Translate this page into:
Expression analysis of fiber-related genes for staple length in upland cotton (Gossypium hirsutum L.)
⁎Corresponding author at: The Advanced Seed Institute, Plant Precision Breeding Academy, Zhejiang Provincial Key Laboratory of Crop Genetic Resources, College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, China. furqan.ahmad@mnsuam.edu.pk (Furqan Ahmad) 11816100@zju.edu.cn (Furqan Ahmad)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Background
Cotton fibers, derived from the seed coat and used in the global textile industry for fabric production, are generally recognized as the predominant form of individual unicellular cells. The investigation of cellular proliferation and differentiation may be efficiently conducted by using cotton fiber production as a model system. Expression profiling techniques are very helpful to determine the tissue (stem and roots) and stage (0, 05, 10, 15 and 20 DPA) specific gene expression in cotton fiber. This profiling technique is helpful in the development of a new plant (variety) through transformation, resulting in the development of a new plant with desirable fiber characteristics.
Methods
The expression profiling was carried out in upland cotton genotypes with variable staple length including: i) long staple length, ii) medium staple length and iii) short staple length) at five different days post anthesis (DPA) phases of fiber elongation (0, 5, 10, 15 and 20 DPA) through real-time PCR. Two internal controls (ubiquitin and 18sr RNA) genes were used for data normalization. Current research has focused on genetic basis of fiber regulation and understanding the molecular basis of fiber development in upland cotton (G. hirsutum L.) genotypes (‘CYTO-179′, ‘CIM-616′ and ‘CIM-707′) with variable staple length medium, long and short respectively. The present work aimed to investigate the expression levels of three fiber genes, namely PEPc, XTH, and GA-20 Oxidase, throughout several developmental phases.
Results
All three genes have same expression at 15 DPA fibers in all three genotypes but PEPc transcripts are high at 15 DPA stage in 'CIM-707' (long staple length genotype). These fiber development genes may be transformed to the plants with long fiber length through breeding programs. The molecular analysis of fiber development has significant importance in understanding the process of plant cell fate determination, which in turn may contribute to the improvement of fiber production in the long run. The primary impetus behind investigating the molecular mechanisms behind fiber creation is in the pursuit of enhancing cotton fiber quality and yield via prospective endeavors in genetic engineering and breeding.
Keywords
Fiber elongation phase
RT-PCR
DPA
GA-20 Oxidase
PEPc
Long staple length
1 Introduction
Cotton is widely recognized as a prominent cash crop in many regions around the globe, and has a position of considerable economic importance as one of the foremost crops worldwide. The cultivation of this crop is crucial for countries that mostly manufacture textiles from natural fibers (Jan et al., 2022). Cotton fiber plays pivotal role in making clothes and yarn for local and commercial textile industries. It is also used for extracting oil after the crushing of cotton seeds (Lee and Fang, 2015; Ute et al., 2019). The unique fibers of cotton are attributed to the presence of seed trichomes, making it a singular botanical specimen. Given that cotton fibers are generated from individual epidermal cells inside the ovule, they provide a very suitable model system for investigating this process (Arpart et al., 2004; Avci et al., 2013; Shahzad, 2021; Tian and Zhang, 2019). A solitary epidermal cell resembling a trichome elongates to become a cotton fiber with an approximate length of one millimeter, without undergoing division. The use of cotton fiber as a model system for investigating the phenomenon of cell elongation in the absence of mitosis is justified by its natural high rate of elongation at the individual cell level (Ruan et al., 2001).
Due to the long fiber length, cotton is very important for both trade and industry purposes and has been used for years across the globe. It is the most popular plant for its use in clothing and decoration due to its porosity and decoration aesthetics (Iqbal et al., 2016). It is further used in local industries due to its staining, simply washed and absorbency properties.
The expansion of fiber cells plays a critical role in facilitating the elongation of fibers, since it governs the augmentation of staple length (Beasley and Ting, 1973). The literature outlines five discrete but interrelated stages involved in the formation of cotton fiber. The process may be divided into five distinct phases. The first stage involves the development of fiber cells. Following this, the second stage entails the synthesis of the primary cell wall, which is characterized by fast elongation. The third stage marks the transition from primary to secondary cell wall synthesis. Subsequently, the fourth stage involves the biosynthesis of the secondary cell wall. Finally, the fifth step signifies the attainment of maturity (Qin et al., 2019; Tian and Zhang, 2019). All phases are very important because they fastened the expansion and elongation rate of fiber cells, results in the increase in staple length and other fiber characteristics. The elongation stage is more critical because it contributes the ultimate staple length. The process of cell wall expansion starts immediately after the cell initiation stage with the deposition of cellulose, hemicellulose fibrils and other water soluble minerals (Martínez-Sanz et al., 2017; Kim et al., 2018). This rapid elongation phase starts with the breakdown non covalent bonds in between the developing fiber cells with the activation of expansins, xyloglucans endotransglycolyses/hydrolyase and other vacuolar invertases genes. The process of cotton fiber elongation and Arabidopsis thaliana shoot elongation has been shown to be regulated by vacuolar invertases, expansins, and xyloglucans endotransglycolyses/hydrolyase, as reported in recent studies (Benitez-Alfonso et al., 2013; Fan et al., 2022; Fang, 2018). The complex mechanisms of cotton fiber elongation and growth are influenced by a multitude of genes and pathways (Han et al., 2016; Jan et al., 2022; Lee et al., 2010; Levy et al., 2007; Liu et al., 2015; Naoumkina et al., 2016; Pu et al., 2008; Ruan et al., 2001; Saladié et al., 2006; Song et al., 2018; Fang, 2018; Li et al., 2017).
