Translate this page into:
Expression profiling of DUF599 genes revealed their role in regulating abiotic stress response in solanum tuberosum
⁎Corresponding authors. xzhuang@szu.edu.cn (Xiazi Huang), sfli@szu.edu.cn (Shuangfei Li)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The proteins containing the domain of unknown functions (DUFs) have significant roles in stress response and the growth of plants. A comprehensive genome-wide analysis and expression profiling of DUF599 was conducted to identify their roles in potato growth and response to stressed conditions. A total of nine DUF599 genes were identified, located on five chromosomes in the potato genome. The phylogenetic analysis divided StDUF599 genes into three groups in accordance with gene conserved motifs and gene structure distribution patterns. The StDUF599 promoters comprised several cis-acting factors responsive to plant hormones and abiotic stresses. The present study revealed that StDUF599 genes also possessed MBS, LTR, ABRE, and anaerobic induction responsive elements, indicating their importance in coping with abiotic stresses. StDUF599 genes were the target of several families of micro-RNAs also identified in this study. Purifying selection pressures lead to the duplication of the StDUF599 genes. Expression analysis of StDUF599-6 and StDUF599-9 in various tissues illustrated their vital role in developmental processes. It was found that StDUF599-7 and StDUF599-9 were highly expressed against heat and salt stresses. Expression profiling revealed that the StDUF599-8 gene has a significant role against GA3 and IAA. Lastly, this article forecasted that the DUF599 genes could enhance plant tolerance against several abiotic stresses.
Keywords
Abiotic stress
Duplication
Gene expression
miRNAs
Phylogeny
Phytohormones
1 Introduction
Several abiotic stress factors negatively affect plant growth and decrease yield (Sade et al., 2018). For instance, environmental deformations cause 70 % yield losses annually (Zaynab et al., 2020). Salinity, temperature, and drought are the significant abiotic stress factors influencing the geographical privilege of plants, curbing crop production, and menacing food safety. In general, plants do not possess structures that are directly involved in discerning environmental changes but could show responses to environmental aberrations (Hamant and Haswell, 2017). Under stress conditions, plants demonstrate comprehensive defence retributions at cellular and molecular levels to counter cell damage (Kim et al., 2014). Easy accessibility to plant genomes empowers us to characterize and identify several gene families against abiotic stress factors (Zaynab et al., 2021b). Various members of these families have been well recognized in plant genomes, with a reasonable number of them have been functionally characterized, including NAC, bHLH, MAPK, bZIP and HSF transcription factor families (Waseem et al., 2021). Moreover, plant genomes possess numerous genes expressed against stresses, comprise highly conserved domains, and carry out crucial tasks in different metabolic processes of plants during stress. Genes possessing these potential hypothetical domains are recognized as the domain of unknown functions (DUFs) (Zaynab et al., 2021c). Researchers have reported that DUF family members perform an imperative role against abiotic stress factors in different plants (Zhao et al., 2021). They reported different gene families, such as DUF936, DUF810, DUF1618, DUF866, and DUF221 in rice and gene families DUF724 and DUF581 in Arabidopsis have a significant role against abiotic stress (Waseem et al., 2021).
Specific genes were also linked to abiotic stresses, including OsDUF810, OsDSR2, and SIDP361, which have been regulated through dehydration and nutritional status during the developmental stage. The reduction in MsDUF gene expression in Medicago sativa was noticed when plants were treated with NaCl, abscisic acid, gibberellic acid, and PEG6000, indicating its negative regulatory role against stress responses (Albornos et al., 2017). The wild-type Arabidopsis contains the TaSRHP gene that possessed the DUF581 domain, induced during salinity stress. Enhanced TaSRHP gene expression illustrated resistance to drought and salinity. Likewise, enhanced expression of CiDUF4228-3 in the genome of Caragana intermedia enabled plants to maintain their normal growth under low temperature, dehydration, and drought, suggesting its critical tasks under stressed conditions (Leng et al., 2021). A previous Arabidopsis study exhibited that the ESK1 gene (AT3g55990), a DUF231 family member, works as a pessimistic regulator for cold acclimatization in plants (Xin et al., 2007). The OsSIDP366 gene (DUF1644) significantly regulates the retaliation against drought and salt stress factors in rice. Moreover, the transgenic rice overexpression of the OsSIDP366 gene shows tolerance against drought and salt (Li et al., 2016). In addition, the competent characterization of DUF genes regarding tolerance against various stresses is presently delineated in model plants only. In contrast, extensive investigations on the DUF family members in different plant species have seldom been reported. This indicates that genes comprising DUF-domain may directly or indirectly participate in plant tolerance mechanisms.
Potato is an imperative economic crop, extensively utilized as food almost everywhere (Zaynab et al., 2021a). However, potato yield is also affected by various environmental stresses (Zaynab et al., 2021c). Therefore, to understand the function of DUFs, we executed a detailed study of the DUF599 gene members in the potato genome. Further, we investigate the structure of genes, their location on chromosomes, phylogenetic relationships of genes, conserved domains of proteins, and gene expression profiling in various tissues. Moreover, we analyzed the expression of StDUF599 genes in response to heat, salinity, and several hormonal stresses. The present study has the ability to dispense a base for functional ratification of these DUF599 genes and their functions in the growth and developmental mechanisms of potato plants under stressed environments.
2 Materials and methods
2.1 Identification of DUF599 genes in S. tuberosum
For the identification of DUF599 genes in the S. tuberosum genome, the HMM (Hidden Markov Model) and BLASTP methods were used. Thus, the potato (S. tuberosum) genome was retrieved from the Phytozome 12.0 (http://phytozome.jgi.doe.gov) database (Goodstein et al., 2012). In addition, the amino acid sequences of 9 AtDUF599 genes were attained from TAIR (https://www.arabidopsis.org/) (Goodstein et al., 2012; Rhee, 2003) database. The 9 AtDUF599 amino acid sequence data were utilized for the BLASTP query in the potato genome. In addition, the HMMER 3.1 program (http://www.hmmer.org/) (Finn et al., 2015) was utilized to find out the DUF599 genes with standard limitations. The HMM files with Pfam ID (PF04654) were attained through the Pfam site (http://pfam.xfam.org/) protein domain dataset (El-Gebali et al., 2019). Finally, nine StDUF599 gene members were identified after using two procedures in the potato genome (Supplementary Table S1). Similarly, six DUF599 genes were also found in Z. mays using the same process. Moreover, its genome sequences were attained using Phytozome JGI 12.0 database (http://phytozome.jgi.doe.gov). The physico-chemical properties were resolute by ProtParam (https://web.expasy.org/protparam) (Gasteiger et al., 2005). Therefore, the StDUF599 proteins' subcellular localizations have been forecasted through CELLO v2.5 (http://cello.life.nctu.edu.tw/). We used the TBtools software (V1.068; https://github.com/CJ-Chen/TBtools) to determine the StDUF599 genes structure. Moreover, StDUF599 proteins conserved motifs were identified using the MEME (https://memesuite.org/meme/db/motifs) website with the following parameters; such as motifs number = 10 (Bailey et al., 2009).
