Translate this page into:
Ganoderma multistipitatum sp. nov. from Chir pine tree (Pinus roxburghii Sarg.) in Pakistan
⁎Corresponding authors. ash.dr88@gmail.com (Aisha Umar), tajammalkhan5@gmail.com (Muhammad Tajammal Khan)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Background
Ganoderma species are in state of flux in Pakistan as well as in the world. The diversity of this wood-inhabiting basidiomycete in Pakistan is still poorly known. A few species have been introduced from the country; even the country is rich with many Ganoderma species, which still need identification.
Methods
Ganoderma multistipitatum sp. nov. is described from Lahore, Punjab Pakistan by using the morpho-anatomical (via naked eye and compound microscope) characters and molecular data from the ITS regions of DNA. The specimens used in this study were collected from the Botanical Garden of Government College University Lahore. The G. multistipitatum sp. nov. is, to date, only known from Lahore on Chir pine tree.
Results
Morphologically, this species is characterized by laccate orange, brown basidiomata, maroon, brown multi stipitate, and anatomically by ellipsoid basidiospores (10.24–11.2 × 5.3–5.4 µm) and trimitic hyphal system. The G. multistipitatum sp. nov. is 21st species from the genus Ganoderma known from Pakistan. This new species is also described with photographs, line drawings and compared with nearby taxa of the phylogenetic tree constructed by software MEGA10. This species resembled with typical G. lucidum, but the ITS DNA sequences of this species were much far away from G. multistipitatum sp. nov. ones.
Conclusion
The phylogeny based on ITS sequences supported this as a new distinct species, which finally compared with closely matrixed G. multipileum and related allies. The aim of this study was to introduce the new species from genus Ganoderma, which is a major contribution in biodiversity of the world.
Keywords
Ganodermataceae
New species
ITS
Morphology
Phylogeny
Pinus roxburghii
1 Introduction
Ganoderma P. Karst. is a cosmopolitan genus and causes root and basal stem rot diseases (da Silva Coelho-Moreira et al., 2018; Luangharn et al., 2019). Ganoderma is a basidiomycete genus, which belongs to the family Ganodermataceae (Richter et al., 2015; Tchotet Tchoumi et al., 2018). Chir pine (Pinus roxburghii) is widely planted for timber in its native area, being one of the most important trees. These trees are medicinal and found in Himalayan regions of Bhutan, Nepal, Kashmir, Sikkim, Tibet, and northern areas of Pakistan (Tripathi et al., 2008). This tree belongs to family Pinaceae commonly known as Chir Pine (Bissa et al., 2008). Pinus roxburghii has many ethnobotanical and medicinal uses (Kaushik et al., 2013).
Taxonomy of Ganoderma is polyphyletic based on pleomorphic morphology of basidiomata and geographical distribution (Wasser et al., 2006). Different phenetic plasticity of morpho-anatomical features hinder the correct identification of species (Hong et al., 2004). Morphological and molecular approaches have been adopting for the Ganoderma species delineation and identification. Sequences of ITS rDNA have been commonly analysed. This analysis proved efficient in delimiting and resolving the species within Ganoderma species complexes (Mukhtar, 2019). The ITS fungal barcode is highly conserved intra-specifically, but acts as a variable for different species used in taxonomy (Fryssouli et al., 2020). The difference of ITS regions less than 2% nucleotides is considered valuable for intraspecific variation in genus Ganoderma, which facilitate the separation of certain species (Badotti et al., 2017).
Regarding the advancement in taxonomy of Ganoderma species, many new records and species are still being discovered and discovering from Asia (Umar et al., 2021a,b; Gafforov et al., 2020; Mukhtar, 2019). This genus is poorly studied in many regions of Asia like Pakistan, from which only G. ahmadii Steyaert, G. lucidum (Curtis) P. Karst., G. applanatum (Pers.) Pat., G. flexipes Pat., G. resinaceum Boud., G. tornatum (Pers.) Bres., G. tsugae Murrill, G. australe (Fr.) Pat., G. boninense Pat., G. chalceum (Cooke) Steyaert, G. curtisii (Berk.) Murrill, G. lipsiense (Batsch) G.F. Atk., G. praelongum Murrill, G. multicornum Ryvarden, G. multiplicatum (Mont.) Pat., and T. colossus (Fr.) Murrill [as G. colossum (Fr.) C.F. Baker] are known (Irshad et al., 2012; Steyaert, 1972; Ahmad, 1972; 1956) without morphological description and molecular data, but these estimations solely relied on morphological criteria of identification. Recently G. leucocontextum (Umar et al., 2021a), G. multipileum Ding Hou., G. gibbosum (Blume & T. Nees) Pat. (Umar et al., 2022), and G. pakistanicum A. Umar (Umar et al., 2021b) are published with morpho-anatomical and molecular data basis. The description of this new species brings a major contribution in the diversity of genus Ganoderma explored from Pakistan. Therefore, a convincing update is needed in taxonomy of Ganoderma species from Pakistan with the support of morphological and molecular data.
