Translate this page into:
Exploring the impact of paracetamol on the transcriptome of Streptococcus pneumoniae D39
⁎Corresponding authors. sulman.shafeeq@ki.se (Sulman Shafeeq), mushahid@ksu.edu.sa (Shahid Mahboob)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Introduction
Paracetamol (acetaminophen) is a medicine used for the treatment of fever, pain, and inflammation during pneumococcal infection.
Objectives
To see how paracetamol affects the transcriptional profile of Streptococcus pneumonia across the genome.
Methods
In this study, microarray analysis was performed for transcriptional profiling.
Results
Transcriptome data showed differential expression of several genes in S. pneumoniae D39 wild-type incorporated with paracetamol in the growth medium. Furthermore, these genes were categorized using Clusters of Orthologous Groups (COG) functional categorization on the basis of the suspected functions of the respective proteins. The majority of differentially expressed genes are in COG categories E (Amino acid transport and metabolism) and I (Lipid transport and metabolism). Analysis of protein–protein interaction networks exhibited compactly connected networks between fatty acid transport/biosynthesis and antibiotic biosynthesis genes. Moreover, pathways enrichment analysis revealed that fatty acid metabolism and biosynthesis genes were significantly affected under the criteria we've established.
Conclusion
These results suggest the fatty acid biosynthesis and metabolism genes to be potential target of paracetamol in S. pneumoniae D39 wild-type.
Keywords
Pneumococcus
Paracetamol
Transcriptomics
Protein-protein interaction networks
Pathway enrichment analysis
1 Introduction
An opportunistic pathogen of humans, Streptococcus pneumoniae, inhabits the mucosa of nasal portions and causes sinusitis, otitis media, sepsis, meningitis and pneumonia which results about a million people deaths every year worldwide (O'Brien et al., 2009). Multi-resistant pneumococcal strains’ emergence and dissemination has been on the rise since mainstream drugs usage worldwide (Kim et al., 2016). Generation of resistant pneumococcal clones results through adaptation to drug pressures enforced although they reside within the human upper respiratory tract (Kim et al., 2016). Most pneumococcal antimicrobial resistances have basic causal factors that have been uncovered (Kim et al., 2016). The escalating rates of resistance to antibiotics to all currently available treatments, along with an almost empty pharmacological pathways for new drugs, has caused a panic in drug discovery efforts around the world (Brockhurst et al., 2019). Though different drugs have shown potency against most pneumococcal infections, the existing options are limited against some pneumococcal isolates (Kim et al., 2016).
Several factors are deemed liable for the development of resistance among intrusive pneumococcal disease cases including modern drug usage (foremost risk factor) (Levine et al., 1999), age (predominantly children below 5 years of age) and pediatric serotypes (serotypes found commonly in children), hospitalization, attending day care, female gender, living in an urban area, HIV infection and immunosuppression (Levine et al., 1999). At present, >40 % of pneumococcal isolates lack significant conjugate vaccine coverage and are penicillin resistant in several countries (Torné et al., 2014). Paracetamol is deemed as the first-line remedy for an acute sore throat. Nonetheless, in primary care, antibiotics are still normally prescribed as first-line treatment for sore throat. Therefore, we believe that it would be very interesting to give a try to a less-problematic and routinely used medication paracetamol (acetaminophen/APAP) and investigate the response of pneumococcus to paracetamol at molecular level. Moreover, paracetamol has been shown to enhance biofilm formation in human pathogen Staphylococcus aureus. This research elucidates the impact of paracetamol on the whole transcriptome of pneumococcus and highlights the putative targets of paracetamol in pneumococcus by differential network analysis and pathway enrichment analysis.
2 Methods
2.1 Bacterial strains and growth experiments
S. pneumoniae D39 wild-type strain was used for our research. Growth of S. pneumoniae D39 was performed as mentioned before (Afzal et al., 2015). S. pneumoniae from −80 °C stocks were first plated on blood agar plates overnight. Next day, bacteria were taken from the blood agar plates and inoculated in GM17 (0.5 % glucose + M17) and grown overnight. Fresh cultures from the overnight grown bacteria were used for our experiments.