2.2 Phylogenetic Tree, Synteny, chromosomal location, and selection pressure analysis
A phylogenetic tree was constructed among Z. mays, A. thaliana, and S. tuberosum protein sequences to study the evolutionary connection of StDUF599 genes. This study utilized MEGA 7 software (https://megasoftware.net/home) (Kumar et al., 2018) for sequence alignment. The neighbor-joining method was used to build a phylogenetic tree with 1,000 bootstrap repetitions. Moreover, the iTOL program (https://itol.embl.de/) was used to beautify the tree. In addition, the synteny relationships among Z. mays, A. thaliana, and S. tuberosum was executed through the Circoletto software tool (http://tools.bat.infspire.org/circoletto/) (Chen et al., 2020). The data associated with the chromosomal location were obtained through the GFF (general feature format) file of the potato genome (Potato Genome Sequencing Consortium (PGSC) Ref seq v4.03) (Potato Genome Sequencing Consortium, 2011). MEGA 7 software was used to align coding sequences for the duplicated genes. The Ka/Ks value of StDUF599 duplicated genes was estimated employing the KaKs calculator 2.0 tool (https://sourceforge.net/projects/kakscalculator2/). Additionally, divergence time (t) = Ks/2r was calculated with r = 2.6x10−9 (Li et al., 2019).
2.3 cis-regulatory elements and miRNA prediction analysis
For the investigation of the cis-elements in the StDUF599 gene promoters, the 2 Kb upstream regions from the start codon were extracted. Moreover, the PlantCARE database site (http://bioinfermatics.psb.ugent.be/webtools/plantcare/html/) was used to study the cis-factors of every promoter and figured out through TBtools (Bailey et al., 2009; Lescot, 2002). The psRNATarget database (http://plantgrn.noble.org/psRNATarget/) (Dai et al., 2018) with default parameters was utilized to predict miRNAs. The figure of the interaction network between the StDUF599 genes and miRNAs was created through the Cytoscape tool (v3.8.2; https://cytoscape.org/download.html).
2.4 Expression profiling of StDUF599 genes
The StDUF599 gene expression of different tissues was obtained from the publicly accessible source with BioProject ID: PRJEB2430. The transcriptome data of StDUF599 genes under abiotic (heat and salt) and hormones (abscisic acid, Gibberellic acid, and indole-3- acetic acid) stress were accessed from NCBI with accession number SRA029323. The expression of tissues (stem, flower, stolon, shoot apex, leaf, petiole, young tuber, tuber cortex, stamen, root, tuber pith, tuber peel, tuber cortex, tuber sprout, mature tuber, and whole in-vitro plant) and abiotic (heat and salt) stress treatments are depicted with details in Supplementary Table S5. Abiotic stress conditions were carried out by 24 h treatment of plants with heat (35 °C) and salt (50 mM NaCl). The hormone stress response of plants was induced by treatment through ABA (abscisic acid, 50 µM), IAA (indole-3-acetic acid, 10 µM), and GA3 (gibberellic acid, 50 μM) for 24 h. The abundance of transcripts was computed through FPKM (Fragment per kilobase million).
3 Results
3.1 Identification and characterization of StDUF599 genes
BLASTP and HMM methods were utilized in the present study to identify DUF599 genes in the S. tuberosum genome. Finally, nine DUF599 genes with Pfam ID (PF04654) were identified after using two methods. These genes were named “StDUF599-1 to StDUF599-9″, and Table 1 shows details of all nine StDUF599 genes. The length of the amino acid (AA), MW (molecular weight), pI (isoelectric point), subcellular localization, and exon–intron numbers were investigated in this analysis. The amino acid numbers for StDUF599 proteins varied from 124 (StDUF599-7) to 252 (StDUF599-5); isoelectric point values were in the range of 5.62 (StDUF599-8) to 9.54 (StDUF599-6); while their molecular weights varied from 14.61 (StDUF599-7) to 28.50 kDa (StDUF599-5) (Table 1). The number of exons varied from one (StDUF599-4, and StDUF599-7) to two (StDUF599-1, StDUF599-2, StDUF599-3, StDUF599-5, StDUF599-6, StDUF599-8, and StDUF599-9) and number of introns were from zero (StDUF599-4 and StDUF599-7) to one (StDUF599-1, StDUF599-2, StDUF599-3, StDUF599-5, StDUF599-6, StDUF599-8, and StDUF599-9). Furthermore, only two genes lack introns, and seven others have one intron. The subcellular localization results showed that all StDUF599 proteins were present on the cell membrane (Table 1). Meanwhile, nine genes in A. thaliana (AtDUF599-1 to AtDUF599-9) and six in Z. mays (ZmDUF599-1 to ZmDUF599-6) were also identified in this study (Supplementary Table. S1).