In the past, discrepancies were observed in taxonomy and classification of genus Ganoderma, so the objective of this study is the identification of new Ganoderma sp. by ITS marker, which facilitates the correct identification of the specimens. The morpho-anatomical studies and phylogenetic tree supported this work in species identification. Here we explored and introduced the 21st Ganoderma species, which highly contributed to the biodiversity of genus Ganoderma.
2 Materials & Methods
2.1 Studied materials
The specimens studied here were collected in 2019 by the fourth author from Botanical Garden of Government College University, Lahore, Punjab, Pakistan (31°49′81″N 73°30′44″E), elevation 217 m a.s.l. This garden is also called “Secretariat of Pakistan Botanic Gardens Network” covered with the major plant groups. Pteridophytes (Nephrolepis, Pteris, Adiantum), Gymnosperms (Cycads), Pinus roxburghii, Ginkgo biloba, Cupressus, and many other important tree plants of Angiosperms. The garden is also enriched with native trees of Punjab (Amin, 2013). This Ganoderma species grows on Pinus roxburghii. The average annual rainfall of Lahore is 607 mm and the temperature 24°C (Shirazi and Kazmi, 2016).
2.2 Morphological examination
Microscopic structures observed from cross sections of the basidiome that were soaked in KOH (5%), stained with Congo red (1%), and viewed under a MX4300H compound light microscope. Data of anatomical features were recorded at a magnification of 100X. Basidiospores were presented as length × width (Nagy et al., 2010) and measured without taking into account the apical umbo when not shrunk. The morphological descriptions of the microscopic features were in part following Torres-Torres & Guzmán-Dávalos (Torres-Torres and Guzmán-Dávalos, 2012).
2.3 DNA extraction
Modified CTAB procedure was followed to extract total genomic DNA from dried specimens (Doyle and Doyle, 1987). The ITS1 + 5.8S + ITS2 rDNA region (henceforth referred to as the ITS region) was used to study the target specimens. This region was amplified by using primers ITS1 and ITS2 (White et al., 1990). Reaction mixtures (20 µl) contained 0.5 µl template DNA, 8.5 ml distilled water, 0.5 µl of each primer, and 10 ml PCR mix [DreamTaqGreen PCR Master Mix (2 X), Fermentas]. Amplification conditions were 35 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 1 min, followed by a final extension at 72 °C for 10 min. Amplified PCR products were purified and sequenced by TSINGKE Co. ltd. (China).
2.4 Data analysis by sequence alignment and molecular phylogeny
The ITS data set comprised DNA sequences of specimens from Pakistan and related species. The consensus of sequence was generated from both ITS1 and ITS2 in BioEdit vers. 7.2.5 (Hall, 1999) and then homology searches were performed at the NCBI web site using BLASTn. Additionally, ITS sequences of related species were downloaded from GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and literature. All sequences were automatically aligned with MAFFT and manually adjusted using BioEdit (Tamura et al., 2011). The phylogenetic tree was constructed MEGA10 software to run the ML (maximum likelihood phylogenetic analysis) with 1000 bootstrap replicates (Tamura et al., 2011); branches with less than 50% bootstrap support were collapsed to matrix the species in which Amaurodema rude selected as an outgroup. The consensuses of both sequences were finally deposited in GenBank for authenticity and reference.
3 Results
3.1 Phylogenetic inference
The ITS dataset enclosed 130 sequences of 47 taxa additionally two sequences of Ganoderma multistipitatum (GenBank No. ON032992, ON032991) and one from Amaurodema rude (outgroup), which was analyzed by maximum likelihood (ML) method. Our new species sequences have maximum query with un-described Ganoderma sp., during the initial blast. But in phylogeny, these un-described Ganoderma species of initial blast were matrixed away from the sequences of this new species. So, these species were removed in the final phylogenetic tree. Cut the ambiguous ends of the ITS regions. Reference sequences from GenBank of closely related species were also combined in phylogeny. The multiple sequence alignment was performed by an online program MAFFT (Katoh and Standley, 2013). The resulting alignment was manually edited in BioEdit (Tamura et al., 2011), and the evolutionary model for nucleotides that fit best in data sets were selected.