2.2 RNA extraction, cDNA preparation and hybridization
Wild-type S. pneumoniae D39 was grown in chemically defined media (CDM) (in replicates) with 0 and 5 mM paracetamol were used for microarrays analysis. Paracetamol was purchased from Sigma Aldrich. For RNA isolation, cells in their corresponding mid-exponential phase (grown for about 6–7 h) were harvested. The extraction of RNA and preparation of cDNA was executed as elucidated before (Afzal et al., 2015). All other aspects of the microarray experiment were carried out as depicted earlier (Afzal et al., 2015).
2.3 Microarray data analysis
The microarray acquisition and analysis software, GenePix® Pro 6, was utilized to perform a pre-analysis on spotted microarray slides. The data was then normalized and processed using the Microprep software program, which was developed in-house (van Hijum et al., 2003). Statistical analysis were executed as depicted previously (van Hijum et al., 2005). Cyber-T integration of a variant of t-test was performed (https://bioinformatics.biol.rug.nl/cybert/index.shtml) and FDRs (False Discovery Rates) were measured as mentioned before (van Hijum et al., 2003). A fold shift cut-off 1.5, FDR < 0.05 and Bayesian p-value of < 0.001were applied to categorize differentially expressed genes. PePPER software package was utilized to perform additional computer research on the data in order to forecast regulatory networks and data mining (de Jong et al., 2012).
2.4 Analysis of protein–protein interaction (PPI) network
The protein–protein interaction (PPI) network was built and visualized via STRING with the default threshold of a combined score > 0.4 (Szklarczyk et al., 2017). Moreover, nodes denote biological molecules and the nodes are connected by the edges to show their interaction. The important nodes in the PPI network were identified using their connection degrees.
2.5 Functional enrichment analysis
Functional annotation analysis was executed using the Search Tool for the identification of associated Genes to further investigate biological processes of genes expressing in numerous ways in involvement of paracetamol (Szklarczyk et al., 2017). With p-values < 0.05, GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways were considered enriched.
3 Results
3.1 Transcriptomic response of S. Pneumoniae to paracetamol
Paracetamol is one of the WHO’s (World Health Organization) essential medicines, that are considered to be the most effective and the safest medicines required in a health system and usually used for mild to moderate pain relief (Lee, 2017). It could be used in conjunction with opioid pain medications for intense pain, such as cancer suffering and post-surgery pain (Scottish Intercollegiate Guidelines Network, 2008). In this study, we tested the exposure of the paracetamol on the whole transcriptome of wild-type of S. pneumoniae D39. Comparisons of microarrays of S. pneumoniae D39 grown in CDM with (5 mM) and without paracetamol was performed. A variety of genes/gene clusters was expressed in numerous ways in the involvement of paracetamol (Table 1). 54 genes were positively regulated during involvement of paracetamol, as opposed to negative regulation of 20 genes. Based on the proposed functions of the related proteins, these genes are additionally classified into COG functional classes (Table 2).
aD39 Tag (spd)
bFunction
cRatio
Upregulated genes
0420
Formate acetyltransferase, PflB
1.8
0458
HrcA
1.5
0459
GrpE
1.6
0460
DnaK
1.9
0461
DnaJ
1.5
0458
Heat-inducible transcription repressor HrcA
1.5
0701
CiaR
1.6
0702
CiaH
1.7
0775
Function Unknown
2.4
0868
Protease maturation protein, putative
2.0
0913
Hypothetical protein
2.0
1375
NADPH-dependent FMN reductase, putative
1.7
1402
Non-heme iron-containing ferritin
1.7
1439
Ribosomal protein S15, RpsO
1.6
1506
Acetyl xylan esterase, putative
1.8
1834
Alcohol dehydrogenase, iron-containing
4.2
1932
Maltodextrin phosphorylase, MalP
1.8
1933
4-alpha-glucanotransferase, MalQ
1.6
1965
PcpA
1.5
2033
YfiA
1.5
2069
SpoJ
2.2
Down-regulated Genes
0161
Hypothetical protein
−1.9
0195
rplW
−1.5
0197
rpsS
−1.5
0262
Mannose/fructose/sorbose family PTS system
−1.5
0263
ManM
−1.6
0264
ManL
−1.5
0317
Cps2C
−1.6
0334
AliA
−2.1
0378
Enoyl-CoA hydratase/isomerase family protein
−2.0
0379
MarR family Transcriptional regulator
