Translate this page into:
Evaluation of the relationships between HLA-G 14 bp polymorphism and two acute leukemia in a Saudi population
⁎Corresponding author at: Department of Zoology, College of Science, Building 05, King Saud University, Riyadh 11451, Saudi Arabia. lmansour@ksu.edu.sa (Lamjed Mansour)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The non-classical Human Leukocyte Antigen G (HLA-G) is an immunomodulatory molecule, with low polymorphism frequency and restricted tissue distribution. Its higher expression was associated with immunoinhibitory properties causing tolerance for cancer progression. The rs371194629 is a 14 bp insertion/deletion polymorphism (Ins/Del) in exon 8 of the 3′ untranslated region (3′-UTR) of the HLA-G gene, has been associated with the stability and the expression level of HLA-G mRNA. In this study, we evaluated the relationship between the HLA-G 14-bp Ins/Del polymorphism and two common blood cancers including acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML) in a population from Riyadh in Saudi Arabia. The study groups include 145 patients diagnosed for ALL, 98 patients diagnosed for AML and 115 healthy individuals. For all these individuals, the 14 bp Ins/Del polymorphism was genotyped using PCR methodology. mRNA gene expression of the HLA-G was assessed using quantitative RT-PCR. Logistic regression model analysis for HLA-G 14 bp Ins/Del polymorphism shows no statistically significant correlation with AML and ALL. For the HLA-G mRNA expression, while slightly increased level among healthy individuals compared to patients was observed, no statistically significant differences were obtained. Although our results did not show association between HLA-G with the two leukemia cancer diseases, the role of HLA-G gene awaits further investigations including large number of subjects with more clinical data combinations.
Keywords
Acute lymphocytic leukemia
Acute myelogenous leukemia
HLA-G 14-bp Ins/Del
Genetic polymorphism
Saudi Arabia
- HLA
-
Human Leukocyte Antigen
- ALL
-
Acute Lymphoblastic Leukemia
- AML
-
Acute Myeloid Leukemia
- Ins/Del
-
Insertion/Deletion
- UTR
-
Untranslated Region
- CML
-
Chronic Myelocytic Leukemia
- CLL
-
Chronic Lymphocytic Leukemia
- NK
-
Natural Killer Cells
Abbreviations
1 Introduction
Cancer is becoming more frequent and lethal in both developed and developing countries, with global economic impacts (Siegel et al., 2021). Blood cancers account for nearly 10% of all newly diagnosed cancer cases in the world (Eid et al., 2021). The most common type of pediatric cancer and the most leading cause of cancer mortality in children is acute lymphoblastic leukemia (ALL) (Inaba et al., 2013). ALL affects both children and adults, with a peak of occurrence between 1 and 4 years (Malard and Mohty, 2020). While the most common type of leukemia malignancy among adults is acute myeloid leukemia (AML). It is characterized by the proliferation of myeloid blasts or progranulocytes that do not differentiate normally (Newell and Cook, 2021). Exogenous or endogenous exposures, genetic predisposition, and randomness all play a role in causation (Alkhouly et al., 2013; Inaba et al., 2013). Also, chromosome abnormalities and genetic alterations that impact lymphoid precursor cell formation and proliferation are directly involved in the abnormal proliferation. However, the genetic factors for these leukemia diseases remain unknown.
According to circumstantial evidence, the host immune system can actively inhibit or tolerate tumor development. The production of immunosuppressive cytokines or the modification of antigen-presenting molecules on neoplastic cells is commonly blamed for tumor escape from host immune surveillance (Alkhouly et al., 2013). HLA-G is a non-classical HLA molecule of the class Ib and is located between HLA-A and HLA-F located on chromosome 6p21.3. It has eight exons and seven introns (Castelli et al., 2008).
The HLA-G protein has special features including the reduced allelic polymorphism, restricted tissue expression, and the existence of seven expression isoforms that could be membranous (G1, G2, G3 and G4) or soluble proteins (G5, G6 and G7) (Paul et al., 2000).
HLA-G has been reported for its suppressive function as it decreases the human immune response through a variety of mechanisms, including the inhibition of natural killer (NK) cells and T lymphocytes' cytotoxic activities (Hofmeister and Weiss, 2003). HLA-G plays a key role in fetal maternal immune tolerance by inhibiting maternal immunity through its expression in trophoblast cells at fetus-maternal interface during pregnancy (Moreau et al., 1998). Thus, reduced placental expression of HLA-G has been associated with pathological conditions such as recurrent spontaneous abortion (Peng et al., 2008). Outside the placenta and in physiological conditions, HLA-G expression has been documented in few immune-privileged tissue, such as the thymus and the cornea (Le Discorde et al., 2003). However, an ectopic expression of HLA-G molecules has been associated with pathological conditions such as cancer, autoimmune and inflammatory diseases (Zaborek-Łyczba et al., 2021).
