Translate this page into:
Expression changes of multiple marker genes and analysis of circulating tumour cells and dehydrogenase for prognostic and pharmacodynamic measures
⁎Corresponding author. nicholaszam1990@gmail.com (Nicholas Daniel Amalorpavanaden) nicholasdaniel@mukuba.edu.zm (Nicholas Daniel Amalorpavanaden)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Objectives
The main objectives of the studies are to analyze melanoma markers from the tumour and blood samples.
Methods
Lactate dehydrogenase (LDH) was assayed from the serum sample to determine metastatic melanoma. The prevalence of circulating tumour cells was explored in tumour subjects and compared with non-tumour cases. Expression analysis of TGF-β and cathepsin-K was determined from the non-melanoma and melanoma cases using western blotting analysis.
Results
The amount of LDH was elevated in most metastatic patients (p < 0.0001). About 10% of patients were associated with the micrometastatic stage and 16.3% of patients were linked with multiple melanomas. The amount of ctDNA considerably declined at <0.01% level after one month of clinical development. The amount of ctDNA in the blood in an initial stage indicates resistance to specific targeted therapy. Expression of S100 level also decreased after two weeks of treatment. TGF-β showed maximum variation in mRNA expression between diseased and healthy cases, whereas, an increased amount of cathepsin-K biosynthesis was observed in tumour tissues (p < 0.001).
Conclusions
MDH, ctDNA, TGF-β and S100 could be potential markers to predict early determination of tumour and non-tumour tissues. Monitoring of ctDNA and S100 is useful to analyze the health benefits of melanoma patients.
Keywords
Melanoma
Cathepsin-K
Circulating tumour DNA
Basal cell carcinoma
1 Introduction
Melanoma is an important form of skin cancer and the occurrence of this disease is increasing in recent years. About 80% of melanoma cases are cured with surgical procedures (Chapman et al., 2011). Many methods have been suggested to classify melanoma. The molecular sub-typing method has been used to sub-type patients using genetic aberrations. Analysis of tumour biopsy after treatment and before treatment is a very difficult task and other approaches are sought for exact determination. Circulating tumour cells (CTCs) are useful as predictive, prognostic and pharmacodynamic biomarkers. Melanoma was the first solid tumour determined using polymerase chain reaction and the lack of standard methodology hampered the accurate quantification of CTCs in melanoma cases (Nezos et al., 2009). CellSearch platform is useful for the determination of CTC number in breast, lung, colorectal and prostate carcinomas (Hou et al., 2012) and the cutoff number of CTC varied based on tumour type (CTC/7.5 mL blood) and prevalence of CTCs. Rao et al. (2011) developed a melanoma-specific CellSearch CTC kit for the determination of melanoma using Melcam and high molecular weight–melanoma associated antibodies.
In recent years, the tumour microenvironment has been considered one of the important factors in tumour development (Hou et al., 2012). In the tumour microenvironment, the interactions between stromal cells, cancer cells, tumour-infiltrating immune cells and stromal cells are very critical for cancer development, including, invasion, immune surveillance evasion and metastasis (Margolis et al., 2019). Both epigenetic and genetic variations that critically interfere with the normal physiological function of the cells are the main reason for tumour development (Zhang et al., 2016). Alternative splicing is one of the important posttranscriptional regulatory mechanisms involved in the transcription of a single form of prem-RNA into mature RNA with different structures and sequences, which are present in about 95% of genes in the genome of human and influence the development of various pathological conditions, including cancer (Song et al., 2018). In tumour cells, splicing is highly altered in most cases (Song et al., 2018; Bradley and Anczuków, 2023). Alternative splicing variations may lead to differences in transcriptome variations in various cancer-related genes and further influence the development of cancer and therapeutic resistance (Chen et al., 2018).