−1.6
0380
FabH
−1.8
0381
AcpP
−1.9
0382
FabK
−2.2
0383
FabK
−2.2
0384
FabK
−1.9
0385
FabK
−2.2
0386
AccB
−2.3
0387
FabK
−2.3
0388
AccC
−2.0
0389
AccD
−2.1
0390
AccA
−1.9
0404
IlvB
−2.4
0405
IlvN
−2.2
0406
IlvC
−2.4
0407
Function Unknown
−2.3
0408
Function Unknown
−2.3
0409
IlvA
−2.1
0447
GlnR
−2.3
0448
GlnA
−1.8
0646
Function Unknown
−1.6
0655
LivG
−1.5
0686
Efflux transporter
−1.5
0749
IlvE
−1.6
0750
Function Unknown
−1.9
0751
Function Unknown
−1.8
0752
Function Unknown
−1.7
0753
Pcp
−1.5
0900
Asd
−1.6
0901
DapA
−1.6
1098
GlnP
−1.7
1099
GlnQ
−1.5
1158
NADP-specific glutamate dehydrogenase, GdhA
−1.6
1217
Function Unknown
−1.6
1524
Transcriptional regulator, GntR family protein
−2.1
1525
ABC transporter, ATP-binding protein
−2.4
1526
Function Unknown
−3.6
1611
Function Unknown
−1.6
1650
Iron-compound ABC transporter, permease protein
−1.5
1651
Iron-compound ABC transporter, ATP-binding protein
−1.5
1652
Iron-compound ABC transporter, iron-compound-binding protein
−1.6
1671
AmiA
−1.5
1726
Pneumolysin, Ply
−1.5
1728
Function Unknown
−1.6
2045
MreC
−1.6
Functional categories
Total
Up
Down
C: Energy production and conversion
2
–
2
E: Amino acid transport and metabolism
11
11
–
F: Nucleotide transport and metabolism
–
–
–
G: Carbohydrate transport and metabolism
5
3
2
H: Coenzyme transport and metabolism
1
1
–
I: Lipid transport and metabolism
8
8
–
J: Translation, ribosomal structure, and biogenesis
4
2
2
K: Transcription
6
3
3
L: Replication, recombination, and repair
–
–
–
M: Cell wall/membrane/envelope biogenesis
4
4
–
O: Posttranslational modification, protein turnover, chaperones
5
1
4
P: Inorganic ion transport and metabolism
5
4
1
Q: Secondary metabolites biosynthesis, transport, and catabolism
2
1
1
R: General function prediction only
4
2
2
S: Function unknown
5
4
1
T: Signal transduction mechanisms
2
1
1
U: Intracellular trafficking, secretion, and vesicular transport
–
–
–
V: Defense mechanisms
1
1
–
Others
9
8
1
Total number of genes
74
54
20
The expression of glnA-glnR, glnPQ and gdhA was downregulated in the administration of paracetamol. In the vicinity of a nitrogen source, the expression of these genes has been shown to be downregulated (Kloosterman et al., 2006). GlnR regulon genes play a role in pneumococcal pathogenesis, with glnA participating to blood colonization and resilience, and glnP crucial for lung survival and probably essential for effective transfer from the lungs to the blood (Kloosterman et al., 2006). An important iron operon spd-1650–2 (piuABC) was found to be downregulated under our tested conditions. This system is among three major iron transport systems in pneumococcus and codes for an ABC transporter (Brown et al., 2002). This ABC transporter has specific roles in respiratory colonization and disease and is believed to be important for virulence in S. pneumoniae (Kadioglu et al., 2008).
A mannose-specific phosphotransferase system (manLMN) and a couple of genes (malPQ) programming for maltose utilization proteins were also expressed in numerous ways in the involvement of paracetamol. Maltose genes are positively regulated during involvement of paracetamol in the medium, whereas the mannose transporter genes are downregulated. Several gens were upregulated in the presence of paracetamol including a group of genes normatively encoding chaperones and heat-shock proteins and few genes participated in production and conversion of energy. Moreover, some amino acid utilization and transport genes were negatively regulated in the presence of paracetamol in the medium.
fab genes (genes for biosynthesis of fatty acids) were negatively regulated in involvement of paracetamol. A fab gene cluster is located in pneumococcal genome along with another system for synthesis of unsaturated fatty acids and enoyl-ACP reduction (Marrakchi et al., 2002). The study of the regulatory mechanisms and interactions of the fab genes in the involvement of paracetamol is necessary because they are important in modulating lipid homeostasis of the bacterial membrane, and are the potential candidates for new antibacterial therapies. Furthermore, changes in the expression of genes encoding for biosynthesis of fatty acid may result in modifications in the cell membrane that promote cell survival in involvement of paracetamol.