The investigation of the impacts of plasmodesmata-dependent mechanisms on turgor pressure and cell elongation may be conducted in cotton fibers. The role of plasmodesmata has been identified as the primary factor contributing to a notable augmentation in cotton staple length (Li et al., 2021). During the process of fiber growth, plasmodesmata experience a change in permeability, shifting from an open state during the period of 0–9 days post-anthesis (DPA) to a closed state between 10 and 15 DPA, and then returning to an open state from 16 DPA onwards. The emergence of this pattern may be influenced by the specific cultivars and species of cotton (Hernández-Hernández et al., 2023). Plasmodesmata serves as the initiators of intercellular interactions. These structures promote the intercellular movement of many chemicals, such as transcription factors, hormones, and carbohydrates, therefore establishing symplasmic continuity within a tissue.
Multiple genes and pathways are involved in the intricate processes of cotton fiber elongation and growth (Jan et al., 2022). A large number of genes/transcriptional factors (i.e., MYB-MIXTA, GhACT_LI1, GhACTIN1, GhADF1, GhFIM2, and GhPFN2; the tubulin-related GhTUA9; and the kinesin-encoding GhKCH1) have been identified in Arabidopsis (a model plant) and other plants which are appeared to have evolved unique roles in fiber elongation and cell wall development. The application of exogenous hydrogen peroxide (H2O2) in wild-type (WT) plants resulted in an elevation in the synthesis of GhCaM7 and early elongation of fibers. Conversely, the overexpression of GhCaM7, a calmodulin protein found in cotton, induced higher levels of reactive oxygen species (ROS) and facilitated the advancement of fiber elongation at an earlier stage (Tang et al., 2014; Cheng et al., 2016; Hernández et al., 2023).
Genes involved in cell wall synthesis, such as expansin and xyloglucan endotransglucosylase (XTH) were identified during the rapid elongation stage of cell wall development in both upland and Pima cotton. The process of cell growth in acidic conditions is facilitated by expansins proteins, as shown by the research conducted by McQueen-Mason et al. (1992). A significant number of GhEXPA1 copies were identified in cotton fiber throughout various stages of growth (Harmer et al., 2002). The enzymatic process of cell wall cellulose formation necessitates the presence of sucrose synthase (SuS), an enzyme responsible for facilitating the conversion of sucrose into fructose and UDPglucose. The SuS gene has significant promise in enhancing the quality of cotton fibers, since previous research has shown that the expression of the SuS gene leads to modifications in both the structure and quality of cotton fibers. The promotion of fiber elongation is facilitated by the upregulation of SuS gene expression, which in turn increases turgor pressure inside the fiber cells. Glucose and fructose, which are types of carbohydrates, are carried into the fiber tissues (Ahmed et al., 2020).
Real time polymerase chain reaction (RT-PCR) analysis is very important technique to explore the transcript level of tissue and time-based genes at small level in a large variety of crops. In cotton, reverse transcriptase RT-PCR analysis is used to identify the transcripts of a potential gene related to long staple length (Qin et al., 2019). This profiling technique is very helpful to identify the novel genes and their specific regulatory regions for the long fiber (Imran and Liu, 2016; Li et al., 2017). After the concluded transcriptional profiling data, we can examine the genes related to long staple length along with their activation and expression stage during the fiber development. After that, the selected genes can be used to enhance the fiber propertied especially fiber length through the process of transformation into the commercially cultivated varieties.
In the present study, three lines of upland cotton (G. hirsutum L.) were used to see the expression of three genes PEPc, XTH and GA-20 Oxidase during the different fiber morphogenesis phases (0, 05, 10, 15 and 20 DPA).
Because cotton fiber length is a key factor for yarn quality. Our work may help understanding the mechanisms behind the regulation of long fiber and their respective genes expression into the commercially cultivated cotton varieties. This would help in the future breeding venture. The simple and basic intention of this study to find out the novel genes with high expression during the different stages of cotton fiber development. After the expression profiling, transformed these genes through a specific breeding program to develop a transgenic cotton variety with long fiber qualities.
2 Materials and methods
2.1 Plant material
Non-transgenic cotton varieties with a different range of fiber length i) 'CIM-707' ii) 'CYTO-179' iii) 'CIM-616' were selected for planting in the field of MNS- University of Agriculture, Multan during the cotton growing season 2022–23. Seeds of these cotton varieties were collected from Central Cotton Research Institute Multan, Punjab, Pakistan. Here, 'CIM-707' is a long staple length, 'CYTO-179' is a medium staple length and 'CIM-616' is a short staple length genotype and used to calculate the transcript abundance of long staple length related genes through RT-PCR. Fiber tissues were collected from the fully matured cotton plants in the field. All selected plants with cotton bolls were tagged before the anthesis. The main purpose of tagging to select the plants at their accurate stages i.e., 0, 5, 10, 15 and 20 DPA of fiber development.