Skin cancer is one of the common types of cancers, squamous cell carcinoma (SCC), basal cell carcinoma (BCC) and melanoma (Srivastava et al., 2023). Melanoma accounts for about 1% of all skin cancer types and this cancer type is involved in the development of skin cancer-related fatalities. Melanoma is a dangerous and aggressive form of skin cancer that develops from melanocytes (Isola et al., 2016). Early diagnosis of melanoma is critical for the survival of this disease and to improve the 5-year survival rate. The five-year survival rate of metastatic melanoma is merely 27% and the primary melanoma is 99% (Siegel et al., 2021). Global incidence rates increased in recent years in the US and predicted about 100,000 new cases in 2021 and the predicted mortality was about 7000 (Siegel et al., 2021). There is a number of factors associated with melanoma, including, fair skin, male, age, number of moles and UV exposure (Shain and Bastian, 2016). The common genetic disorders mainly associated with melanoma are the CDK4 and CDKN2A genes involved in cell cycle regulation, xeroderma pigmentosum and the MC1R gene involved in skin pigmentation (Goodwin et al., 2023; Shan et al., 2022). In this study, cutaneous melanoma biomarkers were analyzed for the earlier determination of melanoma and the result was compared with non-tumour cases.
2 Materials and methods
2.1 Experimental groups
A total of 290 samples were collected from cancer patients and non-tumour cases admitted in hospitals. All samples were collected between March 2021 and November 2021. The selected patients were characterized using clinical summary and blood parameters analysis. Fourteen samples were collected from non-cancer cases. Multiple markers were screened from the tissue samples and blood from the selected groups.
2.2 Determination of LDH levels
The blood sample (0.5 mL) was collected from the selected cases and centrifuged for 5 min at 3000 rpm. The centrifuged serum sample was used for the determination of lactate dehydrogenase (LDH). Experiments were performed in triplicates and the mean value was used for data processing. The number of patients for the determination of LDH was selected based on pathogenic conditions and clinical history (Todorovic et al., 2021).
2.3 Circulating tumour DNA analysis from cutaneous melanoma patients
To determine circulating tumour DNA (ctDNA), the blood sample was collected from the metastatic patients before therapeutic intervention and ten weeks of treatment from the tumour and non-tumour cases. The patients were treated either immunotherapy (Ipilimumab or Pembrolizumab) or BRAF inhibitor. The collected blood sample (1 mL) was mixed with EDTA, centrifuged for 5 min at 2000 rpm at 28 ± 1 °C. The plasma was pooled and centrifuged at 15,000 rpm for 15 min. The final supernatant was aliquoted and stored at –70 °C. DNA was extracted from the sample using a DNA extraction kit (Qiagen, Germany) and the circulating melanoma-derived DNA was quantified as described previously. DNA was divided into various reactions containing 1000 genomic equivalents in an experiment. ctDNA fractions were analyzed by calculating all mutant observations and the total genomic equivalents. The final result was compared with other treatments’ outcomes and the lower limit of sensitivity was about 0.1%.
2.4 Analysis of cytokines
A plasma sample was used for the determination of cytokines as suggested previously. The cytokine level was expressed as pg/mL. The amount of IL-1RA, IL-7, IFN-γ and TNF-α levels were determined from a total of selected 14 melanoma cases (LD1, LD3, LD14, LD22, LD23, LD28, LD39, lD40, LD54, LD59, LD60, LD63, LD65, and LD69) and three healthy control (HL1, HL2 and HL3). The patients were selected based on clinical history and other treatment conditions (Kleiner et al., 2013).
2.5 Analysis of S100B protein
In our study, the amount of S100B protein was assayed from the serum sample. The amount of S100B protein level over 0.15 μg/L and the detection limit was 0.05 μg/mL (Hauschild et al., 1999).
2.6 Analysis of protein marker expression and western blotting
Tissue samples were collected from the disease-confirmed cases and non-tumour cases. The selected samples were not associated with any contagious disease, immunodeficiency disease or disorders or were not under any immunosuppressive treatment. The tissue sample was collected from the subjects and stored at −70 °C until analysis. These samples were lysed using a lysis buffer containing 0.25% Triton X-100. The amount of protein in the sample was determined using Bradford method. A total of 20 μg lyzed sample was isolated using 10% SDS-PAGE and electroblotted on polyvinylidene difluoride membranes. The blank regions were blocked with skimmed milk containing 0.1% Tween 20 for 12 h at 4 °C. Further, the protein-specific antibodies, rabbit anti-TGF-β2, mouse anti-cathepsin K were incubated with proteins at 1:200 dilutions. It was washed several times and the membrane was incubated with horseradish peroxidase-conjugated secondary antibodies, incubated for 1 h and developed in dark for 10 min and visualized.