A gene coding for alcohol dehydrogenase (AdhE) was positively regulated during involvement of paracetamol. S. pneumoniae D39 strain is susceptible to alcohols which positively regulates AdhE (Luong et al., 2015). Colonization, virulence and hemolytic activity of S. pneumoniae, as well as the pro-inflammatory cytokine secretion, inflammation and host cell myeloperoxidase activity were substantially reduced in ΔadhE compared to D39 wild-type (Luong et al., 2015), suggesting AdhE to be a pneumococcal virulence factor (Luong et al., 2015). These genes that expressed in numerous ways could be used as potential vaccination candidates or as therapeutic targets.
The expression of the ciaR-H operon was altered in the presence of paracetamol. The CiaRH system is a two-component signal transduction system (TCS) and CiaR acts as repressor the competence genes in S. pneumoniae (Guenzi et al., 1994). The CiaRH system is vital for providing resistance against cell wall inhibitors and helps maintaining cell integrity (Mascher et al., 2006). Similar findings were observed when pneumococcal transcriptomic response to penicillin was studied (Rogers et al., 2007). These findings imply that stimulation of the CiaRH system and (consequent down-regulation of competence genes) may be a mechanism by which S. pneumoniae fights itself against penicillin-induced cell wall damage.
In our gene expression analysis, we see significant upregulation of genes involved in the modulation of misfolded proteins (hrcA, grpE, dnaK and dnaJ). This result may be expected as a drug (paracetamol in our case) can induce as protein mistranslation and that is why the expression of the genes involved in the regulation of misfolded proteins will be enhanced.
3.2 PPI network analysis of the differentially expressed genes
PPI networks have been utilized to study health and disease of a certain body and can be very handy for comparison of systems across diverse conditions. The STRING database was used to build PPI networks in order to better understand the connections of differentially expressed genes. As shown in Fig. 1A, the hub genes with node degree greater than or equal to 10 fatty acid biosynthesis and metabolism genes. These genes include spd-0378, acpP, fabH, fabK, fabD, fabF, fabG, accB, accC, fabZ, accD and accA. Another network that was prominent in our analysis was the one consisting of genes involved in the biosynthesis of antibiotics. This network includes fabD, fabG, accA, accB, accC, accD, spd-1834 (adhE), dapA, asd, ilvA, ilvB, ilvC, ilvE and ilvN. Both fatty acid biosynthesis and metabolism genes and antibiotics biosynthesis genes’ networks have been shown in separate figures (Fig. 1B and 1C, respectively).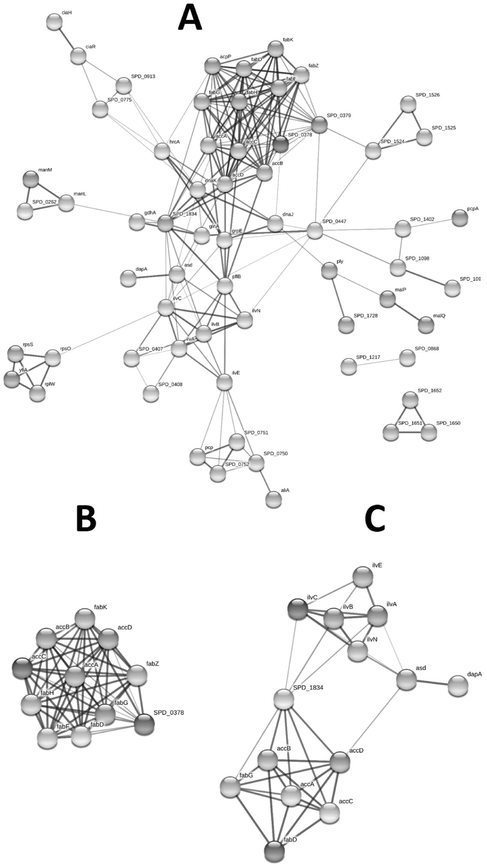
Protein-protein interaction (PPI) differential network analysis of the pneumococcal transcriptome in response to 5 mM paracetamol in the medium (A). PPI differential network analysis of the fatty acid genes (B) and the antibiotic biosynthesis genes (C).