2.2 RNA extraction
Total RNA was extracted and purified from the bolls of cotton plants at different i.e., 0, 05, 10, 15, and 20 DPA stages of fiber development. All samples (bolls) were washed with DEPC (0.01 % w/w) treated water to avoid any adulteration and contamination. All samples were collected and preserved in liquid nitrogen cylinder at −196℃ for total RNA extraction and expression analysis in the laboratory (Khatoon et al., 2018; Nadeem et al., 2021; Iqbal et al., 2016). The determination of purity was conducted by using a Nanodrop ND-1000 spectrophotometer, while the assessment of RNA quality was performed using a 0.8 % agarose gel that was stained with ethidium bromide at a concentration of 0.5–1 mg/mL, as described by Ahmed et al. (2020). Ethidium bromide was used for running the buffer during the separation of DNA fragments by agarose gel electrophoresis. RNA extraction from fiber samples was performed using a plant RNA extraction reagent (Invitrogen, USA), following a previously established protocol as detailed in the studies conducted by Iqbal et al. (2016) and Nadeem et al. (2021).
One µl of 1 mM dNTPs, one µl of Taq polymerase, one ul of 10 mM primer, and twelve µl of 10x PCR buffer, and 1 µl 10 mM primer 2 were used to prepare PCR master mixture. To make a final volume of 25 µl, 1 µl of DNA template (25 ng) and 6 µl ultra-pure (U.P) water were added. The U.P water is free from any microbial activity, used in RNA extraction and cDNA synthesis. The cycle conditions for the PCR were as follows: 5 min of initial denaturation at 95 °C, 30 s of further denaturation at 95 °C, 60 s of annealing at 57 °C, 30 s of extension at 72 °C, and 7 min of final extension at 72 °C. Following analysis, a melt curve was created and held at 95 °C for 30 s.
2.3 First strand cDNA synthesis
The whole RNA extract was purified with the use of DNase1 (1 mg). A quantity of one milligram of RNA was used as the template for the process of first-strand cDNA synthesis, using a commercially available cDNA synthesis kit (Fermentas cat#1622). Following a synthesis period of 30 min at a temperature of 42 °C the first strand of complementary DNA (cDNA) was subjected to an incubation period of 10 min at a temperature of 70 degrees Celsius. The cDNA was then used at a concentration of 2 µl per 25 µl in a reaction. A 10 µl of RNA sample of all fiber developmental phases (0, 5, 10, 15 and 20 DPA) was used to transcribe first strand cDNA. First strand of cDNA was reverse transcribed with the usage Revert Aid Reverse Transcriptase. All three fiber-related genes (PEPc, XTH and GA-20 Oxidase) transcript expression was determined by qRT-PCR (BioRad iCycler iQ5). The expression levels were normalized with an internal control ubiquitin. The RT-PCR reaction mixture (25 µl) consisted of SYBR green supermix (12.5 µl), forward primer (1 µl), reverse primer (1 µl), template cDNA (3 µl), and U.P water (7.5 µl).
The sequence of cotton fiber length related genes (PEPc, XTH and GA-20 Oxidase) Gene Bank Accession number (EU032328.1, EF546794 and AY895169) was retrieved from the National Centre for Biotechnology Information (NCBI) GenBank database. The primers were designed by using online available program Primer 3 (Premier Biosoft International, USA and AmplifX (Jullien, 2013) were used (Iqbal, 2017). An internal control gene ubiquitin with gene bank accession number (CF932135) was used as reference gene for RT-PCR analysis. The gene-specific primers F PEPc (5′ CACCGACCTACTACACGAGGTGTG 3′) and R PEPc (5′ AGAAGCCTCAAAAGGCATTCCTTG 3′), F XTH (5′ CCAAAATTCAGGCTGTGGAT 3′) R XTH (5′ TTGTTCCCTGTCACCCTTTC 3′), F GA-20 Oxidase (5′ CTTGCTTGGGGACTCTCTTG 3′) R GA-20 Oxidase (5′ ACGAAACTGCTTGCATACCC 3′) and an internal control F ubiquitin (5′ TGAATATTGTAATCAGCC 3′) R ubiquitin (5′ GAGCTCGGATACGATTGA 3′) were used to analyze mRNA transcript expression.
For the calculation of relative gene expression level, 2^△△ Ct method was used. By using this formula the temperature of a specific PCR cycle was calculated by taking the difference of temperature of interest gene to the temperature of reference gene (Ct interest gene -Ct reference gene). Here “Ct” is the cycle temperature. These values were calculated after the running of all fiber samples with their respective primers in a 96 welled plate.
3 Results
The objective of this study was to assess the relative expression levels of three genes associated with fiber formation at different time points (0, 05, 10, 15, and 20 days post-anthesis) in upland cotton. The genes under consideration included Ubiquitin, GA-20 oxidase, PEPc, and XTH. The relative gene amplification and their expression was observed at five different stages of fiber development, i.e., i) fiber cell wall initiation, ii) rapid elongation or primary cell wall synthesis, iii) transition between primary cell wall synthesis and secondary cell wall synthesis, iv) secondary cell wall synthesis (SCW) and v) maturation. Expression analysis was conducted to check the transcripts of three gene families (PEPc, GA-20 Oxidase and XTH) in fiber tissues through reverse transcriptase RT-PCR (Table1). Approximately the quantity of total RNA was in ranged of 0.2 µg to 0.5 µg of all fiber samples (Fig. 1). A dilution series of DEPC treated water (0.01 %) was used to balance the concentration of all fiber samples (Fig. 2). The initial step in generating cDNA involved the synthesis of the first strand using an enzyme called reverse transcriptase. Subsequently, the extracted RNA was subjected to evaluation through separation on a 0.8 % agarose gel and staining with ethidium bromide at a concentration of 0.5–1 mg/mL. The purity of the RNA was further verified using a Nanodrop ND-1000 spectrophotometer (Fig. 3). For PCR data normalization, an internal control (Ubiquitin) is used with all long staple length related genes in RT-PCR (Fig. 4). For the achievement of stable level of expression, all three long staple genes primer pairs were validated with a 10 times dilution series of template concentrations (Fig. 5).