2.7 RT-qPCR
RT-qPCR analysis was used for the determination of TGF-β and cathepsin-K mRNA levels. RNA was initially extracted from the sample using RNeasy Kit (Qiagen). The sample was treated with DNAase to remove DNA from the sample and 5 μg RNA was used for the reverse transcription process. The expression level of the target gene mRNA and expression of β-actin mRNA was analyzed using Gene Expression Master mix and gene-specific final PCR products using Real-Time PCR system. In our study, target gene expression was normalized to the expression of β-actin in transcribed cDNA sample.
2.8 Statistical analysis
Analysis of variance was used for the determination of the significance variance of biomarkers between tumour and control cases in relation to age and disease progression. The p-value <0.05 was considered statistically significant.
3 Results
3.1 LDH level in cancer cases
The amount of LDH levels was determined from the serum sample to analyze the enzymatic expression changes. The patients were characterized based on age group and the amount of LDH levels showed significant variation between different age groups. The amount of LDH was elevated in most the metastatic patients (p < 0.0001). About 10% of patients were associated with the micrometastatic stage and 16.3% of patients were associated with multiple melanomas (Table 1).
Age group
Lactate dehydrogenase (IU/L)
Mean
Lower range
High range
31–40
48
1502
289
41–50
52
709
149
51–60
21
1620
319
61–70
45
1541
402
71–80
37
1386
374
3.2 Circulating tumour DNA (ctDNA) in cutaneous melanoma patients
In certain patients, the amount of ctDNA was high because of tumour deposits. In other cases, the amount of ctDNA considerably increased for two weeks and the level declined (p < 0.01%) after one month of clinical development. After two weeks of treatment, the level of ctDNA increased, however further treatment considerably reduced the ctDNA level. After one week of treatment, 1940 ± 10.1 ctDNA copies were detected and it declined continuously after ten weeks of treatment. Likewise, the expression of the S100 level decreased after 14 days (Table 2).
Treatment (week)
DNA fragment/ml
S100 ng/mL
1
1940 ± 10.1
20.3 ± 1.1
2
2180 ± 29.4
21.2 ± 0.1
3
1720.2 ± 10.1
11.8 ± 0.09
4
400 ± 19.2
9.7 ± 0.2
5
149.2 ± 16.2
4.3 ± 0.1
6
58.1 ± 3.3
3.9 ± 0.5
7
52.9 ± 0.6
2.1 ± 0.1
8
13.1 ± 1.1
2 ± 0.2
9
5.8 ± 0.5
1.3 ± 0.01
10
1 ± 0.1
0.49 ± 0.2
3.3 Analysis of cytokines
Analysis of cytokines was performed using these experimental and control groups and assayed by the immunoassay method. Cytokines levels of melanoma patients were compared with healthy controls. IL-1RA level was elevated in melanoma patients than in control (healthy individuals). The amount of IL-7 (pg/mL) level was decreased in tumour samples than in non-tumour cases. The level of other biomarkers, IFN-γ (pg/mL) and TNF-α (pg/mL) were elevated in melanoma patients than in control (p < 0.001) (Table 3).