3.3 KEGG pathways analysis of the differentially regulated genes
These genes pertain to several metabolic pathways including the fatty acid biosynthesis and metabolism pathways (Table 3). The pathway involved in the biosynthesis of antibiotics was also significantly affected under our tested conditions. Moreover, several pathways involved particularly in leucine, valine and isoleucine biosynthesis, pyruvate metabolism, 2-Oxocarboxylic acid metabolism, propanoate and butanoate metabolism, biosynthesis of secondary metabolites and quorum sensing were significantly altered in under our tested conditions (Table 3). These results indicate that paracetamol have diverse effects on several pathways in pneumococcus. Enrichment analysis was performed using the pathway enrichment tool (STRING, version 10.5, https://www.string-db.org/). The observed gene count indicates how many genes match the predicted metabolic pathways. FDR, false discovery rate.
Pathway name
Observed gene count
FDR
Fatty acid biosynthesis
11/13
6.07E-09
Fatty acid metabolism
10/11
1.40E-08
Biosynthesis of antibiotics
15/113
0.00056
Valine, leucine, and isoleucine biosynthesis
5/9
0.00096
2-Oxocarboxylic acid metabolism
5/10
0.0011
Pyruvate metabolism
6/19
0.0015
Propanoate metabolism
5/12
0.0016
Butanoate metabolism
4/7
0.0023
Metabolic pathways
26/345
0.0023
Pantothenate and CoA biosynthesis
4/11
0.0071
Biotin metabolism
3/5
0.0088
Biosynthesis of secondary metabolites
13/138
0.0099
Microbial metabolism in diverse environments
10/91
0.0106
Quorum sensing
7/53
0.0168
Biosynthesis of amino acids
8/76
0.0299
Monobactam biosynthesis
2/4
0.0465
Nitrogen metabolism
2/4
0.0465
4 Discussion
Growing drug resistance is a major challenge in treating pneumococcal throughout the last few decades. Presently, 15–30 % of the pneumococcal strains are categorized as resistant to multiple drugs (Lynch and Zhanel, 2009). Most common drug treatments of pneumococcus involves macrolides, β-lactams, or fluoroquinolones individually or in group (Weiss et al., 2004; Waterer and Rello, 2006). The use of antibiotic combinations broadens the range of bacteria that can be targeted while also increasing efficacy, decreasing the inception and propagation of resistance bacteria. Understanding the response of pneumococci to commonly used drugs may provide novel therapeutic targets and important insights into the adaptive techniques required to interact with the host environment during infection (Leonard and Lalk, 2018). Using paracetamol as a part of combination therapy may be useful. We investigated the relation of S. pneumoniae to paracetamol to identify the potential candidates for drugs against pneumococcus. This research will aid in the knowledge of pneumococcal physiology and response to a commonly used drug. To unveil pneumococcal adaptation to a commonly used drug, paracetamol, pneumococci were grown in CDM in the presence of 5 mM paracetamol. The response of strain D39 to paracetamol is complex, comprising a network of genes involved in fatty acid metabolism and biosynthesis, nutrition and waste transport, environmental stress detection, and antibiotic production.