Name of gene
Primer pair
Primer sequence (5′-3′)
Primer Length
Amplicon size (bp)
Gene bank accession No.
Ubiquitin
RTUBCF
RTUBCRTGAATATTGTAATCAGCC
GAGCTCGGATACGATTGA18 mer
18 mer132
CF932135
PEP carboxylase
RTPEPCF
RTPEPCRCACCG ACCTACTACACGAGGTGTG
AGAAGCCTCAAAAGGCATTCCTTG24 mer
24 mer227
EU032328
GA 20 Oxidase
GA20F
GA20RCTTGCTTGGGGACTCTCTTG
ACGAAACTGCTTGCATACCC20 mer
20 mer219
AY895169
XTH
RTXTHF
RTXTHR1CCAAAATTCAGGCTGTGGAT
TTGTTCCCTGTCACCCTTTC20 mer
20 mer231
EF546794
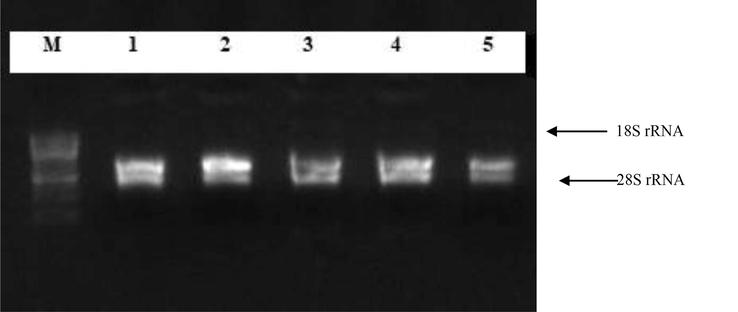
Total RNA isolation from different stages of cotton fiber development. M 1 Kb ladder; Lane 1–5 total RNA extracted from five phases of three long staple length genotypes of cotton (Lane 1: 0 DPA fibers, Lane 2: 5 DPA fibers, Lane 3: 10 DPA fibers, Lane 4: 15 DPA fibers, Lane 5: 20 DPA fibers). This figure is created with Gel Documentation System.
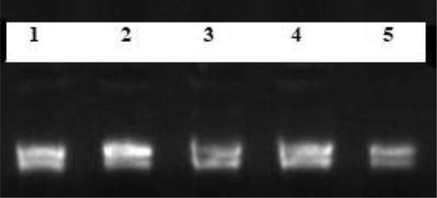
Calibration of total RNA concentration in different fiber tissues. Lanes (1–5) illustrate RNA extracted from 0 DPA, 5 DAP, 10 DPA, 15 DPA and 20 DPA fibers respectively. This figure is created with Gel Documentation System.

Synthesis of first strand cDNA from five different phases of fiber synthesis. M- 0.5 bp ladder; Lane 1–10 cDNA synthesized from different stages of cotton fiber development. (Lane 1–5: 0, 5, 10, 15 and 20 DPA fiber of CIM-707 respectively), (Lane 6–8: 5, 10 and 15 DPA fiber of CYTO-179 respectively), (Lane 9–10: 15–20 DPA fiber of CIM-616 respectively). This figure is created with Gel Documentation System.

Equalization and calibration of various templates concentration with 18S rRNA primer through PCR amplification. This figure is created with Gel Documentation System.
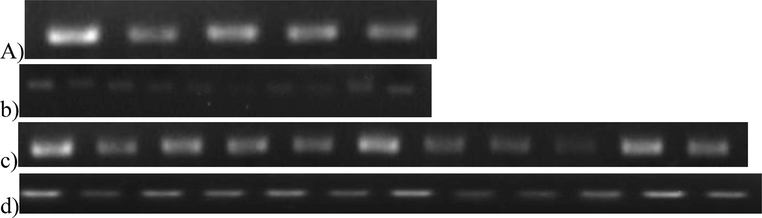
Primer validation and confirmation through a series of fiber cDNA of 10 DPA stage. PCR amplification using primers of (a) 18SrRNA, (b) GA-20 Oxidase, (c) PEPc Carboxylase, d) XTH. This figure is created with Gel Documentation System.