Many studies have been conducted on the HLA-G polymorphism and its functional association with human diseases. Thus far, 94 HLA-G alleles have been identified (IPD-IMGT/HLA, R, 3.47, Marsh, 2022; https://www.ebi.ac.uk/ipd/imgt/hla).
Polymorphism sites in the HLA-G gene's non-coding regions, particularly in the 3′ UTR exon 8, have been reported to influence its expression (Rizzo et al., 2014). The insertion or deletion of a 14-bp (5′-ATTTGTTCATGCCT-3′) polymorphism in the 3′ UTR of the HLA-G gene, also known as rs371194629, is one of the most important polymorphisms (Rousseau et al., 2003). It has been reported in the untranslated exon 8 and was involved in the stability and expression of mRNA. The Del allele stabilizes the mRNA with a consequent higher HLA-G expression, while the Ins allele destabilizes the mRNA and is linked with a lower levels of HLA-G expression (Svendsen et al., 2013). According to Chen et al. (2008), the Ins alleles was associated with lower HLA-G mRNA levels and Del allele with higher level. In recent years, a rising number of studies have investigated the relationship between HLA-G 14-bp Ins/Del polymorphism and malignancies in different populations (Adolf et al., 2021). In Saudi Arabia, few studies have reported the association of HLA-G 14 Ins/Del variant and cancer diseases including breast cancer and colorectal cancer have been reported (Al Omar and Mansour, 2019; Al Omar et al., 2015; Hassan et al., 2019). However, only a handful studies have been done on leukemia. In Saudi Arabia, so far, no studies had investigated the association between HLA-G and any leukemia diseases. In this study, we investigated the genotypic and allelic association of HLA-G 14 bp Ins/Del polymorphism with the two most frequent leukemia malignancies (ALL and AML).
2 Materials and methods
2.1 Ethics statement
The study was approved by the medical ethics committee at King Khaled Hospital University (KKHU) in Riyadh, Saudi Arabia, and was carried out in accordance with King Saud University's ethical guidelines (IRB-No. E-20-5346).
2.2 Patients and healthy groups
A total of 358 human whole blood samples were acquired for this study. The patients in this study were 243 patients, including 145 patients diagnosed with ALL (87 males and 58 females), and 98 patients diagnosed with AML (54 males and 44 females). The control group included 115 healthy individuals of both genders (77 males and 38 females) without history for cancer and another inflammatory or autoimmune diseases. The mean age of patients with ALL was 19.20 ± 14.8 years, and with AML was 27.56 ± 18.5 years, while the mean age of controls was 22.45 ± 20.18 years. All patients were sourced from the Department of Pediatric Hematology-oncology section at King Khaled Hospital University (KKHU) in Riyadh, Saudi Arabia.
2.3 Genomic DNA extraction
3 ml of peripheral blood was taken from patients and healthy controls in ethylene diamine tetra acetic acid (EDTA) coated collection vials from each participant. The QIAamp DNA Blood Mini Kit (QIAGEN, GmbH, Hilden, Germany) was used to extract genomic DNA from peripheral blood, following the manufacturer's instructions.
2.4 Genotyping of the 14-bp Ins/Del
The 14-bp Ins/Del polymorphism (rs371194629) in exon 8 (3′ UTR) of the HLA-G gene (Fig. 1) was genotyped using the primers GE14HLAG:5′-GTGATGGGCTGTTTAAAGTGTCACC-3′ and RHG4: 5′-GGAAGGAATGCAGTTCAGCATGA-3′ (Hviid et al., 1999). Polymerase Chain Reaction (PCR) was done in a volume of 20 μl containing 1 μl of DNA, 10 ul of PCR GoTaq® Master Mixes 2X (Promega, India), 0.5 ul of each primer (10 pmol) and 8 µl of RNase-free water. The PCR reactions were performed in a T100TM Thermocycler (Bio-Rad, Singapore). For amplification, the following thermocycling conditions were used: an initial denaturation phase at 95 °C (5 min), followed by 34 cycles of 94 °C (30 s), 60 °C (30 s), 72 °C (1 min), and 72 °C (5 min) as a final extension. The amplicons, which are either 210 bp in length for the 14-bp Del allele, or 224 bp in length for the 14-bp Ins allele, were migrated in a 3% agarose gel stained with ethidium bromide. The gel was visualized and photographed in a BioDocAnalyze (Biometra GmbH, Germany).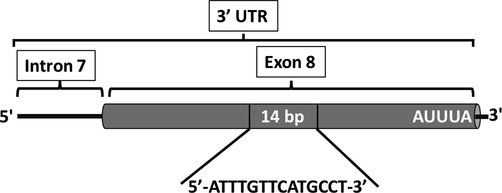
Polymorphism in the HLA-G gene in 3′ UTR. The 14-bp Ins/Del in exon 8.