Fatty acid biosynthesis and metabolism gene cluster was significantly downregulated in the presence of paracetamol. The antibacterial effect of fatty acids on the potential of S. pneumoniae to induce disease is poorly understood. Fatty acid synthase system type II (FASII) manufactures pneumococcal fatty acids which are needed for the cell membrane (Zhang and Rock, 2008), which is encoded by the fab gene cluster. This cluster's transcription is regulated by the MarR-type transcriptional regulator FabT, which suppresses the cluster responsible for fab-transcription when fatty acids attach to it (Jerga and Rock, 2009). The pneumococcus can integrate fatty acids that are produced outside (exogenous) into its membrane via the FakA/B system, in complement to de novo synthesis by the FASII system (Parsons et al., 2015). Exogenous fatty acids have been shown to reduce pneumococcal colonization in investigations, while the molecular mechanism for this antibacterial activity of free fatty acids is unknown (Bomar et al., 2016). The distinct pneumococcal SpFakB3 can be used due to its special polyunsaturates. To bind the fatty acid carbonyl and normalize the protein, pneumococcal FakB3 uses a distinct hydrogen bond network than other FakBs (Gullett et al., 2019). Deletion of fakB3 in S. pneumoniae strain JMG1 led to reduction in linoleate incorporation from human serum confirms the significance of fakB3 in this process. FakB3 (spd-0646) was one of the genes that was downregulated in our transcriptomic analysis in the presence of paracetamol, which might suggest that fakB3can be a very important target for paracetamol and further studies will be needed for deep research. Moreover, the fab genes were shown to be downregulated in pneumococcus in the presence of penicillin (Rogers et al., 2007). These genes may share a stress response to cell wall inhibitors since they respond similarly to paracetamol and vancomycin, with some of them potentially important in shielding the cell from their effects.
Iron has a pivotal role in the pathogenesis of S. pneumoniae. To successfully support infections and survival, pneumococcus has diversified three transporters named ABC, PiuABC, PiaABC, and PitABC, with lipoproteins PiuA PiaA, and PitA as proteins that binds to substrate to uptake iron (Yang et al., 2016). In our microarray findings, piuABC was downregulated in the existance of paracetamol. These findings were in contrast to a gene expression analysis based on microarray in S. pneumoniae which demosnstarted that the fluoroquinolone levofloxacin induced an positive regulation of the piuABC operon (Ferrándiz and de la Campa, 2014). They further suggested that upregulation of piuABC would cause a rise in intracellular iron, which would then activate the Fenton reaction, resulting in an increase in reactive oxygen species (Ferrándiz and de la Campa, 2014).
Genes (glnRA and glnPQ) involved in glutamine synthesis and uptake were among the downregulated ones in the presence of paracetamol. Penicillin therapy increases intracellular glutamine concentrations, according to a recent study (El Khoury et al., 2017). When culture media was supplemented with glutamine, it provided protection against penicillin (El Khoury et al., 2017). The glnA-encoded glutamine synthetase catalyzes the conversion of ammonium and glutamate into glutamine, and its chemical inhibition by the l-methionine sulfoximine (inhibitor) has been demonstrated to make S. pneumoniae susceptible to penicillin, even in penicillin-resistant clinical isolates (El Khoury et al., 2017). Therefore, we believe that paracetamol (by altering glutamine genes express) Interacts with glutamine metabolism, implying techniques that might be employed in standard treatment or to reverse resistance in future.
A couple of sugar systems manLMN and malPQ were also expressed in numerous ways in the presence of paracetamol. ManLMN is typically a major glucose transporter that has alos the ability of transporting a varying number of other carbohydrate substrates including mannose, fructose, galactose and N-acetyl glucosamine (Bidossi et al.,). manLMN is repressed by both the CcpA and CiaR, the response regulator of the conserved TCS CiaRH implicated in competence, autolysis and β-lactam resistance (Halfmann et al., 2007; Carvalho et al., 2011). In S. pneumoniae D39, inactivation of manM encoding the PTS EIIC component, resulted in a mild growth defect in glucose, and more severely reduced growth in N-acetyl glucosamine, mannose, and galactose (Bidossi et al.,). In contrast to D39, ManLMN was found to be essential for growth on five non-preferred carbohydrates in TIGR4, and required to induce expression of downstream metabolic genes (Fleming and Camilli, 2016). malPQ are the maltose utilization genes and have been shown to be regulated by MalR (Afzal et al., 2015). They code for a maltodextrin phosphorylase and a 4-alpha-glucanotransferase, respectively. These maltose genes have been demonstrated to be positively regulated during involvement of cellobiose as well (Shafeeq et al., 2013). Our β-galactosidase assays with PmalP-lacZ in the presence of cellobiose showed that the activity of PmalP was significantly higher in the presence of cellobiose as compared that in glucose (data not shown). This might indicate about the complexity of the role of malPQ in the life-style of pneumococcus and differential expression of malPQ in the presence of paracetamol corroborates our observation. Moreover, both these manLMN and malPQ systems have been shown to be differentially expressed in the presence of penicillin and vancomycin in S. pneumoniae (Rogers et al., 2007; Haas et al., 2004).