Samples
Cytokines
IL-1RA (pg/mL)
IL-7 (pg/mL)
IFN-γ (pg/mL)
TNF-α (pg/mL)
LD1
498.3 ± 12.4
10.2 ± 0.4
14.2 ± 1.1
6.01 ± 0.1
LD3
682.5 ± 20.2
15.2 ± 0.2
13.5 ± 0.8
4.9 ± 0.3
LD14
38.4 ± 1.3
3.2 ± 1.1
3.5 ± 1.4
7.4 ± 1.1
LD22
872.4 ± 12.4
10.9 ± 2.2
10.4 ± 0.3
5.2 ± 0.5
LD23
1075 ± 20.4
8.3 ± 1.1
5.9 ± 1.4
4.1 ± 0.9
LD28
50.4 ± 2.4
15.4 ± 0.9
18.1 ± 0.8
2.9 ± 0.1
LD39
1408 ± 6.4
5.3 ± 0.5
7.3 ± 1.1
6.8 ± 0.5
LD40
49.3 ± 1.4
6.2 ± 0.3
8.2 ± 0.8
3.4 ± 0.5
LD54
58.4 ± 2.5
8.2 ± 1.1
6.4 ± 0.4
4.9 ± 0.1
LD59
682.1 ± 14.3
5.9 ± 0.5
8.1 ± 1.1
1.3 ± 0.7
LD60
509.4 ± 10.5
8.3 ± 0.4
6.5 ± 0.5
5.8 ± 0.6
LD63
408.5 ± 3.5
15.9 ± 1.1
16.7 ± 1.2
3.4 ± 1.1
LD65
98.4 ± 4.8
14.2 ± 0.3
13.2 ± 0.8
7.2 ± 0.32
LD69
105.3 ± 1.4
3.7 ± 1.2
5.8 ± 0.3
5.3 ± 0.1
HL0
146.2 ± 2.9
16.3 ± 1.2
6.3 ± 0.4
1.2 ± 0.2
3.4 Elevated level of S100B protein in cutaneous melanoma patients
The amount of S100B protein was elevated in most of the metastasis melanoma patients than in control cases. In our study, the elevated level was observed in 46.2% of primary and metastasis melanoma patients. S100B protein level was maximum in patients associated with metastasis melanoma (1.3–2.6 μg/L) than primary metastasis (p < 0.001) (Table 4).
S100B protein (μg/L)
Primary malanoma
Metastasis malanoma
LD1
1.4 ± 0.2
LD63
3.1 ± 0.49
LD39
0.9 ± 0.1
LD69
0.42 ± 0.0
LD14
1.5 ± 0.12
LD3
4.2 ± 0.5
LD23
1.82 ± 0.08
LD22
3.1 ± 0.52
LD28
1.01 ± 1.03
LD40
4.8 ± 1.1
LD39
2.08 ± 0.3
LD59
5.7 ± 0.52
LD54
0.59 ± 0.4
LD65
6.1 ± 0.47
LD60
0.34 ± 0.11
HL0
0.27 ± 0.2
3.5 Protein marker expression (TGF-β and cathepsin-K) and western blotting analysis
The expressed level of TGF-β and cathepsin-K was presented in Fig. 1a and b. The expression of proteins analyzed with western blotting was significantly higher than the control group of samples (p < 0.001). Moreover, the expression between the two selected patients was insignificant (p = 0.072). The amount of mRNA level was high than in normal cases (p < 0.0001). TGF-β showed a maximum difference in mRNA expression between diseased and healthy cases. The expression of cathepsin-K was considerably high in tumour tissues.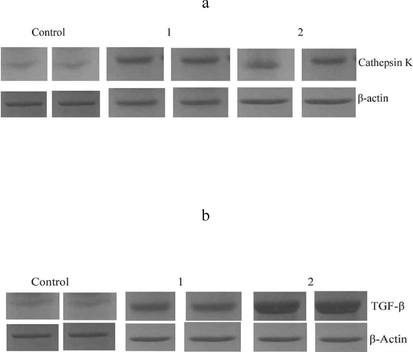
Western blotting analysis of the expression of transforming growth factor (TGF) (a), Cathepsin-K (b) and TGF-β.