For DNA sequencing, six randomly selected samples including the three genotypes were sequenced using the same reverse primer used in PCR reaction. The PCR products were purified using EXO-SAP (New England Biolabs) and sequenced using the BigDye Terminator v.3.1 Sequencing Kit (Applied Biosystems, CA, USA) in an ABI Prism 3730 XL Genetic Analyzer (Applied Biosystems).
2.5 RNA isolation and cDNA synthesis
The PureLink™ RNA Mini Kit (Invitrogen, Thermo Fisher Scientific, USA) was used to isolate total RNA from human whole blood. The quantity and quality of extracted RNA were determined using a NanoDrop™ 8000 Spectrophotometer (Thermo Scientific, USA). The ratio of absorbance at 260 nm and 280 nm is used to assess the purity of RNA. A ratio of about 2.0 of pure RNA. For high-capacity cDNA synthesis, we used a reverse transcription kit (Applied Biosystems™, USA) according to the protocol described by the manufacturer. The kit is designed for real-time, two-step RT-PCR.
2.6 Quantitative RT-PCR analysis of HLA-G mRNA
For qRT-PCR analysis, we performed specific amplification of HLA-G transcripts from cDNA. For specific HLA-G amplification, the following sequences of primers were F: 5′-GTGTTCCGTGTCTCCTCT-3′ and R: 5′-GACAGCGACTCGGCGT-3′. The housekeeping gene used in this analysis was for GAPDH gene using the couple of primers: F: 5′-TCTCCTCTGACTTCAACAGCGAC-3′ and R: 5′-CCCTGTTGCTGTAGCCAAATTC-3′. For each PCR reaction, a total volume of 20 µl was used, which included 10 µl of 2X Fast SYBR®Green Master Mix, 1.5 µl of cDNA, 0.5 of each primer (5 pmol) and 7.5 µl of RNase-free water. The PCR reactions were performed in and ABI PRISM 7500 System Thermal Cycler (Applied Biosystems). The program of amplification was an initial denaturation at 95 °C for 5 min followed by 40 cycles of denaturation at 95 °C for 15 s and annealing at 60 °C for 60 sb and 72 °C for 30 s, and a final extension for 10 mn at 72. For gene expression analysis, relative quantitation of HLA-G mRNA was done by the comparative CT method after normalization with the housekeeping gene (GAPDH). Relative expression level, determined as fold change, was expressed using the 2−(ΔΔCt) method (Schmittgen and Livak, 2008).
2.7 Statistical analysis
We tested the Hardy–Weinberg equilibrium (HWE) for 14 bp Ins/Del allelic and genotypic variations in control subjects. Odds ratios with confidence intervals of 95% were used to measure the strength of the association between the HLA-G 14 bp Ins/Del polymorphism and ALL and AML risk for all genotypes considering all the inheritance models (dominant, co-dominant, recessive, or log-additive). Analyses were performed using SNPStats software (Solé et al., 2006).