Since the tree topologies resulting from the ML analyses is presented, along with the statistical bootstrap values (99%) (Fig. 1). This group formed a clade nearby Ganoderma multipileum, G. martinicense, and G. parvulum. In this study, novel species of Ganoderma found on Pinus roxburghii was described on the appearance of basidiome shape, pileus surface and color along with molecular phylogeny.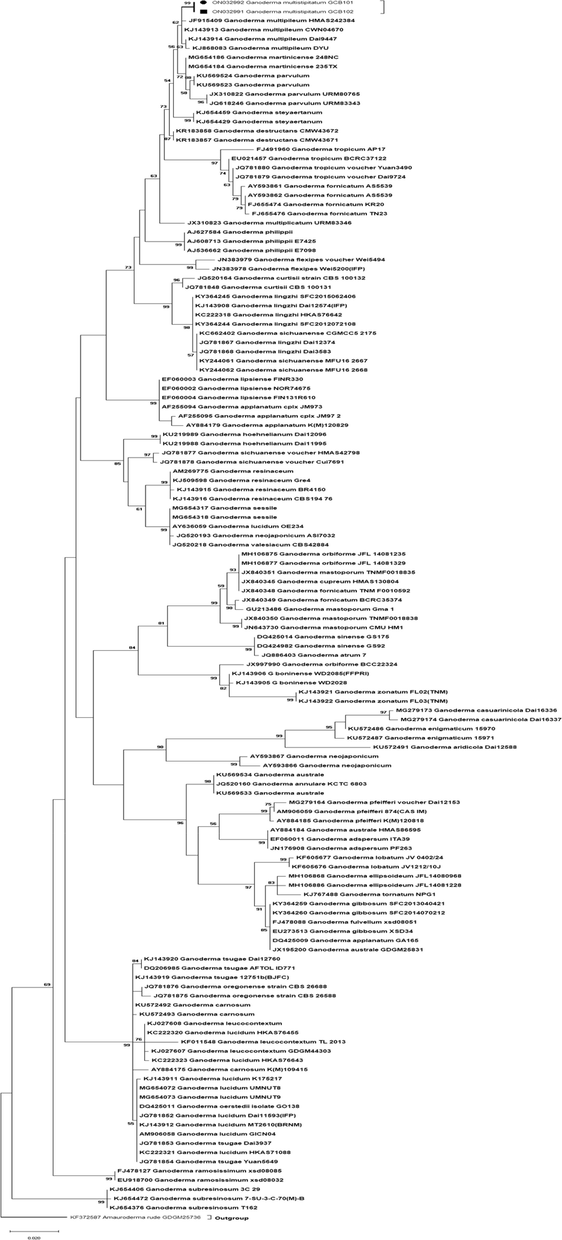
Phylogenetic tree of Ganoderma multistipitatum (ITS rDNA sequences generated by maximum likelihood). Outgroup was Amauroderma rude. Bootstrap values (>50 %) are shown at the branches (New species represented by black dot and box).