A variable level of expression of PEPc gene was observed in all three lines (CIM-707, CYTO-179 and CIM-616) with different ages of fiber development i.e., 0, 05, 10, 15 and 20 DPA. Transcript analysis results showed that an elevated gene expression of PEPc was observed throughout all fiber developmental stages with different DPAs (Fig. 6). A highest amplification profile was detected in elongation stage (15 DPA) of fiber development in both CIM-707 (long fiber length) and CIM-616 (short fiber length). A stable transcript level was also observed in (medium fiber length) at 5 DPA. Further a stable amplification results of PEPc gene was also perceived in all three genotypes of cotton at early stage of primary cell wall synthesis (10 DPA) fibers. At SCW stage of fiber development (20 DPA) nearly a small (negligible) level of expression was also observed in CIM-616. The expression level of PEPc was about to be doubled at 15 DPA stage in CIM-707 fibers than the others DPAs but the transcript level of PEPc was ultimately dropped at the same stage in both medium and short fiber genotypes.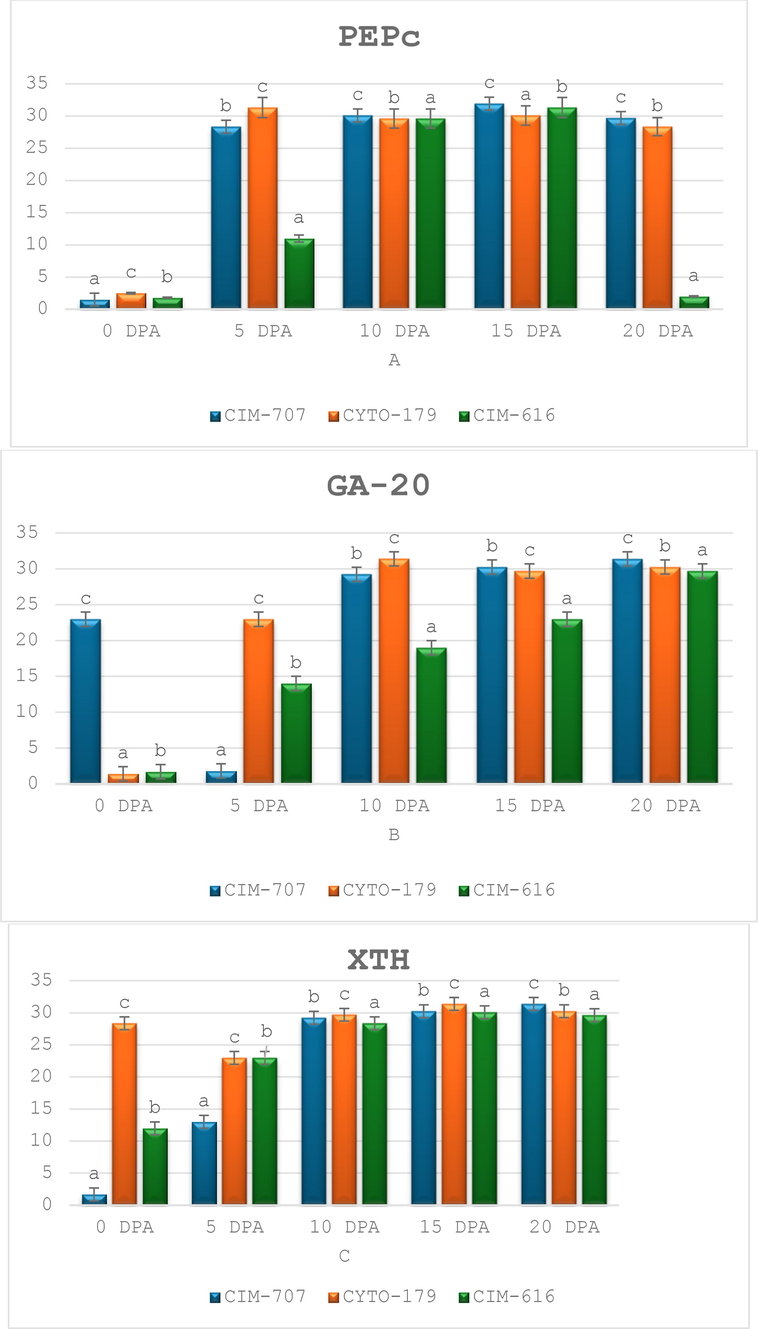
RT-PCR analysis of three fiber related genes a) PEPc, b) GA-20 Oxidase and c) XTH of five different phases i) 0, ii) 05, iii) 10, iv) 15 and v) 20 DPA of fiber development in cotton. With the use of an internal control (18S rRNA), all transcript data was equalized and normalized. This figure is created with MS Excel.
A variable transcript pattern of Oxidase gene (GA-20) was detected at the stages of fiber cell wall synthesis. At rapid elongation stage of fiber expansion, a stable transcript pattern of GA-20 Oxidase was detected in medium staple length genotype. A stable and continuous amplification results of GA-20 Oxidase discovered that the transcript pattern of Oxidase gene was exist at the rapid elongation phase of primary cell wall synthesis at 15 DPA in all three genotypes of upland cotton (Fig. 6). A higher transcript level of GA-20 Oxidase was recognized at primary cell wall synthesis stage at 10 to 15 DPAs and then ultimately declined to SCW stage at 20 DPA and onward. The transcripts of Oxidase gene were about to be doubled at the rapid elongation phase at15 DPA as compared to the fiber cell initiation phase at 0 DPA. The transcript pattern of GA-20 Oxidase showed, a higher level of expression was detected in both genotypes CIM-707 and CYTO-179 at 20 DPA and 10 DPA respectively. In CIM-707, a variable and unstable level of expression of GA-20 Oxidase was noticed at both 0 and 5 DPA stages. Many transcript patterns of GA-20 Oxidase were also detected in CYTO-179 as compared to other lines of upland cotton.