3 Results
The 14 bp Ins/Del HLA-G 3′UTR polymorphism was successfully genotyped for all the 243 patients with acute leukemia, including 145 ALL and 98 AML and 115 healthy controls. Fig. 2A shows a gel image of the genotyping result of the 14 bp polymorphism. Six PCR-genotyped samples from each group were selected for sequencing. Fig. 2B shows an example of electropherograms of the three HLA-G 14 bp Ins/Del genotypes, that confirm the classical genotyping methods. Allele frequencies of the Del and Ins are reported in Table 1. The genotype frequency of the HLA-G polymorphism was in Hardy-Weinberg equilibrium (HWE) for controls, AML and ALL patients. For the genetic and allelic association, we have tested five genetic models (allelic, codominant, dominant, recessive, overdominant and additive). Results are reported in Table 1. The allele distribution between control group and ALL and AML, did not show any significant difference. The Del allele appears slightly higher for all tested groups than the Ins allele, with a frequencies similar to those reported for many populations. For all the tested models of association, no significant differences were observed between controls and the two leukemia cancers. ALL; Acute Lymphoblastic Leukemia; AML: Acute Myeloid Leukemia; CN: Control; OR: odds ratio; CI: confidence interval. Ins: 14 base pairs insertion; Del: 14 base pairs deletion.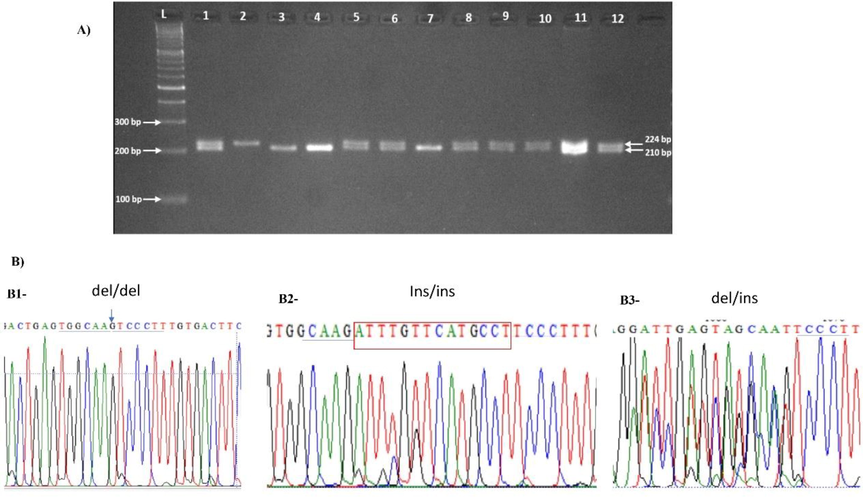
Analysis of the14bp Ins/Del polymorphism using gel electrophoresis (A). For each patient and control. In the gel, Lanes 1, 5, 6, 8–12 are heterozygous; Lane 2 homozygous Ins/Ins and lanes 3, 4 and 7 are homozygous Del/Del. (B) sequencing of the 3′ UTR fragment, showing the electropherogram at the level of the 14 bp Ins/Del. B1, profile sequence of a homozygous Del/Del; B2, Profile of homozygous Ins/Ins and B3, profile of heterozygous Del/Ins.
Genetic model
Allele/Genotype
Controls (N = 115)
N (%)ALL (N = 145)
N (%)OR (95% CI)
P value
AML (N = 98)
N (%)OR (95% CI)
P-value
Allele
Del
0.53
0.59
1.00
0.20
0.54
1.00
0.83
Ins
0.47
0.41
0.79 (0.563–1.130)
0.46
0.959 (0.65–1.40)
Codominant
Del/Del
37 (32.2%)
52 (35.9%)/
0.35
32 (32.6%)
1.00
0.96
Ins/Del
48 (41.7%)
66 (45.5%)
1.02 (0.58–1.79)
42 (42.9%)
0.99 (0.53–1.85)
Ins/Ins
30 (26.1%)
27 (18.6%)
1.56 (0.80–3.05)
24 (24.5%)
1.08 (0.53–2.21)
Dominant
Del/Del
37 (32.2%)
52 (35.9%)
1.00
0.53
32 (32.6%)
1.00
0.94
Ins/Ins + Ins/Del
78 (67.8%)
93 (64.1%)
1.18 (0.70–1.98)
66 (67.3%)
1.02 (0.57–1.82)
Recessive
Ins/Ins
30 (26.1%)
27 (18.6%)
1.00
0.15
24 (24.5%)
1.00
0.79
Del/Del + Ins/Del
85 (73.9%)
118 (81.4%)
1.54 (0.86–2.78)
74 (75.5%)
1.09 (0.59–2.02)
Overdominant
Del/Del + Ins/Ins
67 (58.3%)
79 (54.5%)
1.00
0.54
56 (57.1%)
1.00
0.54
Ins/Del
48 (41.7%)
66 (45.5%)
0.86 (0.52–1.41)
42 (42.9%)
0.96 (0.55–1.65)
Log-additive
–
–
–
1.23 (0.88–1.71)
0.23
1.04 (0.73–1.48)
0.84
Furthermore, we assessed the possible association between HLA-G 14 bp Ins/Del and the risk to ALL and AML by stratified analysis based on gender (Table 2) and age (Table 3). Once again, for both leukemia diseases, no significant association was observed with the studied polymorphism. ALL; Acute Lymphoblastic Leukemia; AML: Acute Myeloid Leukemia; CN: Control; OR: odds ratio; CI: confidence interval. Ins: 14 base pairs insertion; Del: 14 base pairs deletion. ALL; Acute Lymphoblastic Leukemia; AML: Acute Myeloid Leukemia; CN: Control; OR: odds ratio; CI: confidence interval. Ins: 14 base pairs insertion; Del: 14 base pairs deletion.