Gene Name
Transcript Name
Chromosome Name
Renamed
Strand
Gene Start (bp)
Gene End (bp)
Protein (aa)
MW
pI
Subcellular localization
PGSC0003DMG400031770
PGSC0003DMT400081285
ST4.03ch02
StDUF599-1
1
33132572
33133877
240
27770.48
8.48
Plasma Membrane
PGSC0003DMG400000549
PGSC0003DMT400001484
ST4.03ch03
StDUF599-2
−1
46654461
46655258
237
26931.61
9.08
Plasma Membrane
PGSC0003DMG400000550
PGSC0003DMT400001485
ST4.03ch03
StDUF599-3
−1
46651719
46652492
233
26353.03
8.6
Plasma Membrane
PGSC0003DMG400012818
PGSC0003DMT400033369
ST4.03ch03
StDUF599-4
−1
1726382
1726777
131
15127.81
9.18
Plasma Membrane
PGSC0003DMG400014849
PGSC0003DMT400038472
ST4.03ch08
StDUF599-5
−1
2573534
2578039
252
28507.56
8.53
Plasma Membrane
PGSC0003DMG400031231
PGSC0003DMT400080235
ST4.03ch10
StDUF599-6
−1
48797620
48799064
226
25131.57
9.54
Plasma Membrane
PGSC0003DMG400034106
PGSC0003DMT400084511
ST4.03ch12
StDUF599-7
−1
39706885
39707259
124
14611.25
9.02
Plasma Membrane
PGSC0003DMG400029374
PGSC0003DMT400075537
ST4.03ch12
StDUF599-8
1
58025470
58028813
188
20888.59
5.62
Plasma Membrane
PGSC0003DMG400015823
PGSC0003DMT400040908
ST4.03ch12
StDUF599-9
1
40211451
40213089
234
26819.43
9.01
Plasma Membrane
3.2 Phylogenetic relationship of DUF599 genes
The evolutionary relationships among ZmDUF599, AtDUF599, and StDUF599 genes were observed by constructing a phylogenetic tree. The multiple sequence alignment was done by DUF599 protein sequences from A. thaliana, Z. mays, and S. tuberosum (Fig. 1). In the present study results, DUF599 genes of 3 plants (Z. mays, A. thaliana, and S. tuberosum) were grouped into three major clades I, II, and III (Fig. 1). The DUF599 genes from three species were spotted in all three clades indicating that DUF599 gene members were distinguished prior to the partition of their ancestral plant species. The results showed that group I comprised 4 DUF599 family members (1 AtDUF599, 1 ZmDUF599, and 2 StDUF599). Group II comprised 10 DUF599 members (3 AtDUF599, 3 ZmDUF599, and 4 StDUF599). Group III contains 10 DUF599 members (5 AtDUF599, 2 ZmDUF599, and 3 StDUF599) (Fig. 1; Supplementary Table S1). The DUF599 genes clustered in the identical group might continue to perform identical functions. In the present study results, StDUF599 distributed in groups II and III possessed a higher number of StDUF599. The findings revealed that groups II and III have ten DUF599 members each.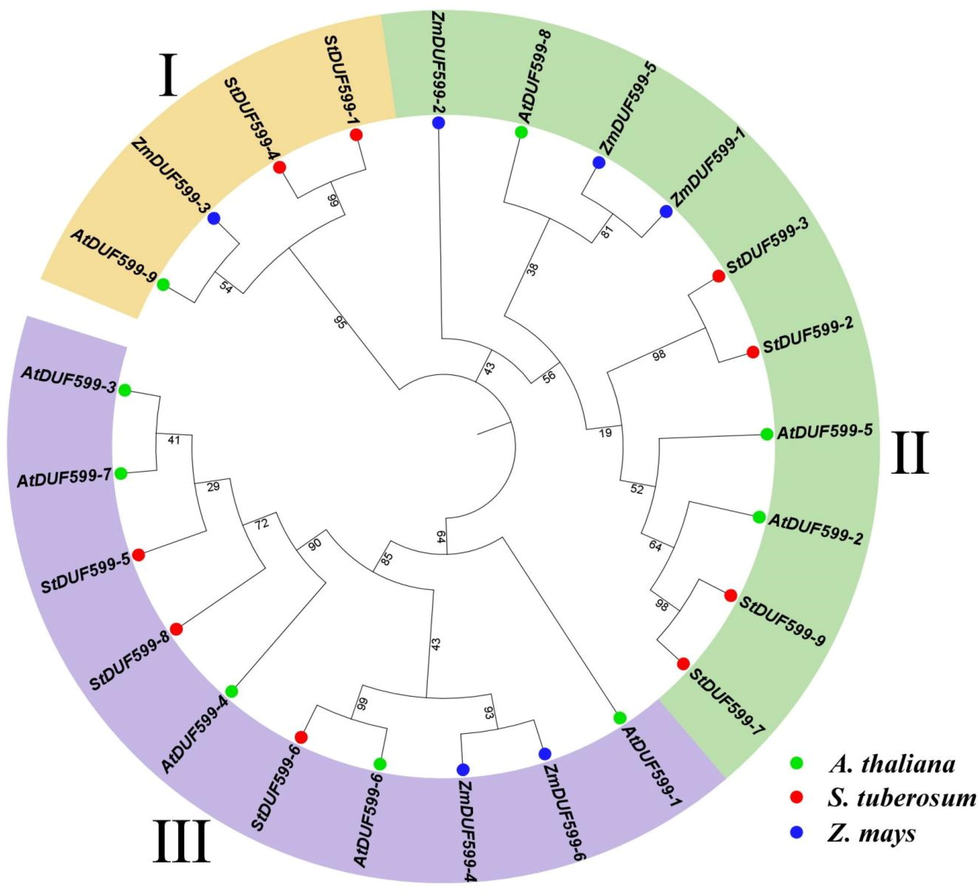
A neighbor-joining phylogenetic tree assessment of DUF599 genes from A. thaliana, Zea mays, and S. tuberosum. Overall, 9 AtDUF599 from A. thaliana (green circles), 6 ZmDUF599 from Z. mays (blue circles), and 9 StDUF599 from S. tuberosum (red circles) were clustered into three major classes, denoted by exclusive colors.
3.3 cis-acting elements analysis and chromosomal locations
The cis-acting elements control gene expression and functions. We investigated cis-acting elements in StDUF599 gene promoters to learn about their regulatory and functional activities. Supplementary Table S2 contains all the information on StDUF599 promoter cis-regulatory elements. We found that the TCA- element, CGTCA-motif, ABRE, TGACG-motifs, P-box, TATC-box, and GARE-motif are the most enriched elements of the four plant hormones, including salicylic acid, methyl jasmonate, gibberellins and abscisic acid (Supplementary Table S2). Furthermore, several phytohormone-correlated components were widely distributed, emphasizing the significance of these genes in phytohormone-arbitrating processes. The MBS, LTR, and ARE were identified as stress-responsive factors (low temperature, drought, and anaerobic induction), suggesting their involvement in stress