We discovered genes of S. pneumoniae that are differently expressed in reaction to paracetamol exposures in our investigation. Several of these genes have been connected to drug resistance or tolerance in the past, demonstrating that their altered expression is part of a stress-protective response in this situation. Such genes could be used as therapeutic targets to improve the effectiveness of paracetamol against this pathogen. Other gene expression changes discovered here could also point to potential paracetamol resistance mechanisms. In this context, more research on these genes is required.
Acknowledgements
We thank Prof. Oscar P. Kuipers and Dr. Anne de Jong, University of Groningen, The Netherlands for their help for microarrays experiments.The authors (MFA, SM) express their sincere appreciation to the Researchers Supporting Project Number (RSP2022436) King Saud University, Riyadh, Saudi Arabia.
Proclamations of Ethics.
Statement of Ethical Guidelines
The manuscript is exempt from ethical committee approval.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Maltose-Dependent Transcriptional Regulation of the mal Regulon by MalR in Streptococcus pneumoniae. PLoS ONE. 2015;10(6)
- [CrossRef] [Google Scholar]
- A Fast and Reliable Pipeline for Bacterial Transcriptome Analysis Case study: Serine-dependent Gene Regulation in Streptococcus pneumoniae. J. Vis. Exp. JoVE.. 2015;98
- [CrossRef] [Google Scholar]
- A. Bidossi L. Mulas F. Decorosi L. Colomba S. Ricci G. Pozzi J. Deutscher C. Viti M.R. Oggioni E.N. Miyaji A Functional Genomics Approach to Establish the Complement of Carbohydrate Transporters in Streptococcus pneumoniae PLoS ONE 7 3 e33320.
- Corynebacterium accolens Releases Antipneumococcal Free Fatty Acids from Human Nostril and Skin Surface Triacylglycerols. mBio.. 2016;7(1)
- [CrossRef] [Google Scholar]
- Assessing evolutionary risks of resistance for new antimicrobial therapies. Nat. Ecol. Evol.. 2019;3(4):515-517.
- [Google Scholar]
- Characterization of Pit, a Streptococcus pneumoniae Iron Uptake ABC Transporter. Infect. Immun.. 2002;70(8):4389-4398.
- [Google Scholar]
- CcpA ensures optimal metabolic fitness of Streptococcus pneumoniae. PLoS ONE. 2011;6(10):e26707.
- [Google Scholar]
- PePPER: a webserver for prediction of prokaryote promoter elements and regulons. BMC Genomics. 2012;13:299.
- [CrossRef] [Google Scholar]
- Penicillin induces alterations in glutamine metabolism in Streptococcus pneumoniae. Sci. Rep.. 2017;7(1):14587.
- [CrossRef] [Google Scholar]
- The Fluoroquinolone Levofloxacin Triggers the Transcriptional Activation of Iron Transport Genes That Contribute to Cell Death in Streptococcus pneumoniae. Antimicrob. Agents Chemother.. 2014;58(1):247-257.
- [CrossRef] [Google Scholar]
- ManLMN is a glucose transporter and central metabolic regulator in Streptococcus pneumoniae. Mol. Microbiol.. 2016;102(3):467-487.
- [CrossRef] [Google Scholar]
- A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol. Microbiol.. 1994;12(3):505-515.
- [CrossRef] [Google Scholar]
- A fatty acid-binding protein of Streptococcus pneumoniae facilitates the acquisition of host polyunsaturated fatty acids. J. Biol. Chem.. 2019;294(44):16416-16428.
- [CrossRef] [Google Scholar]
- Revising the Role of the Pneumococcal vex-vncRS Locus in Vancomycin Tolerance. J. Bacteriol.. 2004;186(24):8463-8471.
- [Google Scholar]
- Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: five out of 15 promoters drive expression of small non-coding RNAs. Mol. Microbiol.. 2007;66(1):110-126.
- [Google Scholar]
- Acyl-Acyl Carrier Protein Regulates Transcription of Fatty Acid Biosynthetic Genes via the FabT Repressor in Streptococcus pneumoniae. J. Biol. Chem.. 2009;284(23):15364-15368.