3.2 Taxonomy of Ganoderma multistipitatum A. Umar, sp. nov.
3.2.1 Diagnosis:
Morphologically, Ganoderma multistipitatum sp. nov. resemble the species in subgenus Ganoderma P. Karst. It is characterized by annual, dimidiate and broadly orange brown laccate basidiome. The unique character of this species is the group of stout stipes give rise to one basidiome. Context was thick with white resinous melanoid bands and white greyish growth zones. Trimitic hyphal system and basidiospores are ellipsoid, guttulated, bitunicate, brown, and with sub free minute inter-walled pillers (Figs. 2-4).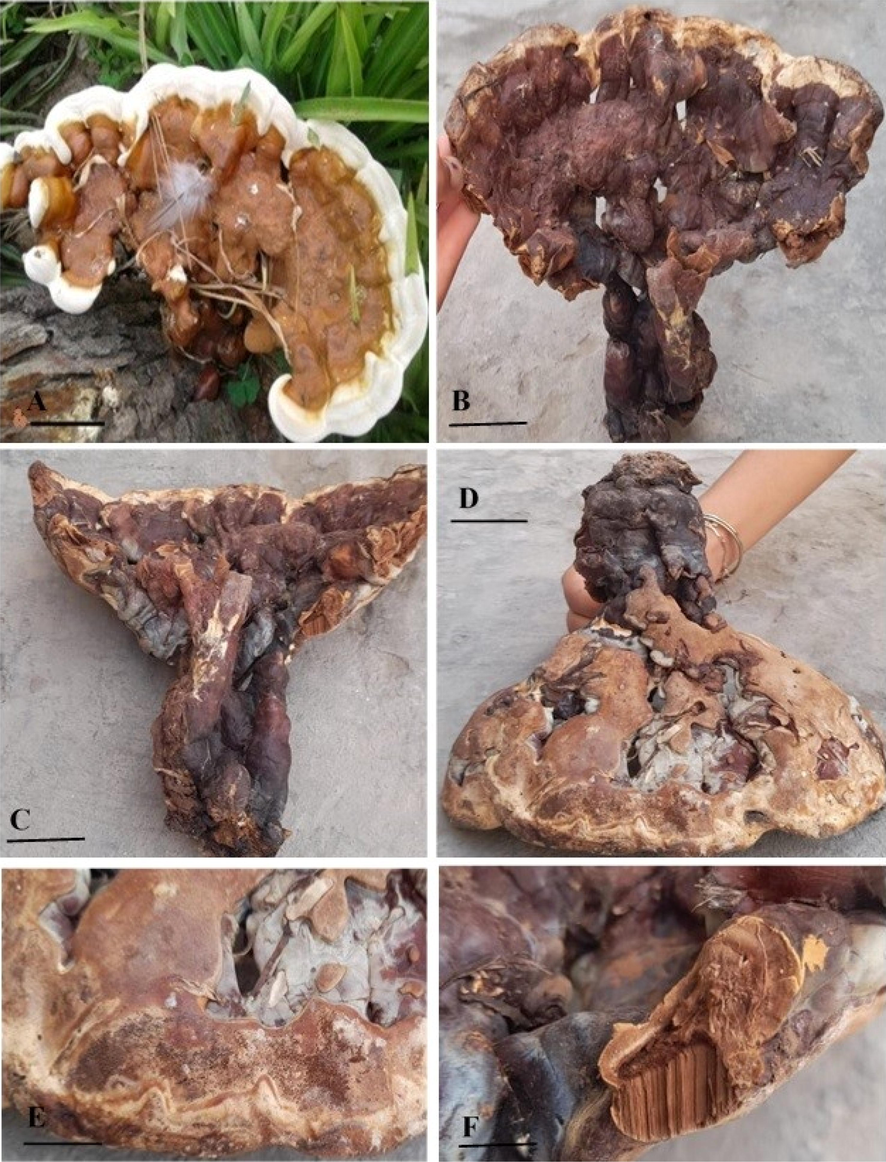
Macroscopic structures of Ganoderma multistipitatum (A–D GCB101). A–B. Basidiomata. C. Group of stipes. D. Lower view of basidiome. E. Pores surface. F. Section of context and tubes. Bars = 2 cm (A–D), 2 mm (E, F). (Photo by Aisha Umar).
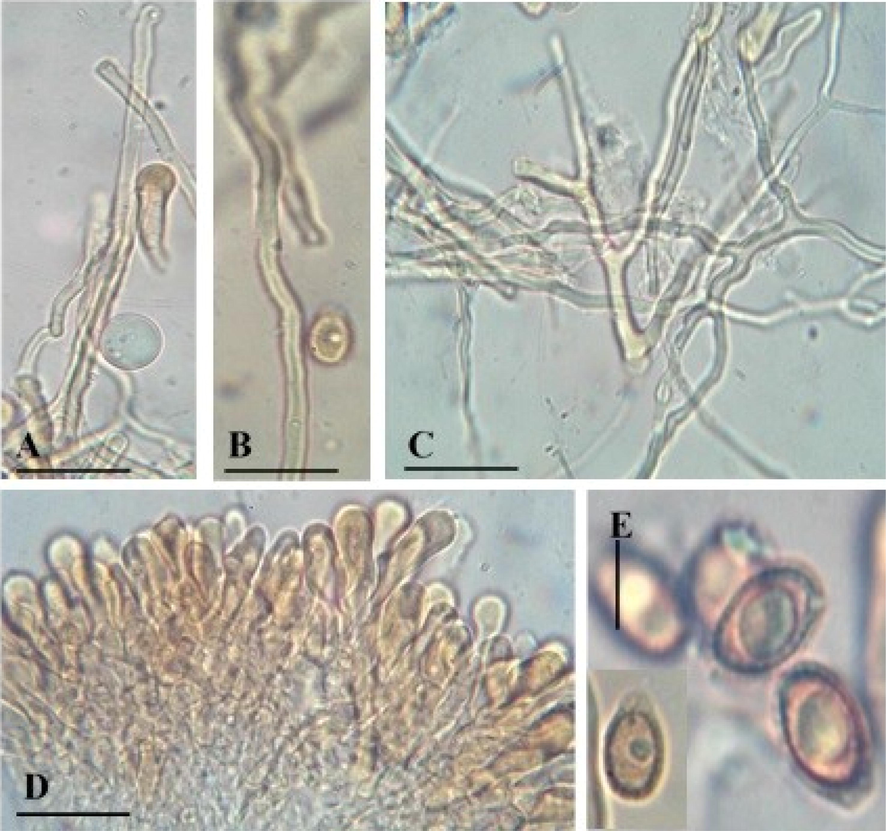
Microscopic structures of Ganoderma multistipitatum (A–E GCB101). A-C. Hyphae (A. Generative), (B. Skeletal), (C. Binding). D. Cells of crustohymeniderm. E. Basidiospores. Bars = 5 μm (A–C), 10 μm (D–E).