stimulation. The MBS (drought stress-responsive element) was found in three StDUF599 genes (StDUF599-1, StDUF599-7, and StDUF599-9). Further, three StDUF599 genes (StDUF599-1, StDUF599-4, and StDUF599-9) contained low-temperature responsive element (LTR) (Supplementary Table S2). Total eight StDUF599 genes (StDUF599-1, StDUF599-2, StDUF599-3, StDUF599-4, StDUF599-5, StDUF599-6, StDUF599-7, and StDUF599-9) had anaerobic responsive elements (ARE) (Fig. 2A; Supplementary Table S2). According to these findings, the expression pattern of StDUF599 genes may differ under phytohormones or abiotic stress. The chromosomal location of StDUF599 genes depicted that all chromosomes have an unequal number of genes. Moreover, a maximum of three genes were present on the chr12 and chr3, while a single gene was present on the chr2, chr8, and chr10 each (Fig. 2B).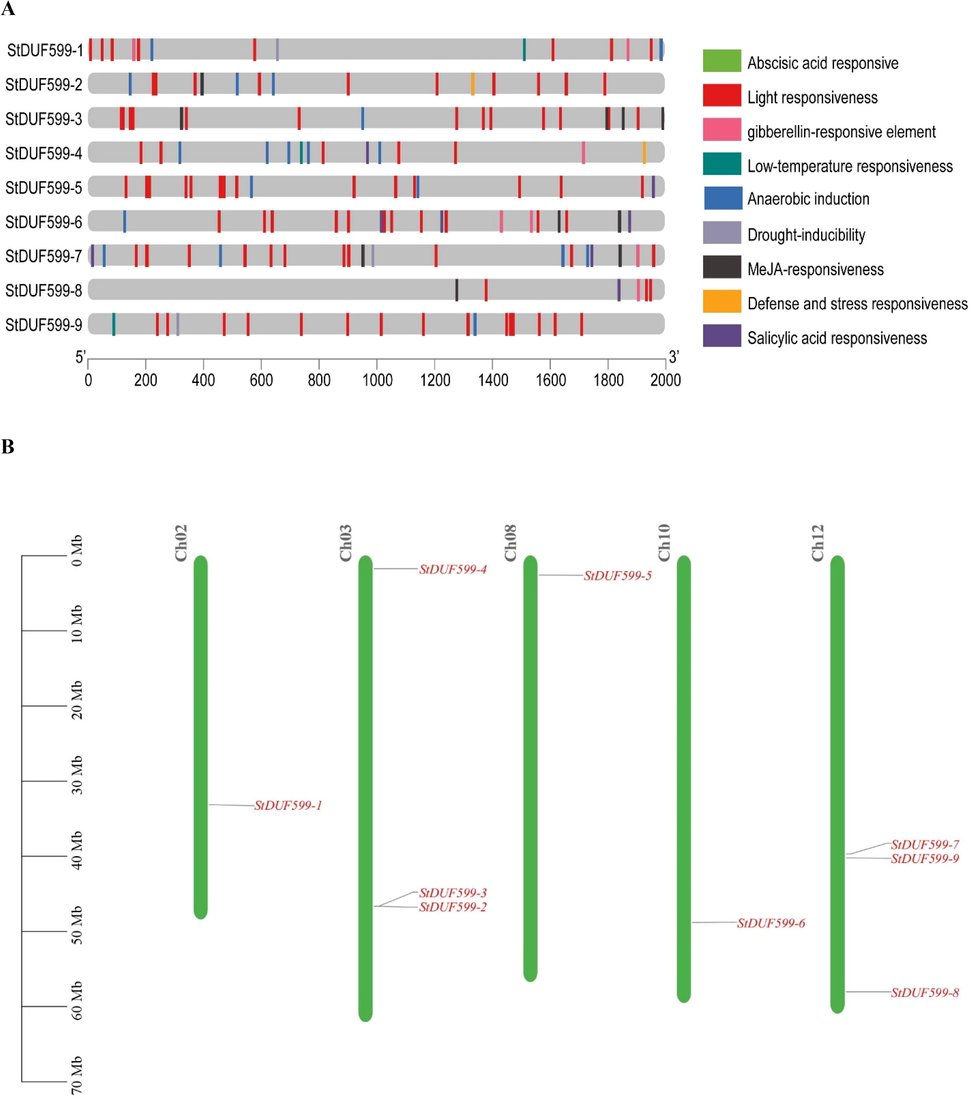
(A) cis-elements in the promoter regions of the StDUF599 genes are linked with different hormone- and stress-responsive elements. Different color boxes show different identified elements. See Supplementary Table S2 for more information. (B) Chromosomal distribution of StDUF599 genes. The green color showed Chromosomes; red color genes.
3.4 Gene duplications and motif analysis of DUF599 genes
This study showed that both segmental and tandem duplication events were present in StDUF599 genes (Table 2). The gene pairs StDUF599-1/StDUF599-4 and StDUF599-5/StDUF599-8 were segmentally duplicated, while the StDUF599-7/StDUF599-9 and StDUF599-2/StDUF599-3 have tandem duplication (Table. 2). Thus, the results illustrated that the segmental duplication events and tandem duplication participated in the DUF599 gene family evolution in potato.
Seq_1
Seq_2
Ka
Ks
Ka_Ks
Time (MYA)
Duplication Type
StDUF599-5
StDUF599-8
0.167201
0.811113
0.206138
155.9833
Segmental
StDUF599-1
StDUF599-4
0.154774
1.093722
0.141511
210.3312
Segmental
StDUF599-2
StDUF599-3
0.070768
0.545704
0.129682
104.9432
Tandem
StDUF599-7
StDUF599-9
0.024876
0.01173
2.120672
2.255855
Tandem
The Ka/Ks value of each duplicated gene pair has been calculated to estimate the rate of molecular evolution. A positive selection effect will be considered if Ka/Ks is greater than 1. If Ka/Ks value is lower than 1, then the purifying selection exists, and if the Ka/Ks value is equal to 1, then neutral selection lies among the duplicated genes. Moreover, our findings demonstrated that most of the duplicated StDUF599 genes usually underwent purifying selection. Moreover, the divergence time (DT) between duplicated gene pairs has also been computed in the present study. When the DUF599 genes exhibited the value of Ks is greater than 0.52, the DT (divergence time) might exceed 100 MYA (million years ago). In our results, the value of Ks for duplicated genes pair StDUF599-1/StDUF599-4 was 1.09, while the expected divergence time was 210.33 MYA (Table 2). The MEME server identified ten conserved motifs in the StDUF599 proteins (Fig. 3A). All StDUF599 genes restrained motif 1 and motif 2. All genes have motif 3 except StDUF599-4 and StDUF599-7. Motifs 1 and 4 have 50 and 49 amino acid residues, respectively; motif 3 contained 47, while 41 amino acids were present in motif 2 (Supplementary Table S3).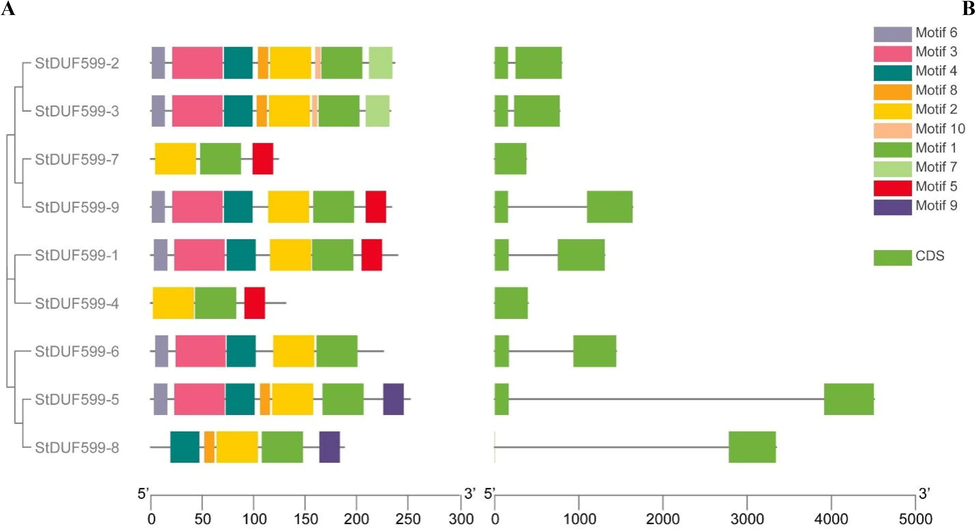
The gene structure and motif analysis of StDUF599 genes. Based on phylogenetic relationships, the StDUF599 were clustered into three major classes. (A) Conserved motif compositions were detected in StDUF599. Different color boxes represent different motifs. (B) Gene structure of StDUF599 genes. The light green color denotes exon and the black horizontal line symbolizes introns.