Genetic model
Allele/Genotype
ALL
(Male)
ALL
(Female)OR (95% CI)
P-value
AML
(Male)AML
(Female)OR (95% CI)
P-value
Allele
Del
0.57
0.61
0.46
0.53
0.55
0.86
Ins
0.43
0.39
0.837 (0.52–1.35)
0.47
0.45
0.95 (0.54–1.67)
Codominant
Del/Del
29 (33.3%)
23 (39.7%)
1.00
0.74
18 (33.3%)
14 (31.8%)
1.00
0.99
Ins/Del
41 (47.1%)
25 (43.1%)
1.30 (0.62–2.72)
23 (42.6%)
19 (43.2%)
0.94 (0.37–2.38)
Ins/Ins
17 (19.5%)
10 (17.2%)
1.35 (0.52–3.50)
13 (24.1%)
11 (25%)
0.92 (0.32–2.66)
Dominant
Del/Del
29 (33.3%)
23 (39.7%)
1.00
0.44
18 (33.3%)
14 (31.8%)
1.00
0.87
Ins/Ins + Ins/Del
58 (66.7%)
35 (60.3%)
1.31 (0.66–2.62)
36 (66.7%)
30 (68.2%)
0.93 (0.40–2.18)
Recessive
Ins/Ins
70 (80.5%)
48 (82.8%)
1.00
0.73
41 (75.9%)
33 (75%)
1.00
0.92
Del/Del + Ins/Del
17 (19.5%)
10 (17.2%)
1.17 (0.49–2.76)
13 (24.1%)
11 (25%)
0.95 (0.38–2.40)
Overdominant
Del/Del + Ins/Ins
46 (52.9%)
33 (56.9%)
1.00
0.63
31 (57.4%)
25 (56.8%)
1.00
0.95
Ins/Del
41 (47.1%)
25 (43.1%)
1.18 (0.60–2.30)
23 (42.6%)
19 (43.2%)
0.98 (0.44–2.18)
Log-additive
–
–
–
1.18 (0.74–1.89)
0.48
–
–
0.96 (0.56–1.63)
0.87
Genetic model
Allele/Genotype
ALL
(Age ≤ 18)
ALL
(Age > 18)OR (95% CI)
P value
AML
(Age ≤ 18)
AML
(Age > 18)OR (95% CI)
P value
Allele
Del
0.59
0.58
0.90
0.58
0.52
0.90
Ins
0.41
0.42
1.03 (0.63–1.66)
0.42
0.48
0.97 (0.60–1.57)
Codominant
Del/Del
33 (36.7%)
19 (34.5%)
1.00
0.95
15 (40.5%)
17 (27.9%)
1.00
0.38
Ins/Del
40 (44.4%)
26 (47.3%)
1.13 (0.53–2.39)
13 (35.1%)
29 (47.5%)
1.97 (0.76–5.11)
Ins/Ins
17 (18.9%)
10 (18.2%)
1.02 (0.39–2.68)
9 (24.3%)
15 (24.6%)
1.47 (0.50–4.33)
Dominant
Del/Del
33 (36.7%)
19 (34.5%)
1.00
0.8
15 (40.5%)
17 (27.9%)
1.00
0.2
Ins/Ins + Ins/Del
57 (63.3%)
36 (65.5%)
1.10 (0.54–2.21)
22 (59.5%)
44 (72.1%)
1.76 (0.74–4.18)
Recessive
Ins/Ins
73 (81.1%)
45 (81.8%)
1.00
0.92
28 (75.7%)
46 (75.4%)
1.00
0.98
Del/Del + Ins/Del
17 (18.9%)
10 (18.2%)
0.95 (0.40–2.27)
9 (24.3%)
15 (24.6%)
1.01 (0.39–2.62)
Overdominant
Del/Del + Ins/Ins
50 (55.6%)
29 (52.7%)
1.00
0.74
24 (64.9%)
32 (52.5%)
1.00
0.23
Ins/Del
40 (44.4%)
26 (47.3%)
1.12 (0.57–2.20)
13 (35.1%)
29 (47.5%)
1.67 (0.72–3.88)
Log-additive
–
–
–
1.03 (0.64–1.64)
0.91
–
–
1.26 (0.73–2.18)
0.41
In order to assess the relationship between HLA-G gene expression and ALL diseases, we analyzed the HLA-G mRNA expression level in 12 ALL and 12 control individuals using relative quantitative RT-PCR. Fig. 3 shows the diagram of the mean fold of expression for healthy control and ALL patients.. In the control group, the level of HLA-G is two times higher than in patients. However, the difference did not reach significance (P = 0.22).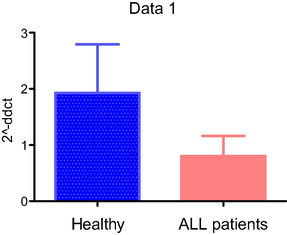
Relative expression of HLA-G from blood samples of ALL patients compared to healthy individuals.