4 Discussion
Cytokines are involved in the regulation and developmental process of tumours. In our study, samples from the melanoma cases showed increased levels of cytokine (TNF-α) than normal non-tumour cases. The amount of cytokine was highly elevated in advanced-stage melanoma cases (Stage IV) than in early stage (Stage I). TNF-α was associated to stimulate cell proliferation and inducing tumour growth, metastatic processes and disease progression (Zhao and Adjei, 2015). S100 is one of the protein markers which have been initially found in the bovine brain and S100A1 and S100B genes were initially identified (Wang et al., 2022). More than 25 S100 genes have been identified in humans, including, S100B, S100A1–S100A16, S100G, S100P, and S100Z (Shang et al., 2008). In the eukaryotic cells, S100 proteins involved in the modulation of various targets can regulate a specific target, leading to a highly complex biology regulatory function. These S100 proteins are involved in cell proliferation, angiogenesis, metastasis, inflammation, invasion and the alteration of various routine biological functions. In many cancer types, the expression of the S100 family gene is modified or affected, revealing that the S100 proteins or genes are associated with tumour development (Bresnick et al., 2015).
The amount of LDH varied between tumour cases and control subjects. Moreover, age group and sex did not show any significant variation in LDH level (p > 0.001). The concentration of LDH and age group were described and analyzed statistically significant. ctDNA has been recognized as one of the important biomarkers to determine advanced melanoma stage treated with BRAF inhibitors (Ascierto et al., 2013). The amount of ctDNA in the blood is useful to predict the significance of ctDNA in melanoma therapy. In a study, the dynamics of ctDNA in melanoma patients subjected to immunotherapies have been established. The levels of ctDNA associated with radiological treatment in melanoma cases have been reported. The amount of ctDNA in the bloodstream is useful to determine the effect of immunotherapy in melanoma cases (Han and Wang, 2011). Monitoring ctDNA helped to predict further required treatment.
In our study, the expression of two proteins involved in tumour growth was analyzed. The important role of TGF-β on tumour development has been established previously. TGF-β/Smad pathway and dysregulation are involved in the development of breast cancer, pancreatic cancer and various types of melanoma (Han and Wang, 2011). The amount of TGF-β changed during the process of carcinogenesis (Gulley et al., 2022). In normal tissues, TGF-β inhibits cell proliferation and blocks G1 phase of the cell cycle and in tumour cells, TGF-β mediated the proliferation of cancer cells and metastatic spread. Expression of TGF-β has been established with isoforms of TGF-β (TGF-β1, TGF-β2 and TGF-β3) and TβR II receptors in cancerous and normal human skin tissue. These earlier investigations revealed elevated levels of TβR II and TGF-β1 in tumour cells (Han and Wang, 2011). The expression of TGF-β was elevated in the tumour cases than non-tumour individuals. The level of mRNA expression varied based on different stages. These previous results with TGF-β were consistent with those of the present finding, which showed a marked elevation of TGF-β expression in the diseased cases compared with healthy tissues. These findings revealed the influence of TGF-β in the pathogenesis of tumours (Gulley et al., 2022).
The expression changes of Cathepsin-K were analyzed in this study and played a significant role in maintaining the balance between proteolysis and protein synthesis (Zheng et al., 2015). It has been previously reported that cathepsin-K is involved in scarring, photo-destruction and carcinogenesis (Rao et al., 2014). In our study, increased expression of cathepsin-K was observed in cancerous tissues than in normal cells. The expression ofcathepsin-K in basal cell carcinoma has been established previously (Quintanilla-Dieck et al., 2008; Ishida et al., 2013).
5 Conclusions
The results of the present investigation revealed the application of biomarkers in monitoring the development of melanoma disease. The prevalence of circulating tumour cells in healthy and tumour subjects was determined. The amount of ctDNA in the blood is useful for the determination of the severity of the disease. The amount of LDH was elevated in most the metastatic patients (p < 0.0001), however, it was not conclusive in certain cases. Expression of S100 level decreased after 14 days of treatment. Monitoring of ctDNA and S100 is useful to analyze the health benefits of melanoma patients. Expression analysis of TGF-β and cathepsin-K was determined from the non-melanoma and melanoma cases using western blotting analysis. TGF-β showed mRNA expression changes between diseased and healthy cases. Cathepsin-K has been considered one of the important markers and the level increased in tumour cells. These biomarkers could be the potential to predict the early determination of tumour and non-tumour tissues.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Phase II trial (BREAK-2) of the BRAF inhibitor dabrafenib (GSK2118436) in patients with metastatic melanoma. J. Clin. Oncol.. 2013;31(26):3205-3211.