3.5 Gene structure organization, miRNA, and synteny analysis
The gene structure analysis revealed that StDUF599-1, StDUF599-2, StDUF599-3, StDUF599-5, StDUF599-6, StDUF599-8, and StDUF599-9 have two exons and one intron, while StDUF599-4 and StDUF599-7 genes contained a single exon (Fig. 3B). Previous studies reported that micro-RNAs are involved in various regulatory mechanisms that carry out plant responses against stresses. The comprehensive details of all genes and target sites for miRNAs are shown in Supplementary Table S4. Our findings illustrated that five stu-miR167 members target two StDUF599-4 and StDUF599-1 genes (Fig. 4; Supplementary Table S4). Moreover, only one gene, StDUF599-3, was targeted by stu-miR8036 (Fig. 4; Supplementary Table S4). Two genes StDUF599-5 and StDUF599-3 were the target of three mi-RNAs members (stu-miR8007b-3p, stu-miR8007a-3p, and stu-miR8007b-5p) and one gene StDUF599-2 was targeted by stu-miR8008. Additionally, two genes, StDUF599-3 and StDUF599-2, were the target of stu-miR7983. The two members of the stu-miR7997 targeted the StDUF599-2 gene, while the stu-miR8019 targeted two other genes, including StDUE599-7 and StDUF599-9 (Fig. 4; Supplementary Table S4). Furthermore, two genes, StDUF599-2, StDUF599-3, and StDUF599-4, were predicted to be targeted by numerous micro-RNAs (Fig. 4; Supplementary Table S4). Synteny analysis among A. thaliana, Z. mays, and S. tuberosum delineated a unique link regarding the expression of genes, their evolution, functions, duplications, and triplications. It was noted that the sequence of the ZmDUF599-5 exhibited synteny with the sequence of the StDUF599-2 gene of S. tuberosum. Furthermore, syntenic relations were present between potato gene StDUF599-5 and A. thaliana gene AtDUF599-1. Gene StDUF599-9 of potato depicted synteny with AtDUF599-2 of A. thaliana (Fig. 5). In comparative synteny analysis, intensity and color of outward and inward ribbons tangling illustrated duplication and conservation events, respectively.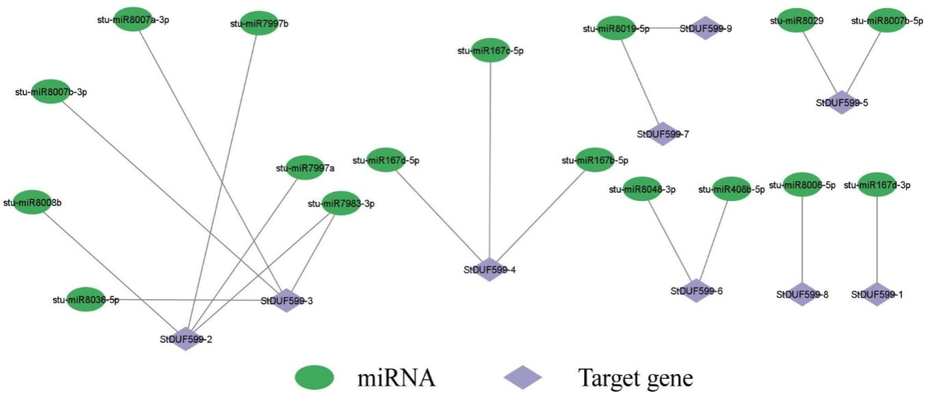
miRNA targeting StDUF599 genes. (A) Network figure of anticipated miRNA targeting StDUF599 genes. Green colors correspond to miRNAs, and purple color represents StDUF599 genes. See Supplementary Table S4 for the detailed data of all predicted miRNAs.
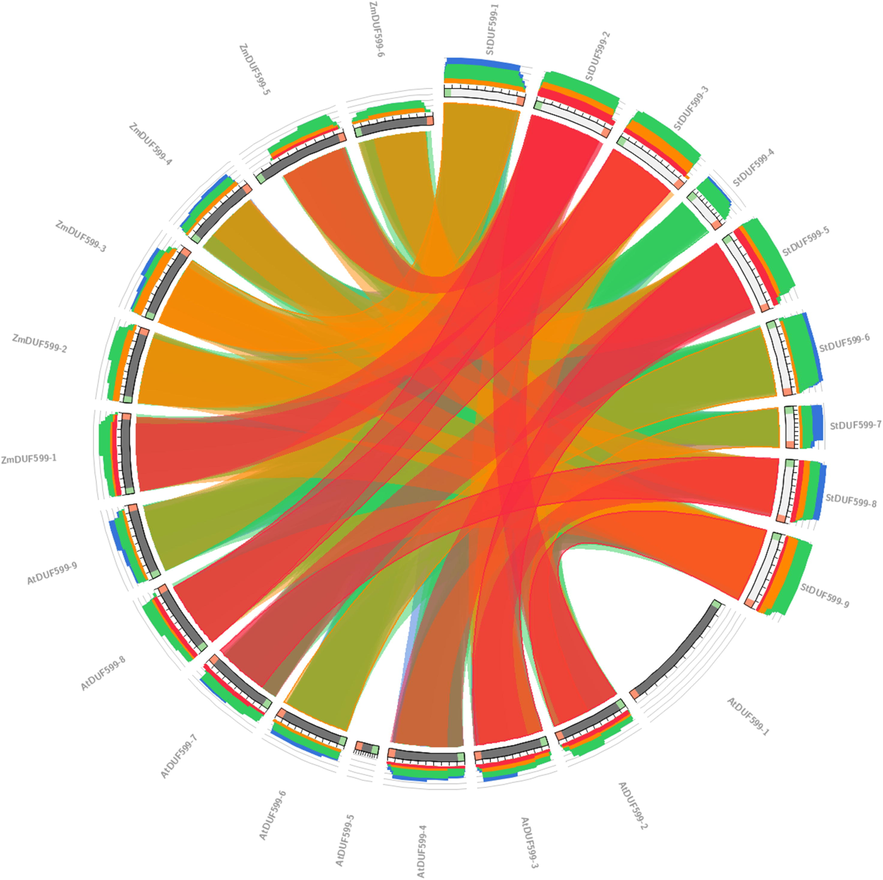
Synteny map among all identified DUF599 sequences of Arabidopsis, Zea mays, and S. tuberosum.