With the passage of time, the activities and expression levels of long staple length related genes also increased, results in the development of long fiber cells. At stage of rapid elongation at 15 DPA, all fiber length genes are fully active especially XTH. Expression profiling results showed that a stable and similar transcript pattern of XTH gene was detected at first two stages of fiber development at 0 and 5 DPA. In CYTO-179, an elevated expression level of XTH gene was found in between fiber cell wall synthesis and rapid elongation stages of fiber development at 10, 15 and 20 DPA. The RT-PCR results showed that the expression of XTH gene was much similar in both long and short staple length genotypes at fiber cell initiation stage at 0 DPA and SCW stage at 20 DPA fibers.
4 Discussion
Fine quality and long staple length fibers are very acceptable for the textile industry. Universally, long staple length fibers are used as raw material for fabrics production in the textile industry. Many long staple length related genes including MYB gene family are activated, results in the long fiber development around the seed trichomes (Shahzad, 2021). Identification and selection of long staple length related genes is very helpful to produce new plants with high fiber length. Transcript analysis of cotton staple length related genes is very helpful to discover the transcripts of three different fiber development related genes at different phases of fiber morphogenesis (Qin et al., 2019).
Many long staple length genes are highly expressed throughout the diverse phases of cotton fiber synthesis (Arpart et al., 2004). Most of them are found to be involved during the fiber expansion and its enlargement, results in the long fiber development. In literature, most of the fiber-related genes including EXP, SUS, LTPs, PEPc, tubulins, GA-20 Oxidase and XTH are believed to be compulsory for the long fiber development (Li et al., 2005; Huang et al., 2013). The successful identification and selection of long staple length related genes along with their upstream regulatory regions (promotors) was done only with expression profiling techniques.
The reverse transcriptase RT-PCR profiling is a very important technique to explore the transcript level of tissue and time-based genes at small level in a large variety of crops. In cotton, reverse transcriptase RT-PCR analysis is used to identify the transcripts of a potential gene related to long staple length. This profiling technique is very helpful to identify the novel genes and their specific regulatory regions for the long fiber (Naoumkina et al., 2016; Li et al., 2017). The transcript analysis data may also help in the investigation and confirmation of the long staple length-related genes along with their stages of activation and amplification throughout the whole process of fiber development. After that, the selected genes can be used to enhance the fiber propertied especially fiber length through the process of transformation into the commercially cultivated varieties (Qin et al., 2019; Ruan et al., 2001, 2004). After the expression profiling analysis, we can change and alter the fiber properties especially fiber length through the overexpression or knockdown mechanisms of fiber-related genes into a commercially cultivated variety, results in the development of a new transgenic plant with long fiber properties (Beasley and Ting, 1973; Cheng et al., 2016; Fang, 2018).
A variable level of expression of PEPc gene was observed in all three lines (CIM-707, CYTO-179 and CIM-616) with different fiber morphogenesis phases i.e, 0, 05, 10, 15 and 20 DPA. Transcript analysis results showed that an elevated gene expression of PEPc was observed throughout all fiber developmental stages with different DPAs (Fig. 6). A highest amplification profile was detected in elongation stage (15 DPA) of fiber development in both CIM-707 (Long fiber length) and CIM-616 (Short fiber length). A stable transcript level was also observed in (Medium fiber length) at 5 DPA. Further stable amplification results of PEPc gene was also detected in all three lines with different fiber staple length (Long, medium and small) at 10 DPA fibers. At SCW (20 DPA) phase of fiber expansion, nearly a small (negligible) transcript pattern was also observed in CIM-616. The expression level of PEPc was about to be doubled at 15 DPA stage in CIM-707 fibers than the others DPAs but the transcript level of PEPc was ultimately dropped at the same stage in both medium and short fiber lines (CYTO-179 and CIM-616).
A variable transcript pattern of Oxidase gene (GA-20) was detected at all fiber synthesis phases. The highest level of expression of GA-20 Oxidase of fiber elongation gene was detected at 10 DPA stage of fiber morphogenesis in CYTO-179. A stable level of transcript and continuous amplification of GA-20 Oxidase discovered that the transcripts of Oxidase gene were noticed throughout the elongation phase (15 DPA) of fiber expansion in all three fiber length genotypes (Fig. 6). The highest level of GA-20 Oxidase transcript was identified at 10 to 15 DPAs and then declined to 20 DPA (SCW stage). The transcripts of this gene were about to be double at elongation phase than initiation phase. The transcript pattern of GA-20 Oxidase showed highest expression in both lines CIM-707 and CYTO-179 at 20 DPA and 10 DPA respectively. A variable and unstable level of expression of GA-20 Oxidase was detected in all three lines at first two stages including 0 and 5 DPA but was much higher at 0 DPA in CIM-707. Many transcript patterns of GA-20 Oxidase were also detected in CYTO-179 as compared to other genotypes of upland cotton (Li et al., 2009).