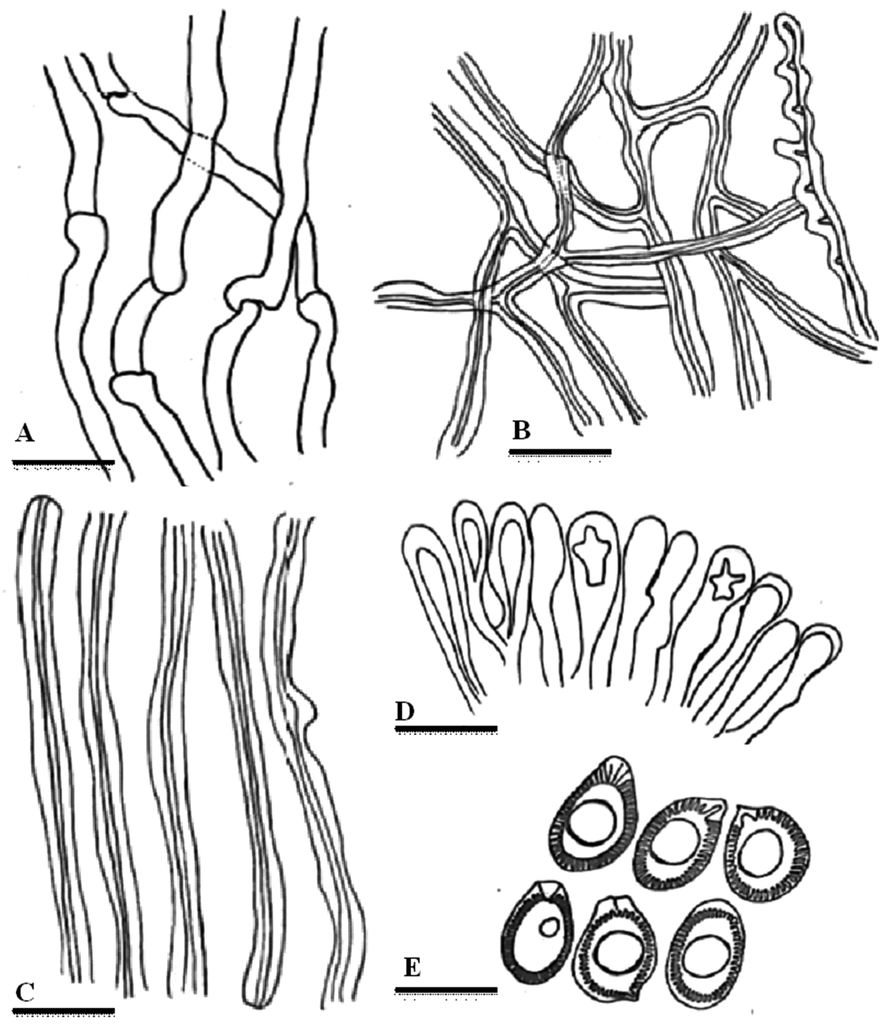
Line drawings of anatomical characters of Ganoderma multistipitatum. A. Generative hyphae. B. Binding hyphae. C. Skeletal hyphae. D. Cells of crustohymeniderm. E. Basidiospores. Bars = 5 μm (A–C), 10 μm (D–E). (Drawen by Aisha Umar)
3.2.2 Holotypus
PAKISTAN. PUNJAB PROVINCE: Lahore, GC University, Lahore, Punjab, Pakistan, (31.4981° N 73.3044° E), elevation 217 m a.s.l, attached to trunk of Pinus roxburghii, 15 July 2018, Aisha Umar, (holotype GCB101). GenBank: ITS = ON032992.