3.6 Tissue-specific expression analysis of StDUF599 genes
The tissue-specific expression of StDUF599 genes was observed in the shoot apex, stem, leaf, flower, stolons, petiole, young tuber, tuber cortex, root, mature tuber, tuber sprout, stamen, tuber pith, tuber peel, whole in-vitro plant using RNA-seq data of S. tuberosum. In general, these results represented that some genes illustrated expression in one tissue (StDUF599-3) and others genes (StDUF599-6 and StDUF599-9) demonstrated expression in all tissues such as young tuber, mature tuber, root, shoot apex, stolon, flower, stem, leaf, root, tuber sprout, tuber cortex, tuber peel, tuber pith, petiole, and the entire in-vitro plant. StDUF599-6, StDUF599-7, StDUF599-8, and StDUF599-9 have higher expression in the stolon. In young tuber, the StDUF599-6, StDUF599-8, and StDUF599-9 genes display higher expression (Fig. 6, Supplementary Table. S5). Moreover, the expression of the StDUF599-1 gene was significantly expressed inside the flower, leaf, stem, and stamen, while the StDUF599-2 gene had high expression in the stamen and flower. In the present study results, the StDUF599-3 gene has only higher expression in flowers, while StDUF599-4 has higher expression in leaf, stem, root, and stamen (Fig. 6, Supplementary Table S5). Gene StDUF599-5 display higher expression in flower, root, and whole in vitro plants. Likewise, the StDUF599-7 gene showed higher expression in stolon, shoot apex, stem, leaf, flower, petiole, root, tuber peel, tuber sprout, and whole in vitro plant. Furthermore, the StDUF599-8 gene showed higher expression in stolon, young tuber, petiole, stem, root, tuber sprout, and whole in vitro plant (Fig. 6, Supplementary Table S5). Moreover, our findings revealed that only a few genes expressed in all observed tissues might significantly participate in the growth process.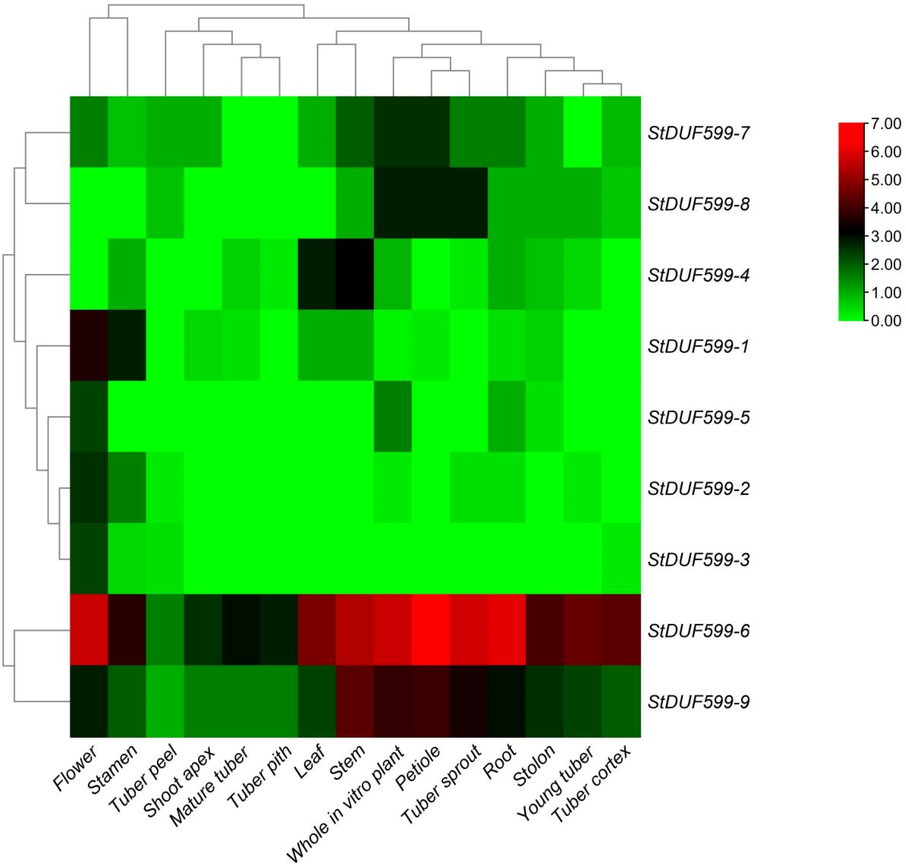
Expression profiling of StDUF599 genes in various developmental tissues. The red, black, and green colors display high to low expression. levels.
3.7 StDUF599 genes expression patterns under heat, salt, and phytohormones stress
RNA-seq-based StDUF599 gene expression was observed under abiotic (heat and salt) and phytohormones (ABA, GA, IAA) stresses. It revealed that StDUF599 genes might carry out significant tasks in potatoes to regulate their response to salt and heat stress. Two StDUF599 genes illustrated high expression against salt stress. After the salt stress, StDUF599-7 and StDUF599-9 showed expression compared to the control (Fig. 7, Supplementary Table S5). At the same time, four StDUF599 genes were expressed against heat stress. When plants were treated to heat, StDUF599-5, StDUF599-6, StDUF599-7, and StDUF599-9 genes were significantly enhanced, while more expression was observed in StDUF599-5 compared to control (Fig. 7, Supplementary Table S5). In the current study, GA3, ABA, and IAA were designated to observe the expression pattern of StDUF599 genes later than phytohormones treatment. Moreover, for the StDUF599 gene's expression profiling against GA3, leaves were treated with GA3. In response to GA3 treatment, the StDUF599-8 gene showed maximum expression levels compared to the control. Similarly, leave tissues were treated with IAA for the StDUF599 gene's expression analysis. Overall, StDUF599-8 was highly expressed when IAA was used. The leave tissues were treated through ABA to observe the expression responses against ABA. In response to ABA, no gene showed expression compared to control (Fig. 7, Supplementary Table S5). Furthermore, StDUF599-1, StDUF599-2, StDUF599-3, and StDUF599-4 did not show expression against any stress, while StDUF599-5 showed significant expression only to heat stress. However, StDUF599-8 was identified to be more active during IAA and GA3 treatment.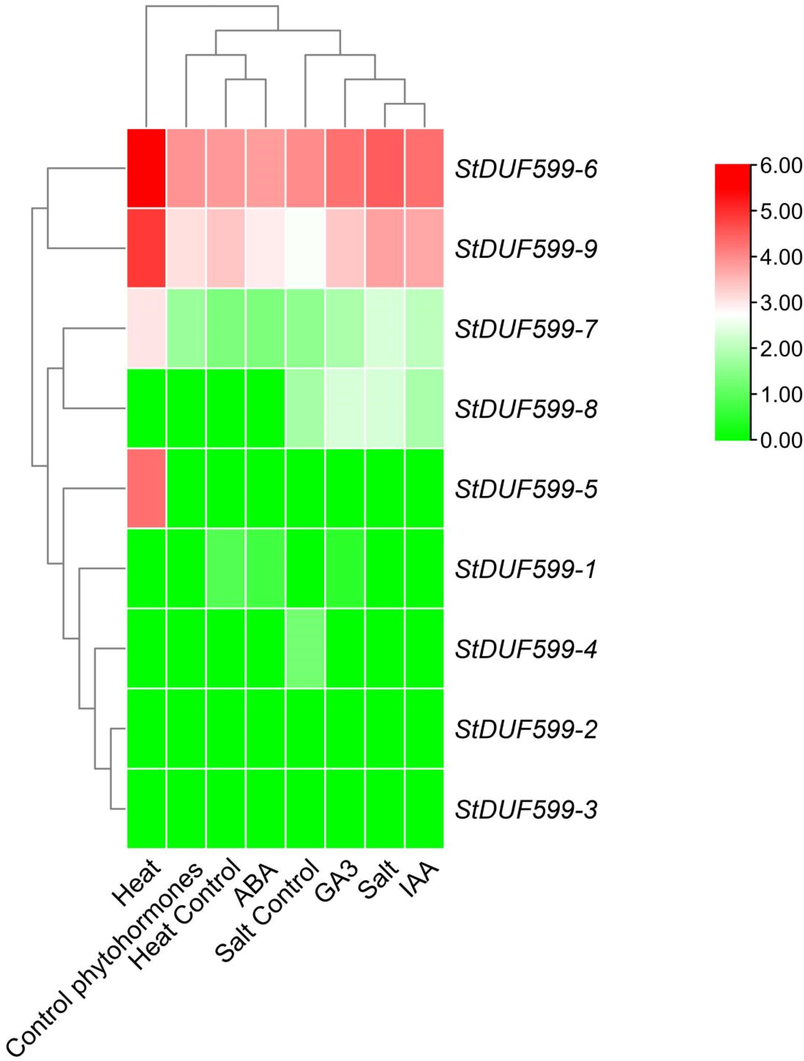
Expression patterns of the StDUF599genes under different abiotic (heat and salt)stress conditions and phytohormones (ABA, GA3, and IAA). The red, black, and green colors display high to low expression levels.