- [Google Scholar]
- RNA splicing dysregulation and the hallmarks of cancer. Nat. Rev. Cancer 2023:1-21.
- [Google Scholar]
- Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med.. 2011;364:2507-2516.
- [Google Scholar]
- Mutually exclusive acetylation and ubiquitylation of the splicing factor SRSF5 control tumor growth. Nat. Commun.. 2018;9(1):1-21.
- [Google Scholar]
- Combination Therapies with CDK4/6 Inhibitors to Treat KRAS-Mutant Pancreatic Cancer. Cancer Res.. 2023;83(1):141-157.
- [Google Scholar]
- Dual inhibition of TGF-β and PD-L1: a novel approach to cancer treatment. Mol. Oncol.. 2022;16(11):2117-2134.
- [Google Scholar]
- Roles of TGFβ signaling Smads in squamous cell carcinoma. Cell Biosci.. 2011;1(1):1-8.
- [Google Scholar]
- S100B protein detection in serum is a significant prognostic factor in metastatic melanoma. Oncology. 1999;56(4):338-344.
- [Google Scholar]
- Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J. Clin. Oncol.. 2012;30:525-532.
- [Google Scholar]
- Cathepsin K expression in basal cell carcinoma. J Eur Acad Dermatol. Venereol. 2013;27(1):e128-e130.
- [Google Scholar]
- Biology, therapy and implications of tumor exosomes in the progression of melanoma. Cancers. 2016;8(12):110.
- [Google Scholar]
- Cytokine levels in the serum of healthy subjects. Med. Inflamm.. 2013;2013 Article ID 434010
- [CrossRef] [Google Scholar]
- Reprogramming lymphocytes for the treatment of melanoma: from biology to therapy. Adv. Drug Delivery. Rev.. 2019;141:104-124.
- [Google Scholar]
- Molecular markers detecting circulating melanoma cells by reverse transcription polymerase chain reaction: methodological pitfalls and clinical relevance. Clin. Chem. Lab. Med.. 2009;47:1-11.
- [Google Scholar]
- Circulating melanoma cells and survival in metastatic melanoma. Int. J. Oncol.. 2011;38:755-760.
- [Google Scholar]
- Cathepsin K in the immunohistochemical diagnosis of melanocytic lesions. Int. J. Clin. Exp. Pathol.. 2014;7(3):1132.
- [Google Scholar]
- Genomic And Tumor Microenvironment Differences Between Cell Cycle Progression Pathway Altered/Non-Altered Patients With Lung Adenocarcinoma. Front. Oncol.. 2022;12:843528
- [Google Scholar]
- Chromosomal mapping, differential origin and evolution of the S100 gene family. Genet Sel Evol.. 2008;40(4):1-16.
- [Google Scholar]
- March. Alternative splicing in cancers: From aberrant regulation to new therapeutics. Semin Cell Dev Biol.. 2018;75:13-22.
- [Google Scholar]
- Unravelling the landscape of skin cancer through single-cell transcriptomics. Translat. Oncol.. 2023;27:101557
- [Google Scholar]
- Four weeks of aerobic training affects cardiac tissue matrix metalloproteinase, lactate dehydrogenase and malate dehydrogenase enzymes activities, and hepatorenal biomarkers in experimental hyperhomocysteinemia in rats. Int. J. Mol. Sci.. 2021;22(13):6792.
- [Google Scholar]
- Increased Set1 binding at the promoter induces aberrant epigenetic alterations and up-regulates cyclic adenosine 5′-monophosphate response element modulator alpha in systemic lupus erythematosus. Clin. Epigenet.. 2016;8(1):1-12.
- [Google Scholar]
- Targeting angiogenesis in cancer therapy: moving beyond vascular endothelial growth factor. The Oncologist. 2015;20(6):660-673.
- [Google Scholar]
- Cathepsin D repairing role in photodamaged skin barrier. Skin Pharmacol. Physiol.. 2015;28(2):97-102.
- [Google Scholar]