3.2.3 Etymology
The species epithet refers to “multistipitatum” represents the “group of stipes” give rise to one basidiome.
3.3 Morphological description
Basidiomata stipitate, verrucose, laccate cinnamon orange brown; Stipe 8.5–8.7 × 5.9–6.2, multistipitate, laccate, maroon brown to blackish brown, soft with hard crust; Margin 1.5–1.6 cm, broader, thick, prominent milky white, obtuse to incurved; Pileus 24–26 × 10–11 cm, 0.5–0.6 mm thickness, glossy, bumps, protuberances, reniform, dimidiate to flabelliform, radially wrinkled, soft flesh with hard crust; Pores 120–153 × 223–255 µm, white (fresh) to light brown (bruised), subcircular; “Cutis/palisadoderm 25.7–32.9 × 7.59–6.9 µm, thick-walled, clavate, palisade elements, yellowish brown”; Tubes 0.9–1.0 mm long, non-stratified, light brown; Context 1.4–1.6 cm long, light brown, dry, fibrous, soft, melanoid bands, white greyish growth zones; ‘Basidiospores 10.24–11.2 × 5.3–5.4 µm, ellipsoid, bitunicate, exosporium hyaline, pigmented, thick, green, inner episporium, echinulae green eusporium, guttulated, highly thick inter-walled pillars, apically truncate, tapering myxosporium end’; ‘Hyphal System 1) generative hyphae (septate, clamped, colorless, thin-walled), 2) skeletal hyphae (thick-walled, colorless, unbranched or few branches with distal end), 3) binding hyphae (arboriform, brown, thick-walled, much-branched’).
3.4 Ecology and habitat
End of June to start of August, this species occurs on trunk of Chir tree forest of Punjab Province.
3.5 Additional specimens examined
PAKISTAN. PUNJAB PROVINCE: Lahore, (31.4981″N 73.3044″E), elevation 217 m a.s.l, attached to a tree trunk of Pinus roxburghii, 25 July 2019, Aisha Umar, (paratype GCB102). GenBank: ITS = ON032991.
4 Discussion
Ganoderma multistipitatum sp. nov. characterized by annual multistipitate, bumps (protuberances), dimidiate to flabelliform, radially wrinkled, corky thick and shiny basidiome with very thick white margins. The light brown context with white melanoid band and trimitic hyphal system was also observed in this species.
This new species was closely related to lowland Taiwan and Chinese G. multipileum (Fig. 1). The main distinguishing features of G. multipileum also observed in Pakistani G. multistipitatum sp. nov. specimens are stalked basidiomata, thin crust, truncate ovoid basidiospores (8.2–9.4 × 4.7 μm) (Wang et al., 2009), with minute inter-walled pillars, heterogeneous context (clay buff to fulvous) with melanoid bands, and concentric growth zones on basidiomata. Enlarged bulbous hyphal ends were observed by Cao et al. (2012) in G. multipileum, but not observed in the specimens of this study. Following Chang (1983), the features of stalked basidiomata and pilei growing together of G. multipileum are not observed in G. multistipitatum sp. nov., whereas group of stipes are useful for the circumscription of this new specimens.
The other closest species of G. multistipitatum sp. nov. in phylogenetic tree of this study was G. martinicense, G. parvulum and G. tropicum (Fig. 1). G. martinicense is relative to G. multipileum, while geographically distant species only known from America (Loyd et al., 2018). G. martinicense has similarity with G. multistipitatum sp. nov. in melanoid bands and concentric growth zones in dark brown context (light brown in our specimens), dark red to black pseudostipe (group of stout stipes in our specimens), and longer coarse ornamented basidiospores (Welti and Courtecuisse, 2010). The G. martinicense has golden yellow pileal surface, whereas light sugar cane brown to brown or maroon, brown pileal surface found in the specimens of this study.