4 Discussion
Potato is a diet source with various nutrients imperative for health and substantial food security worldwide (Zaynab et al., 2017b). Therefore, enhancing potato production will enable us to satisfy the nutritional needs of the world population. Potato exhibits sensitivity toward the various stress factors. Its ability to tolerate different abiotic and biotic stresses is imperative to sustain its productivity and fulfilling food demands in the coming time. Several gene families with the DUF domain have been discovered, and their genome resources are available inside the Pfam database (Bateman et al., 2010). While the origin, variability, and biological duties of these gene families (DUF) were extensively explored in several studies, there is very small information on the biological roles of these genes in potatoes. At present, the database of Pfam possesses 17,939 gene families, of which around 22 % (3,961) are DUFs (Chalupska et al., 2008). DUF599 genes were identified in recent research, and their significant role in potatoes is against abiotic and phytohormones stress. cis-acting factors in DUF599 gene promoter regions are also linked to abiotic stress conditions. The current research identified StDUF599 genes promoter, including stress-associated cis-acting elements, such as LTR, TATC-box, P-box, ABRE, and SARE. ABRE is associated with ABA and salt stress. CGTCA-motif, LTR, and ARE were affiliated with low-temperature stress factors (Maestrini et al., 2009), anaerobic induction response (Geffers et al., 2001), and response to methyl jasmonate (MeJA) (Xu et al., 2018). These findings indicate that cis-acting elements perform a significant role against stress by regulating stress-associated genes. Our results propose that StDUF599 genes may be involved in response to abiotic stress factors. Moreover, genome size, gene duplication, and gene distribution are essential factors in plants' genetic diversity. Moreover, gene duplication is an important feature during the gene families' expansion, diversification, and neofunctionalization (Lavin et al., 2005). Here we also noticed some duplication events, which carry out the critical task of StDUF599 evolution. Furthermore, DUF599 gene distribution at the chromosomal level will provide a deep knowledge to form new varieties with desired traits. In recent decades, extensive research has been executed to recognize miRNAs in plant genomes (Buhtz et al., 2008). The current analysis identified different miRNAs from various families that target several StDUF599 genes. It was earlier observed that miR167 carries out several critical tasks in response to stress conditions (Khraiwesh et al., 2012). Briefly, these researches strengthen our findings and endorse that miRNAs may perform crucial tasks against various stress factors by amending the transcription levels of StDUF599 genes. Waseem et al. (2021) reported the link between ZmDUF and AtDUF proteins and stresses (Waseem et al. 2021). That's why expression profiling of StDUF599 genes is helpful for the functional study of the S. tuberosum. In a recent analysis, nine StDUF599 genes' tissue-specific expression was assessed in various tissues using publicly available RNA-seq data. This study demonstrated that StDUF599 genes illustrated expression in different developmental tissues. Waseem et al. (2021) investigated DUF gene expression of different plant tissues, including leaves, roots, fruits, and flowers, at varying developmental stages of tomato (Waseem et al., 2021). Waseem et al. (2021) reported that DUF221 genes illustrated high expression in specific tissues (Waseem et al., 2021). Moreover, these findings illustrated that these DUF221 genes played a critical role in tissue development (Waseem et al., 2021). Yang Q; (2020) investigated the ATDUF4228 genes expression in various tissues and presented significant variability. According to findings, some ATDUF4228 genes showed high expression in most tissues while others showed lower expression in some tissues. The highly expressed ATDUF4228 genes in tissues have a significant role in plant growth (Yang et al., 2020). These consequences have the same results as our findings, where DUF genes exhibited significant expression at the tissue level, representing that these expressed genes may be involved in potato development and growth. Consequently, more research on DUF genes established their notable roles against different abiotic stress (Leng et al., 2021). Two genes (StDUF599-9 and StDUF599-7) are highly expressed when potato plants treated with salt and heat may have a significant role against abiotic stress. Several hormones participating in signal transduction may alter physiological and biochemical mechanisms (Zaynab et al., 2017a). Hormones like IAA and GA3 are imperative for the immunity of plants. Several studies reported that DUF genes regulate stress response and carry out various developmental and hormonal signalling processes. Furthermore, to observe the DUF599 genes expression in response to hormonal signalling, the leaves of S. tuberosum were treated with IAA and GA3. After the hormone treatments, the expression of the StDUF599-8 gene suggests that DUFs have an essential function in the hormone-induced immune response. According to previous studies, after the treatments of GA3 and IAA, DUF genes upregulation revealed that GA3 and IAA performed a vital function in the immune system (Zaynab et al., 2022). In addition, Yang et al. (2020) revealed that the DUF genes were significantly expressed during the hormonal treatment, revealing that hormones have a significant role in immunity (Yang et al., 2020).