- [CrossRef] [Google Scholar]
- The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol.. 2008;6(4):288-301.
- [Google Scholar]
- Biological and Epidemiological Features of Antibiotic-Resistant Streptococcus pneumoniae in Pre- and Post-Conjugate Vaccine Eras: a United States Perspective. Clin. Microbiol. Rev.. 2016;29(3):525-552.
- [CrossRef] [Google Scholar]
- Regulation of Glutamine and Glutamate Metabolism by GlnR and GlnA in Streptococcus pneumoniae. J. Biol. Chem.. 2006;281(35):25097-25109.
- [Google Scholar]
- Acetaminophen (APAP) hepatotoxicity—Isn’t it time for APAP to go away? J. Hepatol.. 2017;67(6):1324-1331.
- [CrossRef] [Google Scholar]
- Infection and metabolism – Streptococcus pneumoniae metabolism facing the host environment. Cytokine. 2018;112:75-86.
- [CrossRef] [Google Scholar]
- Risk Factors for Invasive Pneumococcal Disease in Children: A Population-based Case-Control Study in North America. Pediatrics. 1999;103(3):e28-e.
- [Google Scholar]
- Ethanol-induced alcohol dehydrogenase E (AdhE) potentiates pneumolysin in Streptococcus pneumoniae. Infect. Immun.. 2015;83(1):108-119.
- [Google Scholar]
- Streptococcus pneumoniae : Epidemiology, Risk Factors, and Strategies for Prevention. Semin Respir Crit Care Med.. 2009;30(02):189-209.
- [CrossRef] [Google Scholar]
- A new mechanism for anaerobic unsaturated fatty acid formation in Streptococcus pneumoniae. J. Biol. Chem.. 2002;277(47):44809-44816.
- [CrossRef] [Google Scholar]
- The CiaRH system of Streptococcus pneumoniae prevents lysis during stress induced by treatment with cell wall inhibitors and by mutations in pbp2x involved in beta-lactam resistance. J. Bacteriol.. 2006;188(5):1959-1968.
- [CrossRef] [Google Scholar]
- Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet. 2009;374(9693):893-902.
- [Google Scholar]
- A thioesterase bypasses the requirement for exogenous fatty acids in the plsX deletion of S treptococcus pneumoniae: Role of PlsX in S. pneumoniae fatty acid metabolism. Mol. Microbiol.. 2015;96(1):28-41.
- [Google Scholar]
- Gene expression profiling of the response of Streptococcus pneumoniae to penicillin. J. Antimicrob. Chemother.. 2007;59(4):616-626.
- [Google Scholar]
- Scottish Intercollegiate Guidelines Network. Control of Pain in Adults with Cancer: A National Clinical Guideline. Scottish Intercollegiate Guidelines Network; 2008.
- Cellobiose-mediated gene expression in Streptococcus pneumoniae: a repressor function of the novel GntR-type regulator BguR. PLoS ONE. 2013;8(2):e57586.
- [Google Scholar]
- The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res.. 2017;45(D1):D362-D368.
- [Google Scholar]
- European enhanced surveillance of invasive pneumococcal disease in 2010: Data from 26 European countries in the post-heptavalent conjugate vaccine era. Vaccine.. 2014;32(29):3644-3650.
- [Google Scholar]
- A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics. 2005;6(1)
- [Google Scholar]
- van Hijum SAFT, Garcia de la Nava J, Trelles O, Kok J, Kuipers OP. MicroPreP: a cDNA microarray data pre-processing framework. ApplBioinformatics. 2003;2(1175-5636; 4):241-244.
- Choosing the right combination therapy in severe community-acquired pneumonia. Crit. Care. 2006;10(1):115.
- [CrossRef] [Google Scholar]
- Clinical Characteristics at Initial Presentation and Impact of Dual Therapy on the Outcome of Bacteremic Streptococcus pneumoniae Pneumonia in Adults. Can Respir J.. 2004;11(8):589-593.
- [Google Scholar]
- Integrated Translatomics with Proteomics to Identify Novel Iron-Transporting Proteins in Streptococcus pneumoniae. Front. Microbiol.. 2016;7
- [CrossRef] [Google Scholar]
- Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol.. 2008;6(3):222-233.
- [CrossRef] [Google Scholar]