Ganoderma parvulum Murrill is morphologically very distinct from G. multipileum (Correia de Lima Júnior et al., 2014) and G. multistipitatum sp. nov. The G. parvulum is known from North and South America, possess light to pale context with thick black resinous bands contrary to Pakistani new species (Correia de Lima Júnior et al., 2014; Cabarroi-Hernández et al., 2019). G. parvulum was synonym of G. stipitatum (Gottlieb and Wright, 1999), because both are same species (Ryvarden, 2004), but preference gives to G. parvulum on G. stipitatum (Murrill) Murrill, as described earlier. The basidiospores size of Brazilian G. parvulum and G. stipitatum is similar but smaller (8–10 × 5–6 μm) than our new species. Free to sub free and very thin pillars in endosporium are found in G. parvulum and G. multistipitatum sp. nov. (Cabarroi-Hernández et al., 2019). The black thick resinose band is present in context of G. parvulum, while white band present in this new species. The clavate shape of cuticular cells are similar in both G. parvulum and G. multistipitatum sp. nov.
Ganoderma tropicum from tropical and subtropical areas of Asia is dissimilar to G. multistipitatum sp. nov. in morphology, because this species has concentric growth zones on the pileus, while absent in this species (Zhou et al., 2015). Both species has melanoid bands in the contextum. The contextum is homogeneous and fulvous in G. tropicum and G. multistipitatum sp. nov. The cuticular cells are mostly clavate, irregular, often with blunt protuberances or outgrowths in G. tropicum, whereas no growth found in G. multistipitatum sp. nov. The basidiospores are coarsely echinulate, ellipsoid to broadly ellipsoid and smaller (8.8–10.7 × 5.5–6.3 µm) than our new Ganoderma species (Zhou et al., 2015).
Careful observations on morpho-anatomy differentiated the Ganoderma multistipitatum sp. nov. from G. multipileum, G. tropicum, G. parvulum, and G. martinicense. Furthermore, these are all phylogenetically distant from the specimens of this study.
5 Conclusion
Ganoderma multistipitatum sp. nov. represents the 21st Ganoderma species (accession numbers ON032992, ON032991) described from Pakistan. The wood-inhabiting basidiomycete diversity is still poorly known in Pakistan. A systematic survey of polypores in Pakistan is strongly needed. Taken the special geographic position of this region into consideration, this kind of survey improves the knowledge about wood-inhabiting polypore diversity of the world.
Acknowledgment
We would like to pay a special thanks to the director of Institute of Botany, University of the Punjab, Lahore Pakistan, who allowed to do this work and facilitated in many ways. The authors extend their appreciation to the Researchers Supporting Project number (RSP2023R193), King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Ahmad, S., 1956. Fungi of Pakistan, mon. 1. Lahore, Biological Laboratories, Government College.
- Ahmad, S., 1972. Basidiomycetes of west Pakistan, mon. 6. Lahore, Biological Laboratories, Government College.
- Guide to botanic garden G.C (Third edition.). University Lahore; 2013.
- Effectiveness of ITS and sub-regions as DNA barcode markers for the identification of Basidiomycota (Fungi) BMC Microbiol.. 2017;17(1):1-2.
- [Google Scholar]
- Antibacterial potential of three naked-seeded (Gymnosperm) plants. Nat. Prod. Res.. 2008;7(5):420-425.
- [Google Scholar]
- The Ganoderma weberianum–resinaceum lineage: multilocus phylogenetic analysis and morphology confirm G. mexicanum and G. parvulum in the Neotropics. Mycokeys.. 2019;59:95-131.
- [Google Scholar]
- Cao, Y., Wu, S.H., Dai, Y.C., 2012. Species clarification of the prize medicinal Ganoderma mushroom “Lingzhi”. Fungal Divers. 56, 49–62. https://doi: 10.1007/s13225-012-0178-5.
- Studies on biology of several species of Ganoderma in Taiwan [Master’s thesis]. Taipei: National Taiwan University; 1983. p. :130.
- Delimitation of some neotropical laccate Ganoderma (Ganodermataceae): molecular phylogeny and morphology. Rev de Biol Trop.. 2014;62:1197-1208.
- [Google Scholar]
- Evaluation of diuron tolerance and biotransformation by the white-rot fungus Ganoderma lucidum. Fungal boil.. 2018;122(6):471-478.
- [Google Scholar]
- A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull.. 1987;19:11-15.
- [Google Scholar]
- A global meta-analysis of ITS rDNA sequences from material belonging to the genus Ganoderma (Basidiomycota, Polyporales) including new data from selected taxa. MycoKeys.. 2020;75:1-143.