5 Conclusion
In the current study, nine DUF599 genes were recognized inside the S. tuberosum (potato) genome after using BLASTP and HMM techniques. The structure of genes, cis-regulatory factors, phylogenetic analysis, and conserved motifs analysis of DUF599 genes were executed to get insights into DUF599 genes in potato. Moreover, the StDUF599 genes expression profiling were analyzed in various tissues to observe their significant role in development and growth. The expression analysis of StDUF599 genes during heat, salt, and hormone treatments revealed their significant role against abiotic stress. Furthermore, our study will provide a base for further functional studies of the DUF599 genes under unfavourable or stressful conditions.
Acknowledgement
This research report is supported by Shenzhen Special Project for Sustainable Development (KCXFZ20201221173211033), Taif University Researchers Supporting Project number (TURSP-2020/94), Taif University, Taif, Saudi Arabia. The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University Saudi Arabia for funding this work through Large Groups Project under grant number RGP.2/28/43.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. All authors have direct intellectual contribution to the manuscript. The authors are in complete agreement to make submission with journal of King Saud University Science.
References
- Three members of Medicago truncatula ST family are ubiquitous during development and modulated by nutritional status (MtST1) and dehydration (MtST2 and MtST3) BMC Plant Biol.. 2017;17:117.
- [CrossRef] [Google Scholar]
- MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res.. 2009;37:W202-W208.
- [CrossRef] [Google Scholar]
- Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J.. 2008;53:739-749.
- [CrossRef] [Google Scholar]
- Chalupska, D., Lee, H.Y., Faris, J.D., Evrard, A., Chalhoub, B., Haselkorn, R., Gornicki, P., 2008. Acc homoeoloci and the evolution of wheat genomes. Proceedings of the National Academy of Sciences 105, 9691–9696. https://doi.org/10.1073/pnas.0803981105.
- TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Molecular Plant. 2020;13:1194-1202.
- [CrossRef] [Google Scholar]
- psRNATarget: a plant small RNA target analysis server (2017 release) Nucleic Acids Res.. 2018;46:W49-W54.
- [CrossRef] [Google Scholar]
- The Pfam protein families database in 2019. Nucleic Acids Res.. 2019;47:D427-D432.
- [CrossRef] [Google Scholar]
- Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M.R., Appel, R.D., Bairoch, A., 2005. Protein Identification and Analysis Tools on the ExPASy Server, in: Walker, J.M. (Ed.), The Proteomics Protocols Handbook. Humana Press, Totowa, NJ, pp. 571–607. https://doi.org/10.1385/1-59259-890-0:571.
- The TATA box and a Myb binding site are essential for anaerobic expression of a maize GapC4 minimal promoter in tobacco. Biochimica et Biophysica Acta (BBA) - Gene Struct. Expression. 2001;1521:120-125.
- [CrossRef] [Google Scholar]
- Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res.. 2012;40:D1178-D1186.
- [CrossRef] [Google Scholar]
- Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2012;1819:137-148.
- [CrossRef] [Google Scholar]
- Mechanisms underlying robustness and tunability in a plant immune signaling network. Cell Host Microbe. 2014;15:84-94.
- [Google Scholar]
- MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol.. 2018;35:1547-1549.
- [CrossRef] [Google Scholar]
- Evolutionary Rates Analysis of Leguminosae Implicates a Rapid Diversification of Lineages during the Tertiary. Syst. Biol.. 2005;54:575-594.
- [CrossRef] [Google Scholar]
- Genome-Wide Analysis of the DUF4228 Family in Soybean and Functional Identification of GmDUF4228–70 in Response to Drought and Salt Stresses. Front. Plant Sci.. 2021;12:628299
- [CrossRef] [Google Scholar]
- PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res.. 2002;30:325-327.
- [CrossRef] [Google Scholar]
- Over-expression of a DUF1644 protein gene, SIDP361, enhances tolerance to salt stress in transgenic rice. J. Plant Biol.. 2016;59:62-73.
- [CrossRef] [Google Scholar]
- Genome-wide identification, characterization and expression analysis of the non-specific lipid transfer proteins in potato. BMC Genomics. 2019;20:375.
- [CrossRef] [Google Scholar]
- Isolation and expression analysis of low temperature-induced genes in white poplar (Populus alba) J. Plant Physiol.. 2009;166:1544-1556.
- [CrossRef] [Google Scholar]
- The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res.. 2003;31:224-228.
- [CrossRef] [Google Scholar]
- Stress-induced senescence and plant tolerance to abiotic stress. J. Exp. Bot.. 2018;69:845-853.
- [CrossRef] [Google Scholar]
- Genome sequence and analysis of the tuber crop potato. Nature. 2011;475:189-195.
- [CrossRef] [Google Scholar]
- The DUF221 domain-containing (DDP) genes identification and expression analysis in tomato under abiotic and phytohormone stress. GM Crops Food 2021:1-14.
- [CrossRef] [Google Scholar]
- Arabidopsis ESK1 encodes a novel regulator of freezing tolerance. Plant J.. 2007;49:786-799.
- [CrossRef] [Google Scholar]
- Glycinebetaine Biosynthesis in Response to Osmotic Stress Depends on Jasmonate Signaling in Watermelon Suspension Cells. Front. Plant Sci.. 2018;9:1469.
- [CrossRef] [Google Scholar]
- Comprehensive genomic analysis of the DUF4228 gene family in land plants and expression profiling of ATDUF4228 under abiotic stresses. BMC Genomics. 2020;21:12.
- [CrossRef] [Google Scholar]
- Proteomic approach to address low seed germination in Cyclobalnopsis gilva. Biotechnol. Lett.. 2017;39:1441-1451.
- [CrossRef] [Google Scholar]
- Rice chitinase gene expression in genetically engineered potato confers resistance against Fusarium solani and Rhizictonia solani. PSM Microbiol.. 2017;2:63-73.
- [Google Scholar]
- CRISPR/Cas9 to generate plant immunity against pathogen. Microb. Pathog.. 2020;141:103996
- [CrossRef] [Google Scholar]
- Mitogen-Activated Protein Kinase Expression Profiling Revealed Its Role in Regulating Stress Responses in Potato (Solanum tuberosum) Plants. 2021;10:1371.
- [CrossRef] [Google Scholar]
- Expression profiling of pathogenesis-related Protein-1 (PR-1) genes from Solanum tuberosum reveals its critical role in phytophthora infestans infection. Microb. Pathog.. 2021;161:105290
- [CrossRef] [Google Scholar]
- ATP-binding cassette transporters expression profiling revealed its role in the development and regulating stress response in Solanum tuberosum. Mol. Biol. Rep. 2021
- [CrossRef] [Google Scholar]
- Genome-Wide Identification and Expression Profiling of DUF221 Gene Family Provides New Insights Into Abiotic Stress Responses in Potato. Front. Plant Sci.. 2022;12:804600
- [CrossRef] [Google Scholar]
- Genome-wide identification of the DUF668 gene family in cotton and expression profiling analysis of GhDUF668 in Gossypium hirsutum under adverse stress. BMC Genom.. 2021;22:395.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2022.102368.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