4 Discussion
The prevalence of leukemia in Saudi Arabia is increasing every year. The number of cases passed from 297,000 to 437,033 between 1990 and 2018. The number of new cases in 2020 was 27885, while it was 24,485 in 2018 (Bawazir et al., 2019; Ferlay et al., 2021). Even though it is a result of screening methods progress, development of the immune therapy methods is necessary in order to improve leukemia control and treatment. HLA-G expression in both tumor and non-tumor cells is one of the mechanisms by which malignant cells escape detection by the host immune system (Amiot et al., 2011). This non-classical HLA-class I molecule was considered as an immunosuppressive checkpoint molecule, associated with immune evasion of malignant cells and was therefore inspected as a potential target for immunotherapy (Jasinski-Bergner et al., 2022). The level of expression of the HLA-G in some pathologic sites was associated in most of cases with unfavorable prognosis (Imani et al., 2018). While the relatively low polymorphism reported in coding or non-coding HLA-G molecule compared to classical HLA-genes, the most relevant polymorphisms were located within the regulatory untranslated regions of the gene, including the 3′ and 5′UTRs. Studies on the association between HLA-G polymorphism in coding and non-coding regions with cancer, infectious or autoimmune diseases have confirmed, in most of them, the biological effect features of this polymorphism (Attia et al., 2020). Controversial results in associations were reported according to the disease and the ethnicity which could be attributed to the etiological processes of cancer, including gene-to-gene or gene-environment interactions, sample sizes, and the age of disease onset. Several studies have found that certain HLA-G gene variants are linked to cancer development (Yie et al., 2007; Zheng et al., 2021). One of the most interesting dimorphic site, was the 14 bp Del/Ins in the 3′UTR region, as it was associated with HLA-G mRNA stability and alternative splicing (Hviid et al., 2003; Rousseau et al., 2003). In this case control study, we have investigated relationships between the allelic and genotypic 14 bp Ins/Del polymorphism of the HLA-G and two main types of acute leukemia; ALL and AML. We have explored different genetic models in this study, however our results showed lack of any association between alleles or genotypes of the HLA-G 14 bp Ins/Del polymorphism and ALL and AML. To the best of our knowledge and instead of the large number of case/ control studies that have explored the association between the HLA-G 14 bp polymorphism and solid cancer diseases. This is the first case/control study exploring the association between HLA-G 14 bp Ins/Del polymorphism and leukemia diseases. In a cohort study, including only patients with chronic lymphoid leukemia. Roberta et al. (2014) showed that the Del/Del genotype was associated with higher expression of HLA-G and reduced overall survival, compared to those with other genotypes and the presence of HLA-G molecules in this leukemia environment, creates a favourable setting for CLL expansion. The authors confirmed that in the presence of Del/Del patients have a poorer overall survival than those sharing either Ins/Del or Ins/Ins genotypes (Roberta et al., 2014). However, in another study, Własiuk et al. (2013) noted that higher levels of HLA-G on the surface and soluble are observed in AML patients especially on the surface of dendritic cells and CD4+ T cells and this high level of expression is not associated with any HLA-G 14 bp genotypes. In addition, we have performed an extensive analysis for the association between this HLA-G polymorphism and AML or ALL in subgroups according to age and gender. For all these analyses, we did not find any association considering these parameters. Considering that the functional feature of the HLA-G molecules are mostly associated with the level of expression, we have studied the mRNA expression and compared between ALL and healthy control. Our results did not show significant difference between these two groups, instead of the higher level among the control group. Unfortunately, we failed to obtain mRNA for AML patients. To compare with our results, we did not find in bibliography any case control study for HLA-G 14 bp genetic association and gene expression for ALL or AML diseases using healthy controls as reference group.