With the increase in days post anthesis (5 to 20 DPA) stages, the level of expression of this fiber-related gene was also increased but at 15 PDA fibers the level of expression was very high. The expression profiling results showed that there is a similar transcript of XTH gene were also detected in all three lines of cotton. Expression profiling results showed that in both CIM-707 and CIM-616 cotton fiber lines, a stable and similar transcript level of XTH gene was detected in first two stages (0 DPA and 05 DPA) of fiber development. In CYTO-179, a non-constant transcript level of XTH gene was found at rapid elongation stage at 10, 15 and 20 DPA in cotton genotypes. Transcripts of XTH genes were discovered in CYTO-179 at 15 DPA fibers but a low transcript level was observed at 5 DPA fibers. The RT-PCR results showed that the expression of XTH gene was much similar in both CIM-707 and CIM-616 at 0 DPA and 20 DPA fibers. Real Time PCR results showed that a variable level of XTH transcripts were detected in all three staple length genotypes in cotton.
5 Conclusions
From the above discussion it is determined that three long staple length related genes exhibited variable expression throughout the different levels of fiber development in cotton fiber tissues. All three staple length related genes are highly expressed at fiber cell initiation stage at 5 DPA, primary cell wall synthesis or rapid elongation stage at 10 and 15 DPA fibers. The comparative expression studies proved that all of these fiber developing genes showed a variable but stage specific expression in all fiber tissues. Highly expressed staple length related genes can also be used for changing the fiber characteristics through the process of gene transformation. The comparative expression study on fiber developing genes can be used to enhance the fiber length of cotton commercial cultivars to support the national economy.
CRediT authorship contribution statement
Muhammad Nadeem: Conceptualization, Funding acquisition, Data curation, Writing – original draft, Writing – review & editing, Investigation. Muhammad Shahzad: Conceptualization, Data curation, Writing – review & editing, Investigation. Zulqurnain Khan: Conceptualization, Funding acquisition, Writing – review & editing, Formal analysis, Methodology, Project administration, Software. Umar Akram: Conceptualization, Data curation, Writing – review & editing, Visualization, Investigation, Methodology. Muhammad Hasnain: Conceptualization, Data curation, Writing – review & editing, Visualization, Investigation, Validation. M. Ajmal Ali: Conceptualization, Funding acquisition, Writing – review & editing, Validation, Methodology. Mohamed Soliman Elshikh: Conceptualization, Funding acquisition, Writing – review & editing, Validation, Methodology. Furqan Ahmad: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing, Validation, Formal analysis, Methodology, Supervision, Resources, Project administration, Software.
Acknowledgements
The authors are thankful to SINO-PAK Joint Laboratory, Institute of Plant Breeding and Biotechnology, MNS University of Agriculture, Multan, Pakistan, for support from ADP Funded Project entitled National Crop Genomics and Speed Breeding Center for Agricultural Sustainability (ADP-LO21002838 Punjab, Pak). The authors extend their appreciation to the Researchers supporting project number (RSP2024R306), King Saud University, Riyadh, Saudi Arabia.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Overexpression of a Sucrose Synthase Gene Indirectly Improves Cotton Fiber Quality Through Sucrose Cleavage. Front. Plant Sci. 2020:11:476251.
- [Google Scholar]
- Functional genomics of cell elongation in developing cotton fibers. Plant Mol. Biol.. 2004;54:911-929.
- [Google Scholar]
- Cotton Fiber Cell Walls of Gossypium hirsutum and Gossypium barbadense Have Differences Related to Loosely-Bound Xyloglucan. PLoS One. 2013;8
- [CrossRef] [Google Scholar]
- Effects of plant growth substances on in vitro fiber development from fertilized cotton ovules. Am. J. Bot.. 1973;60:130-139.
- [Google Scholar]
- Symplastic intercellular connectivity regulates lateral root patterning. Dev. Cell. 2013;26:136-147.
- [Google Scholar]
- GhCaM7-like, a Calcium Sensor Gene, Influences Cotton Fiber Elongation and Biomass Production. Plant Physiology and Biochemistry. 2016;109:128-136.
- [CrossRef] [Google Scholar]
- The Plasmodesmata-Located-1,3-Glucanase Enzyme PdBG4 Regulates Trichomes Growth in Arabidopsis thaliana. Cells. 2022;11:2856.
- [Google Scholar]
- Fang, D.D. Cotton Fiber: Physics, Chemistry and Biology; 2018; ISBN 9783030008710.
- Isolation and Characterization of Two Persimmon Xyloglucan Endotransglycosylase/Hydrolase (XTH) Genes That Have Divergent Functions in Cell Wall Modification and Fruit Postharvest Softening. Front Plant Sci. 2016;7:1-12.
- [CrossRef] [Google Scholar]
- Characterization of six α-expansin genes in Gossypium hirsutum (upland cotton) Mol. Genet. Genom.. 2002;268:1-9.
- [Google Scholar]
- Hernández, V.; Olivier, C. M.; Annamaria, K.; Arezki, B. A mechanohydraulic model supports a role for plasmodesmata in cotton fiber elongation. 2023.
- A fasciclin-like arabinoGAlactan protein, GhFLA1, is involved in fiber initiation and elongation of cotton (Gossypium hirsutum) Plant Physiol.. 2013;161:1278-1290.
- [Google Scholar]
- Genome-Wide Identification and Expression Analysis of the Malate Dehydrogenase Gene Family in Gossypium arboreum. Pak J Bot. 2016;48:1081-1090.