- [CrossRef] [Google Scholar]
- Species diversity with comprehensive annotations of wood-inhabiting poroid and corticioid fungi in Uzbekistan. Front Microbiol.. 2020;3047
- [Google Scholar]
- Gottlieb, A.M., Wright, J.E., 1999. Taxonomy of Ganoderma from southern South America: subgenus Ganoderma. Mycol Res. 103, 661–673. https://dx.doi.org/10.1017/S0953756298007941.
- BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser.. 1999;41:95-98.
- [Google Scholar]
- Phylogenetic analysis of Ganoderma based on nearly complete mitochondrial small-subunit ribosomal DNA sequences. Mycologia.. 2004;96(4):742-755.
- [Google Scholar]
- Purification and characterization of α-amylase from Ganoderma tsuage growing in waste bread medium. African J. Biotech.. 2012;11:8288-8294.
- [CrossRef] [Google Scholar]
- MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol.. 2013;30:772-780.
- [Google Scholar]
- Ethnobotany and phytopharmacology of Pinus roxburghii Sargent: a plant review. J. Integr. Med.. 2013;11(6):371-376.
- [Google Scholar]
- Elucidating “lucidum”: Distinguishing the diverse laccate Ganoderma species of the United States. PloS One.. 2018;13(7):e0199738.
- [Google Scholar]
- Additions to the knowledge of Ganoderma in Thailand: Ganoderma casuarinicola, a new record; and Ganoderma thailandicum sp. nov. MycoKeys.. 2019;59:47.
- [Google Scholar]
- Morphological characterization of Ganoderma species from Murree Hills of Pakistan. Plant Prot.. 2019;3(2):73-84.
- [Google Scholar]
- Type studies and nomenclatural revisions in Parasola (Psathyrellaceae) and related taxa. Mycotaxon. 2010;112:103-141.
- [Google Scholar]
- An assessment of the taxonomy and chemotaxonomy of Ganoderma. Fungal Divers.. 2015;71(1):1-15.
- [Google Scholar]
- Ryvarden, L., 2004. Neotropical polypores: Part 1: Introduction, Ganodermataceae & Hymenochaetaceae. Fungiflora.
- Analysis of socio-environmental impacts of the loss of urban trees and vegetation in Lahore, Pakistan: a review of public perception. Ecol. Process.. 2016;5(1):1-2.
- [Google Scholar]
- Species of Ganoderma and related genera mainly of the Bogor and Leiden Herbaria. Persoonia.. 1972;7:55-118.
- [Google Scholar]
- MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol.. 2011;28:2731-2739.
- [CrossRef] [Google Scholar]
- Three Ganoderma species, including Ganoderma dunense sp. nov., associated with dying Acacia cyclops trees in South Africa. Australas. Plant Pathol.. 2018;47:431-447.
- [CrossRef] [Google Scholar]
- The morphology of Ganoderma species with a laccate surface. Mycotaxon.. 2012;119:201-216.
- [CrossRef] [Google Scholar]
- Gymnosperms of Nainital. Nainital: Kumaun University; 2008.
- Ganoderma leucocontextum, a new record from Pakistan. Mycotaxon.. 2021;136(2):529-539.
- [Google Scholar]
- Ganoderma pakistanicum sp. nov. (Ganodermataceae, Basidiomycota) from Pakistan. Nova Hedwigia.. 2021;113(3–40):531-543.
- [CrossRef] [Google Scholar]
- Ganoderma multipileum and Tomophagus cattienensis—new records from Pakistan. Mycotaxon.. 2022;137(1):135-151.
- [CrossRef] [Google Scholar]
- Ganoderma multipileum, the correct name for ‘G. lucidum’ in tropical Asia. Bot Stud. 2009;50:451-458.
- [Google Scholar]
- Morphological traits of Ganoderma lucidum complex highlighting G. tsugae var. jannieae: the current generalization. ARA Gantner Verlag, Ruggell; 2006. p. :1-187.
- The Ganodermataceae in the French West Indies (Guadeloupe and Martinique) Fungal Divers.. 2010;43:103-126.
- [CrossRef] [Google Scholar]
- Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenies. PCR protocols, a guide to methods and applications.. 1990;18(1):315-322.
- [Google Scholar]
- Global diversity of the Ganoderma lucidum complex (Ganodermataceae, Polyporales) inferred from morphology and multilocus phylogeny. Phytochemistry.. 2015;114:7-15.
- [Google Scholar]