However, some studies have explored the relationships between these two aspects of HLA-G and some clinical and cellular parameters of variant leukemia. In some studies, the circulating blasts from CML and AML have been reported to express or to secrete HLA-G and the level of expression was associated with higher blast cells in the bone marrow and relapse (Caocci et al., 2017). Moreover, HLA-G expression has been suggested as predictive tumor biomarker to track disease progression or as a therapeutic target to improve immune responses to leukemia (Locafaro et al., 2014). Maki et al. (2008) pointed out that blocking HLA-G1 with a particular antibody enhanced the sensitivity of CLL samples to NK-mediated death, suggesting that HLA-G protects CLL cells from NK-mediated killing and so may contribute to their immunological escape in vivo. On the other hand, some studies reported opposite results. In one of these studies, patients with AML, CML, ALL, and CLL were selected for their HLA-G expression and they found that the HLA-G molecule was not expressed in newly separated human leukemia cells and hence plays no role in their immunological escape (Poláková et al., 2003). In the secreted forms of HLA-G, Giannopoulos et al. (2008) did no found differences in HLA-G mRNA expression or levels in the plasma of patients.
In conclusion, although our study did not show a significant association between HLA-G 14 bp polymorphism or gene expression with AML or ALL in the Saudi population, further investigations including large number of subjects with more clinical data are necessary to state for this association.
Acknowledgement
This project was supported by the Researcher Supporting Project number (RSP-2021/75), King Saud University, Riyadh, Saudi Arabia.
Conflicts of interest
The authors declare that the publishing of this paper does not include any conflicts of interest.
References
- HLA-G and single nucleotide polymorphism (SNP) associations with cancer in African populations: Implications in personal medicine. Genes Dis. 2021
- [Google Scholar]
- Al Omar, S., Mansour, L., 2019. Association of HLA-G 14-base pair insertion/deletion polymorphism with breast cancer in Saudi Arabia. Genet. Mol. Res. 18.
- Genetic association between the HLA-G 14-bp insertion/deletion polymorphism and the recurrent spontaneous abortions in Saudi Arabian women. Genet. Mol. Res.. 2015;14:286-293.
- [Google Scholar]
- HLA-G expression in acute lymphoblastic leukemia: a significant prognostic tumor biomarker. Med. Oncol.. 2013;30:1-9.
- [Google Scholar]
- Biology of HLA-G in cancer: a candidate molecule for therapeutic intervention? Cell. Mol. Life Sci.. 2011;68(3):417-431.
- [Google Scholar]
- The molecular and functional characteristics of HLA-G and the interaction with its receptors: where to intervene for cancer immunotherapy? Int. J. Mol. Sci.. 2020;21(22):8678.
- [Google Scholar]
- The burden of leukemia in the Kingdom of Saudi Arabia: 15 years period (1999–2013) BMC Cancer. 2019;19:1-10.
- [Google Scholar]
- HLA-G molecules and clinical outcome in chronic myeloid leukemia. Leuk. Res.. 2017;61:1-5.
- [Google Scholar]
- HLA-G polymorphism and transitional cell carcinoma of the bladder in a Brazilian population. Tissue Antigens. 2008;72(2):149-157.
- [Google Scholar]
- The 14 bp deletion polymorphisms in HLA-G gene play an important role in the expression of soluble HLA-G in plasma. Tissue Antigens. 2008;72:335-341.
- [Google Scholar]
- Evaluation of MLPA as a comprehensive molecular cytogenetic tool to detect cytogenetic markers of chronic lymphocytic leukemia in Egyptian patients. J. Genet. Eng. Biotechnol.. 2021;19:1-7.
- [Google Scholar]
- Cancer statistics for the year 2020: An overview. Int J Cancer.. 2021;149(4):778-789.
- [Google Scholar]
- The significance of soluble HLA-G plasma levels as well as messenger HLA-G for B-cell chronic lymphocytic leukemia (B-CLL) Leuk. Res.. 2008;32(12):1815-1819.
- [Google Scholar]
- Relationship of HLA-G expression and its 14-bp insertion/deletion polymorphism with susceptibility to colorectal cancer. Genet Mol Res. 2019;18:1-12.