- [Google Scholar]
- Expression profiling of cotton fiber genes and evaluation of some regulatory sequences for gene expression in Dicots. Islamabad, Pakistan: Dept. Biotech. Univ. Quaid-i-Azam; 2017. Ph.D. Thesis
- Expression Analysis of Fiber Related Genes in Cotton (Gossypium hirsutum L.) through Real Time PCR. Pak J Bot. 2016;48:1099-1106.
- [Google Scholar]
- Molecular Regulation of Cotton Fiber Development: A Review. Int. J. Mol. Sci.. 2022;23:5004.
- [Google Scholar]
- Jullien, N. AmplifX Version 1.7.0. Available on-line with updates at http://crn2m.univ-mrs.fr/pub/amplifx-dist. 2013.
- Comparative Analysis of Fiber Morphogenesis Genes of Calotropis procera and Gossypium hirsutum. Int J Agric Biol. 2018;20:288-296.
- [CrossRef] [Google Scholar]
- Comparison and Validation of Fourier Transform Infrared Spectroscopic Methods for Monitoring Secondary Cell Wall Cellulose from Cotton Fibers. Cellulose. 2018;25:49-64.
- [CrossRef] [Google Scholar]
- Xyloglucan Endotransglycosylase/Hydrolase Genes in Cotton and Their Role in Fiber Elongation. Planta. 2010;232:1191-1205.
- [CrossRef] [Google Scholar]
- Cotton as a World Crop: Origin, History, and Current Status. Cotton. 2015;57:1-23.
- [CrossRef] [Google Scholar]
- A plasmodesmata-associated_-1, 3-glucanase in Arabidopsis. Plant J.. 2007;49:669-682.
- [Google Scholar]
- Genome-Wide Identification and Characterization of TCP Transcription Factor Genes in Upland Cotton (Gossypium hirsutum) Sci. Rep.. 2017;7:1-14.
- [Google Scholar]
- Intercellular trafficking via plasmodesmata: molecular layers of complexity. Cell. Mol. Life Sci.. 2021;78:799-816.
- [Google Scholar]
- The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell. 2005;17:859-875.
- [Google Scholar]
- Developmental and molecular physiological evidence for the role of phosphoenol pyruvate carboxylase in rapid cotton fiber elongation. J. Exp. Bot.. 2009;61:287-295.
- [Google Scholar]
- Gossypium barbadense Genome Sequence Provides Insight into the Evolution of Extra-Long Staple Fiber and Specialized Metabolites. Sci Rep. 2015;5:1-14.
- [CrossRef] [Google Scholar]
- Structure of Cellulose Microfibrils in Mature Cotton Fibres. Carbohydr Polym. 2017;175:450-463.
- [CrossRef] [Google Scholar]
- Two endogenous proteins that induce cell wall expansion in plants. Plant Cell. 1992;4:1425-1433.
- [Google Scholar]
- Comparative expression studies of fiber related genes in cotton spp. J. Biores. Manag.. 2021;8:1.
- [Google Scholar]
- RNA-Seq Analysis of Short Fiber Mutants Ligon-Lintless-1 (Li 1) and - 2 (Li 2) Revealed Important Role of Aquaporins in Cotton (Gossypium hirsutum L.) Fiber Elongation. BMC Plant Biol. 2016;15:1-14.
- [CrossRef] [Google Scholar]
- The R2R3 MYB Transcription factor GhMYB109 is required for cotton fiber development. Genetics. 2008;180(2):811-820.
- [Google Scholar]
- Transcriptome Analysis Reveals Differences in the Mechanisms of Fiber Initiation and Elongation between Long- and Short-Fiber Cotton (Gossypium hirsutum L.) Lines. BMC Genomics. 2019;20:1-16.
- [CrossRef] [Google Scholar]
- The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell. 2001;13:47-63.
- [Google Scholar]
- Genotypic and Developmental Evidence for the Role of Plasmodesmatal Regulation in Cotton Fiber Elongation Mediated by Callose Turnover. Plant Physiol. 2004;136:4104-4113.
- [CrossRef] [Google Scholar]
- Characterization of a New Xyloglucan Endotransglucosylase/Hydrolase (XTH) from Ripening Tomato Fruit and Implications for the Diverse Modes of Enzymic Action. Plant Journal. 2006;47:282-295.
- [CrossRef] [Google Scholar]
- A Review on Role of Trichomes in Plant Physiology and Genetic Mechanism Involved in Trichome Regulation in Cotton. Pure and Applied Biology. 2021;10:458-464.
- [CrossRef] [Google Scholar]
- Characterization of the XTH Gene Family: New Insight to the Roles in Soybean Flooding Tolerance. Int J Mol Sci. 2018;19
- [CrossRef] [Google Scholar]
- Tang, W.; Tu., L.; Yang, X.; Tan, J.; Deng, F.; Hao, J.; Guo, K.; Lindsey, K.; Zhang, X. The calcium sensor GhCaM7 promotes cotton fiber elongation by modulating reactive oxygen species (ROS). New Phytologist 2014, 202, 509–520.
- MIXTAs and phytohormones orchestrate cotton fiber development. Curr. Opin. Plant. Biol.. 2019;59:101975
- [Google Scholar]
- Ute T.B., Celik P., Uzumcu M.B. Utilization of Cotton Spinning Mill Wastes in Yarn Production. Text. Indus. Envir. Intech Open. 2019.







