Google Scholar]
- Vijayashree Priyadharsini, Jayaseelan. 2019. “In Silico Validation of the Non‐antibiotic Drugs Acetaminophen and Ibuprofen as Antibacterial Agents against Red Complex Pathogens.” Journal of Periodontology 90(12): 1441–48
- Interleukin-6 (IL-6) induces insulin resistance in 3T3-L1 adipocytes and Is, like IL-8 and tumor necrosis factor-α, overexpressed in human fat cells from insulin-resistant subjects. J. Biol. Chem.. 2003;278(46):45777-45784.
- [Google Scholar]
- The database of interacting proteins: 2004 update. Nucleic Acids Res.. 2004;32(90001):449D-D451.
- [Google Scholar]
- Relationship between estrogen receptor 1 gene polymorphisms and postmenopausal osteoporosis of the spine in Chinese women. Genet. Mol. Res.. 2016;15(2)
- [Google Scholar]
- Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.. 2003;13(11):2498-2504.
- [Google Scholar]
- Resolution of inflammation: a new paradigm for the pathogenesis of periodontal diseases. J. Dent. Res.. 2003;82(2):82-90.
- [Google Scholar]
- Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 Countries, 1990–2016: a systematic analysis for the global burden of disease study 2016. Lancet. 2017;390(10100):1211-1259.
- [Google Scholar]
- Correlation of estrogen receptor alpha gene polymorphisms and bone mineral density in Chinese women with chronic periodontitis. Chin. Med. J. (Engl.). 2010;123(22):3262-3327.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2022.102469.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