- [Google Scholar]
- HLA-G modulates immune responses by diverse receptor interactions. Semin. Cancer Biol. Elsevier. 2003;13(5):317-323.
- [Google Scholar]
- Polymorphism in the regulatory region located more than 1.1 kilobases 5′ to the start site of transcription, the promoter region, and exon 1 of the HLA-G gene. Hum. Immunol.. 1999;60(12):1237-1244.
- [Google Scholar]
- HLA-G allelic variants are associated with differences in the HLA-G mRNA isoform profile and HLA-G mRNA levels. Immunogenetics. 2003;55(2):63-79.
- [Google Scholar]
- HLA-G expression is associated with an unfavorable prognosis of oral squamous cell carcinoma. Asian Pac J Cancer Prev. 2018;19:2527-2533.
- [Google Scholar]
- The human leukocyte antigen G as an immune escape mechanism and novel therapeutic target in urological tumors. Front. Immunol.. 2022;13
- [Google Scholar]
- Expression of HLA-G in human cornea, an immune-privileged tissue. Hum. Immunol.. 2003;64(11):1039-1044.
- [Google Scholar]
- HLA-G expression on blasts and tolerogenic cells in patients affected by acute myeloid leukemia. J. Immunol. Res.. 2014;2014:1-10.
- [Google Scholar]
- NK resistance of tumor cells from multiple myeloma and chronic lymphocytic leukemia patients: implication of HLA-G. Leukemia. 2008;22(5):998-1006.
- [Google Scholar]
- Molecular and Immunologic Aspects of the Nonclassical HLA Class I Antigen HLA-G: Evidence for an Important Role in the Maternal Tolerance of the Fetal Allograft. Am. J. Reprod. Immunol.. 1998;40(3):136-144.
- [Google Scholar]
- Newell, L.F., Cook, R.J., 2021. Advances in acute myeloid leukemia. bmj 375.
- Identification of HLA-G7 as a new splice variant of the HLA-G mRNA and expression of soluble HLA-G5,-G6, and-G7 transcripts in human transfected cells. Hum. Immunol.. 2000;61(11):1138-1149.
- [Google Scholar]
- The expression of human leukocyte antigen G and E on human first trimester placenta and its relationship with recurrent spontaneous abortion. Sichuan da xue xue bao. Yi xue ban= Journal of Sichuan University. Med. Sci. Ed.. 2008;39:976-979.
- [Google Scholar]
- Poláková, K.n., Kŕčová, M., Kuba, D., Russ, G., 2003. Analysis of HLA-G expression in malignant hematopoetic cells from leukemia patients. Leukemia research 27, 643-648.
- HLA-G is a component of the chronic lymphocytic leukemia escape repertoire to generate immune suppression: impact of the HLA-G 14 base pair (rs66554220) polymorphism. Haematologica. 2014;99(5):888-896.
- [Google Scholar]
- HLA-G is a component of the chronic lymphocytic leukemia escape repertoire to generate immune suppression: impact of the HLA-G 14 base pair (rs66554220) polymorphism. Haematologica. 2014;99:888-896.
- [Google Scholar]
- The 14 bp deletion-insertion polymorphism in the 3′ UT region of the HLA-G gene influences HLA-G mRNA stability. Hum. Immunol.. 2003;64(11):1005-1010.
- [Google Scholar]
- Analyzing real-time PCR data by the comparative CT method. Nat. Protoc.. 2008;3(6):1101-1108.
- [Google Scholar]
- SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22(15):1928-1929.
- [Google Scholar]
- The expression and functional activity of membrane-bound human leukocyte antigen-G1 are influenced by the 3′-untranslated region. Hum. Immunol.. 2013;74(7):818-827.
- [Google Scholar]
- Total expression of HLA-G and TLR-9 in chronic lymphocytic leukemia patients. Hum. Immunol.. 2013;74(12):1592-1597.
- [Google Scholar]
- Expression of human leucocyte antigen G (HLA-G) is associated with prognosis in non-small cell lung cancer. Lung Cancer. 2007;58(2):267-274.
- [Google Scholar]
- The HLA-G immune checkpoint plays a pivotal role in the regulation of immune response in autoimmune diseases. Int. J. Mol. Sci.. 2021;22:13348.
- [Google Scholar]
- Interaction between HLA-G and NK cell receptor KIR2DL4 orchestrates HER2-positive breast cancer resistance to trastuzumab. Signal Transd. Targeted Ther.. 2021;6:1-15.
- [Google Scholar]







